Abstract
The attKLM operon encodes a lactonase (AttM) that hydrolyzes acylhomoserine lactone autoinducers, as well as two putative dehydrogenases (AttK and AttL). Here we show that AttK, AttL, and AttM collectively covert gamma-butyrolactone to succinate. Two metabolic intermediates, gamma-hydroxybutyrate and succinic semialdehyde, inactivated the AttJ repressor in vitro and induced attKLM transcription in vivo.
N-Acyl-homoserine lactones (AHLs) are utilized by a variety of proteobacteria as signal molecules that mediate cell-cell chemical communication. These signals are thought to provide information about cell population density, a phenomenon termed autoinduction or, more recently, quorum sensing (8, 18, 20). In general, quorum-sensing bacteria synthesize AHLs by a LuxI-type AHL synthase (12), and in most cases these AHL molecules are freely diffusible across the cell envelope. High population densities cause the accumulation of AHLs, which interact with a cognate LuxR-type AHL-dependent transcription factor (13, 19, 24, 25).
AHLs can be metabolized by a variety of bacteria, a phenomenon sometimes referred to as quorum quenching (23). AHL metabolism has attracted a great deal of interest, partly for possible therapeutic applications. A group of AHL-specific lactonases can inactivate AHLs by hydrolysis of the lactone ring (2, 6, 7, 11, 22). AHL lactonases have been identified in Bacillus spp., Agrobacterium tumefaciens, Arthrobacter sp., and cultured human cells (5, 11, 22). Uncultured bacteria in soil samples also rapidly degrade AHLs (17). The physiological or ecological roles of AHL lactonases in these bacteria are not clear. These enzymes may have evolved to block AHL-mediated cell-cell communication, while it is also possible that they evolved by selection for degradation of non-AHL compounds and that the hydrolysis of AHLs is incidental.
The AttM protein of A. tumefaciens, encoded on the megaplasmid pAtC58, was previously shown to have AHL lactonase activity in vivo (22). Nopaline-type A. tumefaciens strains have an additional AHL lactonase (AiiB) carried on the Ti plasmid (2). A. tumefaciens is thus one of the few bacteria known to encode both an AHL synthase and AHL-degrading enzymes. The attM gene is the last gene of the attKLM operon (21). The products of the attL and attK genes strongly resemble alcohol dehydrogenases and semialdehyde dehydrogenases, respectively (1). The transcription of the attKLM operon is repressed by AttJ, an IclR-type transcription factor encoded by an adjacent, divergent gene (22).
One study suggested that AttM converts gamma-butyrolactone (GBL) to gamma-hydroxybutyrate (GHB), that AttL converts GHB to succinic semialdehyde (SSA), and that AttK convert SSA to succinate (SA) (1). These findings were not confirmed biochemically. That study also suggested that GBL, GHB, and SSA were able to relieve repression by AttJ (1), although the interconversion of these compounds by the three catabolic enzymes clouded this conclusion. In a second study, the AttJ repressor was purified and demonstrated by EMSA to interact with the attK promoter, although the effects of GBL, GHB, and SSA on binding were not tested, and the binding site was not localized (22). Gamma-aminobutyric acid was found to induce the expression of the operon, possibly by metabolism to the direct inducer (1). In the present study, we use biochemical methods to positively identify the reactants and products of each of the three catabolic enzymes. We also disrupted each catabolic gene to learn whether they are essential for each reaction and found a second attK ortholog that can convert SSA to SA. We identified the attKLM transcription start site and used footprinting and promoter resections to identify the AttJ binding site. Inactivation of AttJ binding by GHB and SSA was reconstituted in vitro.
GBL is metabolized to succinate by AttM, AttL, and AttK in vitro.
In an earlier study, it was found that expression of AttL in Escherichia coli enabled the bacterium to multiply at the expense of GHB, whereas the coexpression of AttL and AttM enabled growth on GBL (1). To confirm the reaction carried out by AttM and to determine the reactions carried out by AttL and AttK, we overexpressed each protein separately in E. coli and made clarified extracts of each strain. We incubated putative substrates with each extract and identified the reaction products by mass spectrometry (MS). An E. coli extract expressing AