Abstract
The VirB protein of Shigella flexneri is a positive regulator of the major virulence operons of this enteroinvasive intracellular pathogen. VirB resembles no other transcription factor but is strongly homologous to plasmid partition proteins. We found that the binding of the VirB protein to the promoter region of the icsB virulence gene induced hypersensitivity to cleavage by DNase I over a region to which the H-NS repressor protein binds and completely abolished the protection of this sequence from DNase I by H-NS. In the absence of H-NS, the VirB protein had no additive effect on the ability of the icsB promoter to form an open transcription complex, indicating that VirB is not involved in the recruitment of RNA polymerase to the promoter or in open complex formation. Similarly, VirB did not stimulate promoter function in an in vitro transcription assay but acted as an antagonist of H-NS-mediated repression. A sequence located upstream of the icsB promoter and related to cis-acting elements involved in plasmid partitioning was required for promoter derepression by VirB. Alterations to one heptameric motif within this DNA sequence attenuated VirB binding and derepression of icsB transcription.
Understanding how gene-regulatory circuits have evolved, and may evolve in the future, represents a significant challenge in molecular microbiology. Part of the challenge concerns the need to understand how genes acquired by horizontal transfer become integrated into the existing gene-regulatory networks of the cell. The virulence genes of Shigella flexneri, the etiological agent of bacillary dysentery, represent an interesting case. They are located on a large (∼230-kbp) plasmid that has been acquired by horizontal transfer and encode a type III secretion system and effector proteins that enable S. flexneri to invade epithelial cells in the lower gut of humans (11, 25). Gene expression is controlled at several levels, but control at the level of transcription initiation is an important regulatory step. The key virulence genes are organized within operons on the virulence plasmid, and the promoters of these operons are active under growth conditions that approximate those found at the site of invasion. A temperature of 37°C is a particularly important environmental signal for transcriptional activation (25, 30).
The control of transcription within the virulence regulon is complex and involves several regulatory genes, some of which are located on the virulence plasmid and others that are on the bacterial chromosome (12, 15, 25). The H-NS protein is a key regulatory factor that is contributed by the chromosome (13, 14, 23, 41). It acts throughout the genome as a transcription repressor and displays a preference for binding to planar curves in DNA rather than a specific nucleotide sequence (13, 47, 48). Such curves are frequently composed of A+T-rich DNA, and the widespread association of A+T-rich DNA and intrinsic curvature with many bacterial promoters (38) may explain the broad spectrum of genes repressed by H-NS. The A+T-rich nature of many horizontally acquired virulence genes (27, 34, 36), including those of the S. flexneri virulence plasmid (65 to 70% A+T content), may make these a target for H-NS-mediated repression (11, 56).
The positive regulators VirF and VirB act in a regulatory cascade to derepress virulence gene transcription following the receipt of the correct environmental signals by the bacterium. The AraC-like VirF protein activates the transcription of the virB regulatory gene, and the product of this gene in turn activates the promoters of the structural virulence genes (2, 43). The S. flexneri virulence gene-regulatory cascade is somewhat unusual in having an intermediate regulator, VirB. The advantage of possessing this protein is uncertain but may involve a regulatory checkpoint that comes immediately prior to complete commitment to the expression of the full regulon of virulence genes.
VirB is also an unusual transcriptional regulator from the perspective of its structure. It belongs to no known family of transcription factors (37, 39) and instead resembles DNA binding proteins involved in plasmid partitioning (1, 5, 6, 31, 40, 45, 58). Examples of well-characterized partition factors with strong amino acid sequence homology to VirB include SopB and ParB, expressed by the F factor and the P1 plasmid/bacteriophage, respectively (28, 49, 54). The virulence plasmid of S. flexneri is a composite and has at least two functioning plasmid partition systems (11, 46). There is no evidence that VirB contributes to the maintenance of the modern virulence plasmid. It is tempting to speculate that VirB has been coopted into a regulatory role during the evolution of the S. flexneri virulence regulatory cascade. Whatever its previous biological roles, VirB is now an essential component of the regulatory cascade, and virB knockout mutations result in the silencing of virulence gene expression and the loss of virulence (2). It has been shown previously that DNA binding proteins can be made to act as transcription regulators by careful relocation of their binding sites and that it is the positions of these sites with respect to the promoter that determine whether the influence on transcription will be positive or negative (16, 17). This finding points to a considerable plasticity in transcription factor activity and suggests that a DNA binding protein can be reprogrammed to influence gene expression by judicious placement of its binding site. In this study, we examined the mechanism by which VirB, a close relative of plasmid partition factors, positively influences transcription.
MATERIALS AND METHODS
Bacterial strains and growth media.
All bacterial strains were derivatives of Escherichia coli K-12 or S. flexneri 2a 2457T and are listed in Table 1. Bacteria were grown on Luria-Bertani (LB) agar plates or in LB liquid medium at 30°C or 37°C. Antibiotics were used at the following concentrations: carbenicillin, 50 μg/ml; chloramphenicol, 25 μg/ml; and kanamycin, 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| S. flexneri 2a 2457T derivatives | ||
| BS184 | mxiC::mudI1734 Kmr | 30 |
| E. coli K-12 derivatives | ||
| BL21(DE3) | F−dcm ompT hsdS gal (λ cIts857 ind1 sam7 nin5 lacUV5-T7gene1) | 53 |
| CJD1634 | BE1414/pACYC184 | 4 |
| CJD1635 | BE1414/pAF201 | 4 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tcr)] | Stratagene |
| Plasmids | ||
| pACYC184 | Cloning vector; p15A replicon; Cmr Tcr | New England Biolabs |
| pBAD33VirB | virB gene subcloned behind arabinose-inducible PBAD promoter; Apr | 4 |
| pBluescript | Cloning vector | Stratagene |
| pET22bH-NS | hns gene in pET-22b | This work |
| pET22bVirB | virB gene in pET-22b | 31 |
| pKK232.8 | Promoter cloning vector | 9 |
| pZep08 | GFP reporter plasmid | 20 |
| pZep-proicsB-2 | icsB promoter cloned into pZEP08 | This work |
| pZep-proicsB-4 | 1-bp substitution in box 1 | This work |
| pZep-proicsB-5 | First 3 bp in box 1 replaced | This work |
| pZep-proicsB-6 | Second 3 bp in box 1 replaced | This work |
| pZep-proicsB-7 | Box 1 sequence inverted | This work |
| pZep-proicsB-8 | All box 1 bases replaced | This work |
| pZep-proicsB-9 | Box 2 sequence inverted | This work |
| pZep-proicsB-10 | All box 2 bases replaced | This work |
| pZep-proicsB-11 | 3′ end-flanking CCC replaced | This work |
Plasmid construction.
The icsB promoter region was amplified as a ∼400-bp fragment by PCR using the oligonucleotide primer pair ET1 and ET2 (Table 2). The PCR fragment was digested using restriction enzymes BamHI and EcoRI, and the digested fragment was ligated into the BamHI- and EcoRI-digested plasmid pBluescript. The structure of the resulting construct was verified by DNA sequencing, and the construct was designated pBSproicsB. Subsequently, mutations within the icsB promoter were made by using the QuikChange II site-directed mutagenesis kit (Stratagene). For in vitro transcription, the icsB promoter region was amplified by PCR using the primer pair ET23 and ET24 (Table 2), digested with EcoRI and BamHI, and ligated into EcoRI- and BamHI-digested plasmid pKK232.8 (9). The structure of the resulting plasmid pKK232.8proicsB was confirmed by DNA sequencing. For flow cytometric analysis, the wild-type icsB promoter and its mutant derivatives were amplified by PCR using primers ET25 and ET26 (Table 2), digested with XbaI and NotI, and ligated into XbaI- and NotI-digested plasmid pZep08. The structures of the resulting plasmids pZep-proicsB-1 to pZep-proicsB-12 were verified by DNA sequencing.
TABLE 2.
Oligonucleotide primers
| Primer | Sequencea (5′ to 3′) | Description or use |
|---|---|---|
| ET1 | GGCCGGATCCATGCAATCCCAAATTAGT | Cloning of icsBp into pBluescript and DNase I footprinting PCR primer |
| ET2 | CCGGGAATTCTCAATGAAATTGCTAATT | Cloning of icsBp into pBluescript |
| ET3 | GAAAAGCTGGGTACCGGGCC | DNase I footprinting PCR primer (hybridizes to pBluescript) |
| ET5 | CACTTTATCTTGTGGGATTTCATGCAAAGTAGAGCACTACATATGCTCACGAGG | Box 1 inversion |
| ET6 | CCTCGTGAGCATATGTAGTGCTCTACTTTGCATGAAATCCCACAAGATAAAGTG | Box 1 inversion |
| ET9 | CTTGTGGGATTTCATGATGAAATGAGCACTACATATGC | Substitution of 1 bp in box 1 |
| ET10 | GCATATGTAGTGCTCATTTCATCATGAAATCCCACAAG | Substitution of 1 bp in box 1 |
| ET11 | CTTGTGGGATTTCATGATGCCCCGAGCACTACATATGC | Replacement of first 3 bp in box 1 |
| ET12 | GCATATGTAGTGCTCGGGGCATCATGAAATCCCACAAG | Replacement of first 3 bp in box 1 |
| ET13 | CTTGTGGGATTTCATGCGTAAACGAGCACTACATATGC | Replacement of second 3 bp in box 1 |
| ET14 | GCATATGTAGTGCTCGTTTACGCATGAAATCCCACAAG | Replacement of second 3 bp in box 1 |
| ET15 | CACTTTATCTTGTGGGATTTCATGCGTCCCTGAGCACTACATATGCTCACGAGG | Replacement of all box 1 bases |
| ET16 | CCTCGTGAGCATATGTAGTGCTCAGGGACGCATGAAATCCCACAAGATAAAGTG | Replacement of all box 1 bases |
| ET23 | CCCGGATCCTCAATGAAATTGCTA | Cloning of icsBp into pKK232.8 |
| ET24 | GGGGAATTCATGCAATCCCAAATTAG | Cloning of icsBp into pKK232.8 |
| ET25 | GGGCGGCCGCATGCAATCCCAAATTAG | Cloning of icsBp into pZEP08 |
| ET26 | GGGTCTAGATCAATGAAATTGCTA | Cloning of icsBp into pZEP08 |
| ET29 | CTTTATCTTGTGGGTACTTTAGATGAAACGAGCACTACATATG | Box 2 inversion |
| ET30 | CATATGTAGTGCTCGTTTCATCTAAAGTACCCACAAGATAAAGTGCCTG | Box 2 inversion |
| ET35 | CTTTATCTTGTGGGGCCCAGCGATGAAACGAGCAC | Replacement of all box 2 bases |
| ET36 | GTGCTCGTTTCATCGCTGGGCCCCACAAGATAAAG | Replacement of all box 2 bases |
| ET39 | CTTTATCTTGTATAATTTCATGATGAAACGAGCAC | Replacement of 3′ end-flanking C's |
| ET40 | GTGCTCGTTTCATCATGAAATTATACAAGATAAAG | Replacement of 3′ end-flanking C's |
Boldface indicates changes relative to the wild-type sequence.
Purification of the VirB and H-NS proteins.
VirB protein was overproduced from plasmid pET22bVirB (31), a derivative of pET22b (Novagen). Proteins were expressed in the E. coli K-12 derivative strain BL21(DE3) containing the LacI-expressing plasmid pDIA17. Cells were grown in LB broth containing appropriate antibiotics at 37°C. The expression of C-terminally His-tagged VirB was induced with 0.8 mM IPTG (isopropyl-β-d-thiogalactopyranoside) in exponentially growing 100-ml cultures. Three hours later, the cells were harvested and lysed by sonication. Lysates (∼10 ml) were applied to a His-Bind Quick column (Novagen) preequilibrated with binding buffer (31). The column was then washed with binding buffer followed by wash buffer (31), and the protein was eluted in 1-ml fractions of elution buffer (10% glycerol, 50 mM Tris-HCl [pH 7.9], 0.5 M NaCl, 0.1 mM phenylmethylsulfonyl fluoride, 500 mM imidazole). Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and those containing VirB were pooled and dialyzed three times against storage buffer (100 mM phosphate buffer [pH 8.0], 10% glycerol, 1 mM EDTA, 300 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol [DTT]). VirB was estimated to be 95% pure and was quantified against lysozyme standards. The His-tagged VirB was found to be fully functional in up-regulating the transcription of an mxiC::lacZ fusion (data not shown).
The hns open reading frame of the hns gene was placed into the multicloning site of the pET22bH-NS plasmid by using standard methods, and the protein was overproduced and purified using the same approach described above for VirB. The His-tagged H-NS protein was found to be fully functional in vivo and to have the same DNA binding characteristics as the batch kindly supplied by Sylvie Rimsky and used by Beloin and Dorman (4).
β-Galactosidase assays.
The transcription of mxiC::lacZ, a VirB-dependent gene that is representative of the virulence gene expression in S. flexneri, was monitored by assaying cells grown to stationary phase (optical density at 600 nm, ∼2.4) for β-galactosidase activity according to the method of Miller (32). Assays were performed in duplicate, and the data are expressed as the means of two measurements. The standard deviations were calculated and are indicated in the relevant figures. Experiments were performed on at least three independent occasions.
Flow cytometry analysis.
Flow cytometry analysis was used to monitor gene expression by using a green fluorescent protein (GFP)-based assay (7, 20) in which the gfp gene was placed under the regulatory control of the promoter of interest. Promoter activity determined the level of GFP fluorescence, and this fluorescence was measured by flow cytometry. The bacterial culture (10 μl) to be assayed was harvested in duplicate and immediately fixed at room temperature in 0.25 ml of 4% (wt/vol) formaldehyde (Sigma) freshly prepared in phosphate-buffered saline and then stored at 4°C in the dark until analysis. For flow cytometry, samples were diluted to a concentration of approximately 106 bacteria per ml, transferred into flow cytometer tubes, and analyzed with an EPICS-XL flow cytometer (Beckman Coulter). Approximately 10,000 bacteria per sample were collected, and the relative levels of GFP fluorescence were expressed as the mean level of fluorescence of the populations after analysis with EXPO-32 software (Beckman Coulter). Each assay was performed in duplicate, and the mean values were determined from the results of at least three independent experiments.
Electrophoretic mobility shift analysis.
A 380-bp DNA fragment encompassing the icsB-ipgD intergenic region was amplified by PCR using 5′ end digoxigenin-labeled ET1 and ET2 primers (Table 2). The PCR fragment was gel purified twice by using the High Pure PCR product purification kit (Roche). The digoxigenin-labeled probe was then incubated with increasing concentrations of recombinant VirB protein (0 to 30 μM final concentration) for 45 min at 37°C in a 20-μl reaction mixture containing 20 mM Tris-HCl (pH 7.5), 80 mM NaCl, 1 mM EDTA, 100 ng of bovine serum albumin (BSA), 25 μg/ml poly(dI-dC), 10% glycerol, and 1 mM DTT. Each reaction mixture contained approximately 5 ng of DNA. The protein-DNA complexes were then resolved by electrophoresis through 6% nondenaturing polyacrylamide gels for 3 h at room temperature. After electrophoresis, the digoxigenin-labeled DNA was transferred onto a Biodyne B nylon membrane (Pall) and then immobilized by UV cross-linking. The DNA was then visualized using the chemiluminescent-nucleic acid detection kit per the instructions of the manufacturer (Pierce).
DNase I footprinting.
A fragment of the icsB regulatory region was amplified by PCR using the oligonucleotide primers ET1 and ET3 (Table 2). The PCR fragment was then labeled at both ends with [α-32P]ATP and purified using the High Pure PCR product purification kit (Roche). The labeled fragment was then subjected to EcoRI digestion and gel purified using a 6% polyacrylamide gel, separating into two fragments labeled at one end only. The desired fragment (containing the icsB promoter region) was excised from the gel, eluted into 3 ml of elution buffer (10 mM Tris-HCl [pH 8.8], 300 mM sodium acetate, 1 mM EDTA, and 0.2% sodium dodecyl sulfate), and incubated overnight at 56°C and 200 rpm. The DNA probe was extracted by phenol-chloroform extraction and ethanol precipitated.
Increasing amounts of protein were incubated for 30 min at 37°C with a labeled probe in binding buffer (20 mM Tris-HCl [pH 7.5], 80 mM NaCl, 1 mM EDTA, 100 μg/ml BSA, 10% glycerol, and 1 mM DTT) to a final volume of 50 μl. The addition of 50 μl of a solution containing 10 mM MgSO4 and 10 mM CaCl2 was followed by a further 10-min incubation at 37°C. DNase I was added to each reaction mixture, and the mixtures were incubated for a further minute at 37°C. Transfer to ice and the addition of a stop solution (400 mM sodium acetate [pH 5.2], 2.5 mM EDTA [pH 8.0], 50 μg/ml Saccharomyces cerevisiae tRNA, 5 μg/ml sonicated salmon sperm DNA) stopped the reactions. DNA was extracted with 250 μl of phenol and ethanol precipitated before being electrophoresed through a 6% polyacrylamide sequencing gel. Protected bands were identified by comparison with a DNA sequence ladder generated with appropriate primers.
Potassium permanganate footprinting.
The icsB regulatory-region probe was prepared as described above. Protein-DNA and protein-nucleoside triphosphate (NTP)-DNA mixtures were incubated for 45 min at 37°C to allow the formation of the open and initiation complexes, respectively. KMnO4 was added to 4 mM for 10 s. A stop solution (14 M β-mercaptoethanol, 0.3 M sodium acetate, 8 μg of sonicated salmon sperm DNA) was added to terminate the reaction. The permanganate-treated DNA was ethanol precipitated and then treated with 0.5 M piperidine at 90°C for 20 min. LiCl was added to 0.5 M. DNA was extensively washed twice using 100% ethyl alcohol and once using 70% ethyl alcohol. The extracted DNA was electrophoresed through a 6% polyacrylamide sequencing gel. Cleaved fragments were identified by comparison with a DNA sequence ladder generated with appropriate primers.
In vitro transcription.
In vitro transcription with the supercoiled pKK232.8proicsB plasmid as the template was performed as pseudo-single-round reactions as previously reported by Hsu (24). In short, initiation complexes were formed in a total volume of 50 μl with 9 nM active RNA polymerase saturated with σ70 in the presence of 3 nM template DNA and the initiating NTPs ATP (500 μM) and CTP (50 μM) in the reaction buffer (50 mM Tris-acetate [pH 8.0], 10 mM magnesium acetate, 0.5 mM DTT, 0.1 mM EDTA, 100 μg/ml acetylated BSA, and 80 mM potassium glutamate). [α-32P]UTP (20 μCi) was added, and the reaction mixture was incubated for 20 min at 30°C. Initiation was stopped by the addition of 50 ng/μl heparin. Following a further 5-min incubation at 30°C, an NTP mix was added (each NTP at 500 μM) and a 5-min elongation reaction occurred at 30°C. A chase mixture (each NTP at 2 mM, 2 mg/ml heparin) was added, and the samples were incubated at 30°C for 5 min. The samples were put on ice, and the reaction was stopped by the addition of formamide loading buffer containing a defined radiolabeled loading standard. Transcription products were separated on 10% polyacrylamide gels and visualized by autoradiography.
RESULTS
VirB interaction with the regulatory region of the icsB virulence gene.
The transcription start site of the icsB gene has been mapped previously (42), and the interaction of the H-NS repressor protein with the icsB promoter region has been investigated by DNase I footprinting (4). The H-NS footprint encompasses a region extending from position −120 to +60 with respect to the transcription start site (Fig. 1). An electrophoretic mobility shift assay has shown the VirB protein to bind to the icsB regulatory region (31). Here we examined the interaction of the VirB protein with the icsB regulatory region by DNase I footprinting as a first step in understanding the mechanism of action of VirB in this region.
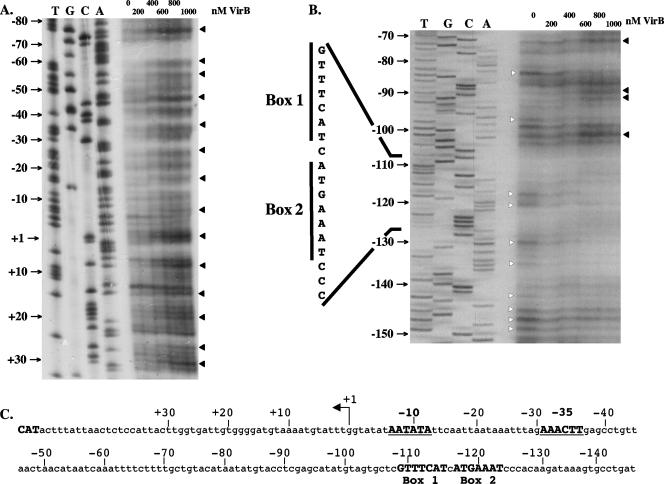
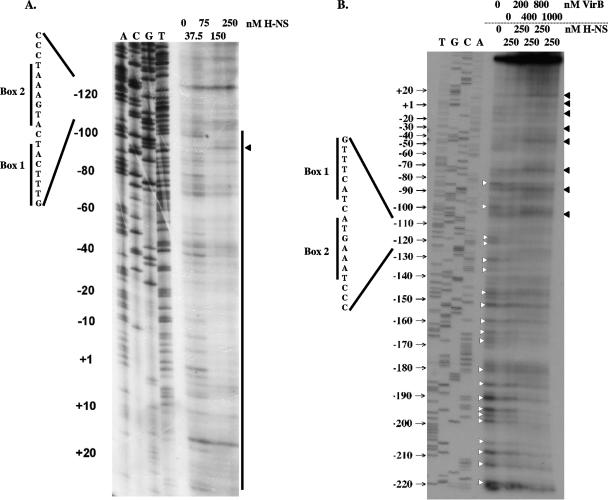
FIG. 1.
Interaction of VirB with the transcription regulatory region of the icsB virulence gene. DNase I footprinting was used to examine the interaction of purified VirB protein with the promoter-proximal (A) and promoter-distal (B) regions of the icsB upstream regulatory region (C). An angled arrow labeled +1 in panel C represents the transcription start site that was determined previously for icsB (42). The −10 and −35 hexamers of the promoter are shown in bold, as are the box 1 and 2 motifs. In panels A and B, white arrowheads indicate bands protected from DNase I digestion by VirB while black arrowheads indicate residues showing hypersensitivity to DNase I in the presence of VirB.
VirB exerted two types of effects on binding to the icsB DNA. Enhanced sensitivity to DNase I cleavage was detected in the region between +30 and −105. In addition, a region of VirB-mediated protection from DNase I digestion was seen upstream from position −105 (Fig. 1). These data suggested that the protein bound to the region extending upstream from residue −105 of the regulatory sequence and that its presence there resulted in a structural distortion of the DNA between positions −105 and +30 such that this region exhibited enhanced sensitivity to cleavage by DNase I. This enhanced sensitivity was consistent with the wrapping of the DNA around the protein, with the concomitant exposure of specific residues to enhanced DNase I cleavage (35, 57). The periodicity of the hypersensitive bands between +30 and −105 (Fig. 1) was consistent with this hypothesis. Interestingly, the regions affected by VirB binding overlapped those shown previously to be protected by H-NS (4).
cis-acting site required for VirB activation.
The pattern of DNA protection from DNase I digestion seen following VirB interaction within the icsB regulatory region suggested that protein binding occurred at and upstream of coordinate −105 (Fig. 1). An inspection of this region revealed the presence of a near-perfect inverted repeat in the DNA sequence, designated box 1 and box 2 (Fig. 1 and 2A). This motif was subjected to site-directed mutagenesis (summarized in Fig. 2B), and the mutated sequences were introduced into a copy of the icsB promoter driving the transcription of a gfp reporter fusion. The recombinant plasmids harboring these constructs were placed in S. flexneri strain BS184, which already contained a fusion of the lacZ reporter to the VirB-activated mxiC gene (Table 1). The VirB protein was expressed from a virB gene that was under the control of the arabinose-inducible PBAD promoter in plasmid pBAD33virB+.
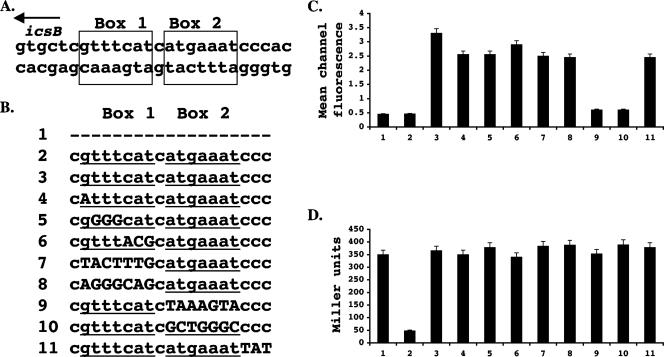
FIG. 2.
Identification and mutagenesis of a cis-acting sequence required for VirB-mediated derepression of icsB transcription. (A) The nucleotide sequences of the box 1 and 2 motifs that make up the inverted repeat in the region that was protected from DNase I digestion by VirB are shown, together with the direction of transcription of the icsB gene (arrow). (B) Summary of the mutations introduced into the box 1 and 2 sequences (underlined). Base substitutions are shown in uppercase letters, and absent bases are represented by hyphens. Construct 1 had no icsB sequences added to the gfp reporter gene in the pZep08 plasmid. The effect of these icsB regulatory sequence mutations on the VirB activation of an icsB::gfp (C) and an mxiC::lacZ (D) reporter fusion is shown. Numbers at the bottom indicate the mutated sequences. In each case (C and D), the data corresponding to sequence 2 were obtained without the addition of an arabinose inducer.
The bacterial cells were grown in LB at 30°C, conditions known to repress the transcription of the S. flexneri virulence regulon (12, 30). The expression of the VirB protein was induced by the activation of the PBAD promoter with arabinose. Basal levels of icsB::gfp and mxiC::lacZ gene expression in the absence of arabinose induction of virB gene expression were established (Fig. 2C and D, lanes 2). The activity of induced VirB within the normal virulence regulon was confirmed by monitoring the expression of the mxiC::lacZ reporter fusion from the large virulence plasmid (Fig. 2D). Simultaneously, the ability of the VirB protein to activate the modified icsB promoters was assessed by measuring the expression of the gfp reporter gene. The data showed that two icsB promoter derivatives were defective in responding to VirB. In both cases, these derivatives had extensive sequence changes in the promoter-distal box 2 arm of the inverted repeat: derivative number 9 had a complete inversion of the 7 bp of this arm, and derivative number 10 had each purine base replaced with a pyrimidine base and vice versa (Fig. 2). In contrast, changes to the base composition of the promoter-proximal box 1 arm had little or no effect on the response of the icsB promoter to VirB. These data identified the promoter-distal arm (box 2) of the inverted repeat as being critical for VirB-mediated activation of the icsB promoter and indicated that the inverted repeat structure per se was irrelevant to the activation mechanism.
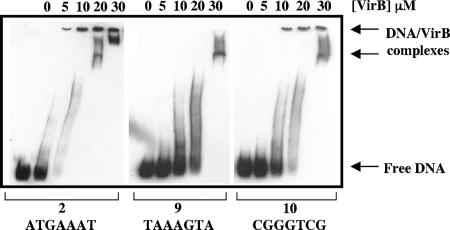
Electrophoretic mobility shift assays were used to examine the effects of the mutations in derivatives 9 and 10 on VirB binding to the icsB regulatory region, as these derivatives had the most marked negative effect on icsB activation. The wild-type sequence was shifted by using 10 μM VirB. In contrast, 20 to 30 μM concentrations of the same purified VirB protein preparation were required to shift the mutant sequences known as derivatives 9 and 10 (Fig. 3). These findings were in keeping with the proposed role for the box 2 sequence 5′-ATGAAAT-3′ in the binding of VirB to DNA. The effect on the electrophoretic mobility of the probe DNA varied with VirB protein concentrations. This pattern was thought to reflect the previously described ability of the protein to polymerize on DNA, resulting in a variety of nucleoprotein complexes (31).
FIG. 3.
VirB requires the box 2 sequence to bind to the icsB regulatory region. Electrophoretic mobility shift assays were carried out with icsB regulatory-region DNA containing the wild-type sequence (2) and mutant derivative sequences (9 and 10). The concentrations of purified VirB protein used are shown above the lanes. Arrows at the right indicate the positions of the free probe and VirB-DNA complexes. The purified VirB protein did not shift the mobility of a nonspecific DNA sequence from within the open reading frame of the S. flexneri icsP gene (31) under these conditions (data not shown).
VirB does not enhance open transcription complex formation.
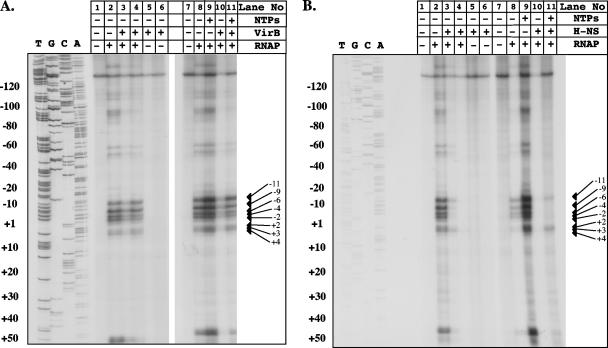
Conventional transcription activators often work by recruiting RNA polymerase to promoters and/or lowering the activation energy barrier for isomerization of the closed transcription complex into an open complex (10, 57). Potassium permanganate footprinting is a useful technique for monitoring the formation of single-stranded regions in A+T-rich DNA (8, 33, 51), and we applied it to an investigation of the effect of VirB on the formation of an open transcription complex at the icsB promoter.
KMnO4 footprinting identified the region of the icsB regulatory region that becomes single-stranded in the presence of RNA polymerase (Fig. 4) and showed that its location was fully consistent with the proposed position of the Pribnow box (Fig. 1) that was based on the previous measurement of the transcription start site (42). RNA polymerase was found to be capable of forming an open complex in the absence of any other factor, and this formation was not enhanced by the addition of VirB protein. The VirB protein had no ability to induce single-stranded DNA formation in the absence of RNA polymerase (Fig. 4A). The addition of NTPs to the in vitro reaction mixture allows RNA polymerase to initiate the early steps in RNA synthesis, and in other systems this procedure has revealed otherwise obscure details of promoter regulatory mechanisms (50). KMnO4 footprinting showed that adding NTPs to the icsB promoter reaction mixture resulted in only a slight enhancement of the band intensities of the previously seen reaction products. The VirB protein did not alter the pattern of open complex formation, and it did not enhance the intensity of the bands seen in its absence. In fact, the presence of 500 nM VirB slightly reduced the intensity of the bands representing reactive thymine residues in the KMnO4 experiment (Fig. 4A, lane 3). These data ruled out a role for VirB as an essential prerequisite in the binding of RNA polymerase to the icsB promoter and the formation there of an open complex.
FIG. 4.
Open complex formation at the icsB promoter does not depend on VirB and is repressed by H-NS. Potassium permanganate footprinting was used to detect single-stranded regions in icsB promoter DNA that were indicative of open transcription complex formation. Experiments were performed to examine the effects of VirB (A) and H-NS (B) on open complex formation. Arrows indicate reactive thymine bases in the region of the Pribnow box of the icsB promoter that reacted strongly with KMnO4. Plus and minus signs above the lanes indicate the presence or absence, respectively, of the reaction mixture components listed at the right. These components were added at the following concentrations: RNA polymerase (RNAP), 500 nM; NTPs, 30 μM; VirB, 500 nM (lanes 3, 5, and 10) and 1 mM (lanes 4, 6, and 11); H-NS, 100 nM (lanes 3, 5, and 10) and 250 nM (lanes 4, 6, and 11).
H-NS inhibits open complex formation at the icsB promoter.
We next examined open complex formation at the icsB promoter in the presence and absence of the H-NS repressor. The results showed that the presence of H-NS strongly inhibited open complex formation and that this inhibition occurred irrespective of the presence of NTPs (Fig. 4B). This finding was fully consistent with the known role of H-NS as a transcription repressor and with the pattern of its binding to the icsB promoter (4).
VirB antagonism of H-NS in the icsB regulatory region.
The interaction of H-NS and VirB at the icsB regulatory region was next monitored by DNase I footprinting. First, the H-NS protein was bound in increasing concentrations to the icsB promoter region and the DNase I protection pattern was determined (Fig. 5A). At 250 nM, the H-NS protein protected a region extending from −110 to at least +25, in agreement with previous data (4). Next, the effect of adding increasing concentrations of VirB to the same icsB DNA sequence that had been prebound by 250 nM H-NS was investigated. The addition of the VirB protein restored DNase I hypersensitivity to the DNA (Fig. 5B) at positions previously noted in DNase I footprints with VirB alone (Fig. 1). These hypersensitive bands occurred at regular intervals between positions −105 and +20, a region that corresponded to that protected by H-NS and lay between the box 1 sequence and the icsB transcription start site. The hypersensitive residues are indicated by black arrowheads in Fig. 5B. The DNA in the vicinity of the box 1-box 2 inverted repeat and extending upstream to position −210 was protected from DNase I cleavage by VirB. The protected residues are indicated by white arrowheads in Fig. 5B. These findings showed that the VirB protein bound to the box 2 region and upstream sequences and remodeled the DNA throughout the region bound by H-NS, completely abolishing H-NS-mediated DNase I protection. This result was consistent with the displacement of H-NS from the icsB promoter by VirB.
FIG. 5.
VirB displaces H-NS from the icsB regulatory sequence. (A) DNase I footprinting was used to reveal the pattern of H-NS-mediated protection of the icsB regulatory region. The vertical line shows the region of protection, and a residue showing hypersensitivity to DNase I is indicated by an arrowhead. (B) H-NS protein (250 nM) was prebound to the same icsB sequence. VirB protein was added in increasing concentrations (0 to 1,000 nM). Bases protected from DNase I cleavage are indicated by white arrowheads, and those showing hypersensitivity to DNase I in the presence of VirB are indicated by black arrowheads. In each gel, a DNA sequencing ladder generated using the same oligonucleotide primer used to generate the probe for footprinting is shown, as are the locations of the box 1 and box 2 motifs described in the legends to Fig. 1 and 2.
VirB activates the H-NS-repressed icsB promoter in vitro.
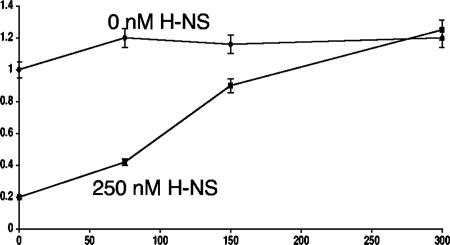
The data obtained so far in this study pointed to a role for VirB as an antagonist of H-NS-mediated transcription repression. This hypothesis was tested further in an in vitro transcription assay in which the influence of VirB and H-NS on the performance of RNA polymerase could be monitored in the absence of input from other cellular components. As expected, the icsB promoter was silent in the absence of RNA polymerase and became active following its addition (Fig. 6). The subsequent addition of increasing concentrations of purified VirB protein resulted in only a very mild enhancement in transcription (Fig. 6). This result was consistent with the outcome of the KMnO4 footprinting experiment showing that VirB did not act to enhance open complex formation at the icsB promoter (Fig. 4). The addition of 250 nM H-NS to the in vitro transcription mixture had a repressive effect on transcription, as expected. However, this repression was relieved by the subsequent addition of increasing concentrations of the VirB protein, elevating the level of the icsB transcript approximately fivefold, to values equal to those seen in the complete absence of H-NS (Fig. 6). This result was in keeping with a regulatory model in which the VirB protein acts as an antirepressor, removing the H-NS protein from the icsB promoter and hence relieving a transcriptional blockade.
FIG. 6.
VirB alleviates H-NS-mediated repression of the icsB promoter in vitro. In vitro transcription was carried out with supercoiled pKK282.3proicsB plasmid DNA in the presence of H-NS with VirB at the indicated concentrations. Radiolabeled transcript was electrophoresed, and the resulting gel was subjected to autoradiography. The bands on the autoradiograph were quantified by densitometry. Data obtained from three separate experiments were averaged and used to plot the graph. Results obtained in the absence and presence of H-NS are represented by diamonds and squares, respectively, and are labeled. The error bars around each datum point show the standard deviation.
DISCUSSION
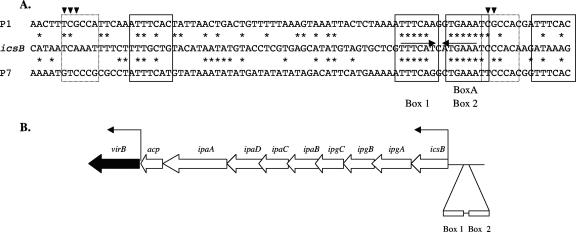
Here we investigated the interaction of the VirB transcription regulatory protein with the promoter of the H-NS-repressed icsB virulence gene. H-NS was shown previously to bind to the icsB regulatory region (4), and this binding was confirmed in the present study. Furthermore, we found that VirB also binds to the icsB regulatory region, where it affects the structure of this DNA segment as shown by enhanced sensitivity to cleavage by DNase I (Fig. 1). Our investigation also reveals a DNA sequence, designated box 1-box 2, in the icsB regulatory region that is essential if VirB is to bind to and activate icsB (Fig. 2). This cis-acting sequence is located at the boundary between the region that is protected by VirB and one where enhanced DNase I cleavage is detected (Fig. 1 and 5). The critical component of the box 1-box 2 element is the sequence motif 5′-ATGAAAT-3′ within box 2, which is essential for normal binding by the VirB protein (Fig. 2 and 3). This cis-acting sequence motif has been implicated previously in the positive control in S. sonnei of the expression of the operon ipaBCD, which is under the control of the icsB promoter and requires the InvE (VirB) regulatory protein (55). The S. sonnei study described the similarity between this sequence and the BoxA motif found within the cis-acting parS sequence that is bound by the ParB partitioning protein of plasmid P1. An examination of the sequence in S. flexneri revealed that the similarity extends significantly beyond the box 2-like heptameric BoxA element (Fig. 7A).
FIG. 7.
(A) Alignment of the icsB regulatory region with the parS sequences of phages/plasmids P1 and P7. The DNA sequence of the promoter-distal portion of the icsB regulatory region that contains boxes 1 and 2 is aligned with the parS sequences of phages/plasmids P1 and P7. The converging horizontal arrows show the inverted repeats associated with boxes 1 and 2. The four heptameric and two hexameric parS motifs involved in ParB protein interaction are boxed by solid- and dotted-line rectangles, respectively. Downward-pointing arrowheads indicate residues within the hexamers that allow ParB proteins to distinguish different parS sequences. The asterisks indicate residues that are conserved between the icsB regulatory region and the parS sequences. (B) Genetic map of the portion of the large virulence plasmid showing the relative locations of the virB gene and the regulatory sequences of the icsB-ipg-ipa-acp operon. The angled arrows represent promoters. The relative positions of the box 1 and box 2 motifs upstream of the icsB promoter are shown. The diagram is not drawn to scale.
The parS sequences of plasmids P1 and P7 contain four heptameric and two hexameric imperfect repeats that are required for the binding of the cognate ParB plasmid-partitioning protein. Each heptamer is an individual binding site for ParB (21). The hexamers are in close contact with the bound protein and determine with which ParB protein the system will interact (19, 45). The icsB regulatory region shows DNA sequence homology throughout this region (Fig. 7A). The icsB gene also shares with ParB-dependent plasmid-partitioning systems a dependency on the integration host factor protein (19), and the integration host factor both binds to the icsB regulatory region and enhances the activity of the promoter (42).
Interestingly, the promoter-proximal box 1 arm of the inverted repeat identified in this study (Fig. 2 and 3) is also equivalent to a parS heptamer and its sequence is very similar to those of the corresponding portions of the P1 and P7 parS regions (Fig. 7A). Nevertheless, site-directed mutagenesis of this region within icsB did not support a role for this motif in VirB-dependent gene activation. Immediately upstream of the BoxA-like element in icsB (called box 2) is a C triplet that corresponds to one of the hexamer-associated sequence elements that is involved in discriminating among different ParB proteins (Fig. 7A). Substituting other bases for these C residues resulted in only a mild derepression of the icsB promoter, suggesting that these residues do not play a critical role in the VirB-icsB interaction (Fig. 2). The strong similarity between the sequence of the icsB regulatory region and the parS sequences of plasmids P1 and P7 is very interesting in the light of the strong amino acid sequence homology between VirB and ParB-like plasmid-partitioning proteins (5, 40, 58). It suggests that the icsB regulatory region contains a degenerate copy of a parS-like sequence that retains the ability to bind the ParB-like protein VirB. Previous work has identified sequences with similarity to that of the element herein designated box 2 in the regulatory regions of other VirB-dependent virulence genes such as virA and spa15 (55). However, these similarities do not extend over the whole parS-like sequence identified in this study. Perhaps the relationship between VirB and its binding site has evolved to a point where only the box 2 element within the cis-acting region is truly essential for protein binding. The retention by VirB of some DNA sequence specificity in its binding site is presumably essential if it is not to act generally as an antagonist of H-NS at every promoter that is repressed by this nucleoid-associated protein. We have performed some experiments to address the specificity issue and discovered that VirB has no influence on the H-NS-repressed proU and bgl operons or the fliC gene (positively, albeit indirectly, regulated by H-NS) in E. coli. Similarly, the expression of VirB does not affect the ability of H-NS to repress a mucoid phenotype in E. coli or S. flexneri (data not shown). Although this is hardly a comprehensive survey, it does suggest that VirB is likely to be specific for the promoters that contain at least close matches to the box 2 element.
The data obtained in this study show that VirB does not act to recruit RNA polymerase to the icsB promoter. Polymerase can utilize the promoter, provided that the H-NS repressor protein is absent (Fig. 4). Furthermore, VirB is not required for RNA polymerase to form an open transcription complex or to initiate transcript elongation, as shown here by KMnO4 footprinting in the presence of RNA polymerase, with and without NTPs (Fig. 4). Thus, VirB does not participate in many of the activities that are associated with conventional transcription factors. Its key function at the icsB promoter is that of an antirepressor that opposes the action of H-NS. DNase I footprinting shows that the two proteins affect the same DNA sequence in different ways: H-NS protects the portion of the icsB regulatory region between positions +20 and −110 from DNase I digestion, whereas VirB has a protective effect beginning in the vicinity of the parS-like box 1 sequence and extending upstream (Fig. 5). Within the segment of DNA that is bound by H-NS, the presence of VirB results in enhanced DNase I digestion with a periodicity that is consistent with DNA wrapping (Fig. 5). Wrapping the DNA would be expected to increase the exposure of certain phosphodiester bonds to DNase I cleavage in the footprinting experiment (35, 57). Our finding that VirB can completely abolish the H-NS-mediated DNase I protection of this sequence (Fig. 5B) is indicative of H-NS displacement by VirB and is consistent with the known weak DNA binding activity of the H-NS protein (52) and its requirement for the maintenance of an appropriate DNA conformation at its binding sites (44, 48).
The strong amino acid sequence homology between VirB and plasmid partition proteins (5, 40) and the strong nucleotide sequence similarity between plasmid-partitioning parS elements and the DNA sequence to which VirB binds (Fig. 7A) suggest either that VirB is a former plasmid partition protein that has been redirected to regulate transcription or that it is ab initio a transcription factor whose similarity to plasmid partition factors is coincidental. Why a dedicated transcription factor should evolve so as to converge structurally with proteins specializing in plasmid partitioning is far from clear. The virulence plasmid in S. flexneri has a mosaic structure and it contains two functional plasmid-partitioning systems (11, 46). It seems reasonable to propose that part of a third system (VirB), redundant in the context of the modern virulence plasmid, could be coopted to perform a gene-regulatory role. If VirB performed a role as a plasmid-partitioning protein earlier in its natural history, that role has clearly been obviated within the context of the modern S. flexneri virulence plasmid. Interestingly, the virulence plasmid does not appear to encode a partner protein for VirB that is equivalent to the ParA proteins that interact with ParB-like molecules in plasmid-partitioning reactions (11). Perhaps the loss of the gene coding for the putative VirB partner protein combined with the existence of other partitioning systems allowed VirB to become available for other tasks within the plasmid.
Bioinformatic studies suggest that positively acting transcription factors and their binding sites are evolutionarily uncoupled, allowing target genes to join and leave the regulons controlled by the regulatory proteins of the cell as they gain and lose appropriate binding sites (3, 22, 26). In this way, VirB dependency could result from the acquisition by icsB of a cis-acting site similar to that used by the protein in its former role as a plasmid-partitioning factor. The parS-like sequence bound by VirB is located at a distance from the virB gene (Fig. 7B). This represents a point of difference from the P1 and P7 parS sequences, which are located immediately downstream of their respective parB genes (1, 18, 29). Thus, the involvement of VirB in the regulation of the icsB promoter is not simply a consequence of the fortuitous colocation of this promoter and the virB gene because the virB gene and the icsB promoter are separated by several kilobases of DNA in the plasmid (11) (Fig. 7B). If the box 1-box 2 motif was formerly located in the promoter region of virB, it has become disconnected from that gene by the insertion of the entire icsB-ipg-ipa-acp operon, an event that would have placed the motif in its present position upstream of icsB (Fig. 7B).
The story of VirB hints at the flexibility of bacterial gene-regulatory circuits and their capacity to coopt DNA binding proteins for new regulatory roles. Protein reassignments of the type discussed here offer bacteria the potential to explore novel regulatory arrangements as part of the evolutionary process. This has important practical implications for antimicrobial strategies that target known gene-regulatory proteins or families of proteins.
Acknowledgments
We thank Chris M. Thomas for useful discussions.
This work was supported by a grant from the Wellcome Trust.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Abeles, A. L., S. A. Friedman, and S. J. Austin. 1985. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J. Mol. Biol. 185:261-272. [DOI] [PubMed] [Google Scholar]
- 2.Adler, B., C. Sasakawa, T. Tobe, S. Makino, K. Komatsu, and M. Yoshikawa. 1989. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3:627-635. [DOI] [PubMed] [Google Scholar]
- 3.Babu, M. M., S. A. Teichmann, and L. Aravind. 2006. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 358:614-633. [DOI] [PubMed] [Google Scholar]
- 4.Beloin, C., and C. J. Dorman. 2003. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 47:825-838. [DOI] [PubMed] [Google Scholar]
- 5.Beloin, C., S. McKenna, and C. J. Dorman. 2002. Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J. Biol. Chem. 277:15333-15344. [DOI] [PubMed] [Google Scholar]
- 6.Bignell, C., and C. M. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1-34. [DOI] [PubMed] [Google Scholar]
- 7.Bongaerts, R. J., I. Hautefort, J. M. Sidebotham, and J. C. D. Hinton. 2002. Green fluorescent protein as a marker for conditional gene expression in bacterial cells. Methods Enzymol. 358:43-66. [DOI] [PubMed] [Google Scholar]
- 8.Borowiec, J. A., L. Zhang, S. Sasse-Dwight, and J. D. Gralla. 1987. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J. Mol. Biol. 196:101-111. [DOI] [PubMed] [Google Scholar]
- 9.Brosius, J. 1984. Plasmid vectors for the selection of promoters. Gene 27:151-160. [DOI] [PubMed] [Google Scholar]
- 10.Browning, D. F., and S. J. W. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57-65. [DOI] [PubMed] [Google Scholar]
- 11.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. d'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 12.Dorman, C. J. December 2004, posting date. Virulence gene regulation in Shigella. Chapter 8.9.3. In R. Curtiss III et al. (ed.), EcoSal-Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 13.Dorman, C. J. 2004b. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 14.Dorman, C. J., and M. E. Porter. 1998. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29:677-684. [DOI] [PubMed] [Google Scholar]
- 15.Dorman, C. J., S. McKenna, and C. Beloin. 2001. Regulation of virulence gene expression in Shigella flexneri, a facultative intracellular pathogen. Int. J. Med. Microbiol. 291:89-96. [DOI] [PubMed] [Google Scholar]
- 16.Dove, S. L., J. K. Joung, and A. Hochschild. 1997. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 386:627-630. [DOI] [PubMed] [Google Scholar]
- 17.Elledge, S. J., and R. W. Davis. 1989. Position and density effects on repression by stationary and mobile DNA binding proteins. Genes Dev. 3:185-197. [DOI] [PubMed] [Google Scholar]
- 18.Friedman, S. A., and S. J. Austin. 1988. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid 19:103-112. [DOI] [PubMed] [Google Scholar]
- 19.Funnell, B. E., and L. Gagnier. 1993. P1 plasmid partition complex at parS. II. Analysis of the parB protein binding activity and specificity. J. Biol. Chem. 268:3616-3624. [PubMed] [Google Scholar]
- 20.Hautefort, I., M. J. Proenca, and J. C. D. Hinton. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 69:7480-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes, F., and S. J. Austin. 1993. Specificity determinants of the P1 and P7 centromere analogs. Proc. Natl. Acad. Sci. USA 90:9228-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershberg, R., and H. Margalit. 2006. Co-evolution of transcription factors and their targets depends on mode of regulation. Genome Biol. 7:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hromockyj, A. E., S. C. Tucker, and A. T. Maurelli. 1992. Temperature regulation of Shigella virulence: identification of the repressor gene virR, an analogue of hns, and partial complementation by tyrosyl transfer RNA (tRNA1Tyr). Mol. Microbiol. 6:2113-2124. [DOI] [PubMed] [Google Scholar]
- 24.Hsu, L. M. 1996. Quantitative parameters for promoter clearance. Methods Enzymol. 273:59-71. [DOI] [PubMed] [Google Scholar]
- 25.Le Gall, T., M. Mavris, M. C. Martino, M. L. Bernardini, E. Denamur, and C. Parsot. 2005. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology 151:951-962. [DOI] [PubMed] [Google Scholar]
- 26.Lozada-Chávez, I., S. C. Janga, and J. Collado-Vides. 2006. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 34:3434-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucchini, S., G. Rowley, M. D. Goldberg, D. Hurd, M. Harrison, and J. C. D. Hinton. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:748-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch, A. S., and J. C. Wang. 1995. SopB protein-mediated silencing of genes linked to the sopC locus of Escherichia coli F plasmid. Proc. Natl. Acad. Sci. USA 92:1896-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, K. A., S. A. Friedman, and S. J. Austin. 1987. Partition site of the P1 plasmid. Proc. Natl. Acad. Sci. USA 84:8544-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurelli, A. T., B. Blackmon, and R. Curtiss III. 1984. Temperature-dependent expression of virulence genes in Shigella species. Infect. Immun. 43:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenna, S., C. Beloin, and C. J. Dorman. 2003. In vitro DNA binding properties of VirB, the Shigella flexneri virulence regulatory protein. FEBS Lett. 545:183-187. [DOI] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 33.Nasser, W., R. Schneider, A. Travers, and G. Muskhelishvili. 2001. CRP modulates fis transcription by alternate formation of activating and repressing nucleoprotein complexes. J. Biol. Chem. 276:17878-17886. [DOI] [PubMed] [Google Scholar]
- 34.Navarre, W. W., S. Porwollik, Y. Wang, M. McClelland, H. Rosen, S. J. Libby, and F. C. Fang. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella enterica serovar Typhimurium. Science 313:236-238. [DOI] [PubMed] [Google Scholar]
- 35.Nickerson, C. A., and E. C. Achberger. 1995. Role of curved DNA in binding of Escherichia coli RNA polymerase to promoters. J. Bacteriol. 177:5756-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oshima, T., S. Ishikawa, K. Kurokawa, H. Aiba, and N. Ogasawara. 17 October 2006. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 13:141-153. [Epub ahead of print.] [DOI] [PubMed]
- 37.Pabo, C. O., and R. T. Sauer. 1992. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 61:1053-1095. [DOI] [PubMed] [Google Scholar]
- 38.Pedersen, A. G., L. J. Jensen, S. Brunak, H. H. Staerfeldt, and D. W. Ussery. 2000. A DNA structural atlas for Escherichia coli. J. Mol. Biol. 299:907-930. [DOI] [PubMed] [Google Scholar]
- 39.Pérez-Rueda, E., and J. Collado-Vides. 2000. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 28:1838-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porter, M. E. 1998. The regulation of virulence gene expression in Shigella flexneri. Ph.D. thesis. University of Dublin, Dublin, Ireland.
- 41.Porter, M. E., and C. J. Dorman. 1994. A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J. Bacteriol. 176:4187-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porter, M. E., and C. J. Dorman. 1997. Positive regulation of Shigella flexneri virulence genes by integration host factor. J. Bacteriol. 179:6537-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porter, M. E., and C. J. Dorman. 2002. In vivo DNA-binding and oligomerization properties of the Shigella flexneri AraC-like transcriptional regulator VirF as identified by random and site-specific mutagenesis. J. Bacteriol. 184:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prosseda, G., M. Falconi, M. Giangrossi, C. O. Gualerzi, G. Micheli, and B. Colonna. 2004. The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol. 51:523-537. [DOI] [PubMed] [Google Scholar]
- 45.Radnedge, L., M. A. Davis, and S. J. Austin. 1996. P1 and P7 plasmid partition: ParB protein bound to its partition site makes a separate discriminator contact with the DNA that determines species specificity. EMBO J. 15:1155-1162. [PMC free article] [PubMed] [Google Scholar]
- 46.Radnedge, L., M. A. Davis, B. Youngren, and S. J. Austin. 1997. Plasmid maintenance functions of the large virulence plasmid of Shigella flexneri. J. Bacteriol. 179:3670-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rimsky, S. 2004. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 7:109-114. [DOI] [PubMed] [Google Scholar]
- 48.Rimsky, S., F. Zuber, M. Buckle, and H. Buc. 2001. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 42:1311-1323. [DOI] [PubMed] [Google Scholar]
- 49.Rodionov, O., M. Lobocka, and M. Yarmolinsky. 1999. Silencing of genes flanking the P1 plasmid centromere. Science 283:546-549. [DOI] [PubMed] [Google Scholar]
- 50.Schneider, R., A. Travers, T. Kutateladze, and G. Muskhelishvili. 1999. A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol. Microbiol. 34:953-964. [DOI] [PubMed] [Google Scholar]
- 51.Severinov, K., and S. A. Darst. 1997. A mutant RNA polymerase that forms unusual open promoter complexes. Proc. Natl. Acad. Sci. USA 94:13481-13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shin, M., M. Song, J. H. Rhee, Y. Hong, Y. J. Kim, Y. J. Seok, K. S. Ha, S. H. Jung, and H. E. Choy. 2005. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Eσ70 as a cofactor for looping. Genes Dev. 19:2388-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 54.Surtees, J. A., and B. E. Funnell. 2001. The DNA binding domains of P1 ParB and the architecture of the P1 plasmid partition complex. J. Biol. Chem. 276:12385-12394. [DOI] [PubMed] [Google Scholar]
- 55.Taniya, T., J. Mitobe, S. Nakayama, Q. Mingshan, K. Okuda, and H. Watanabe. 2003. Determination of the InvE binding site required for expression of IpaB of the Shigella sonnei virulence plasmid: involvement of a ParB BoxA-like sequence. J. Bacteriol. 185:5158-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Venkatesan, M. M., M. B. Goldberg, D. J. Rose, E. J. Grotbeck, V. Burland, and F. R. Blattner. 2001. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect. Immun. 69:3271-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner, R. 2000. Transcription regulation in prokaryotes. Oxford University Press, Oxford, England.
- 58.Watanabe, H., E. Arakawa, K. Ito, J. Kato, and A. Nakamura. 1990. Genetic analysis of an invasion region by use of a Tn3-lac transposon and identification of a second positive regulator gene, invE, for cell invasion of Shigella sonnei: significant homology of invE with ParB of plasmid P1. J. Bacteriol. 172:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]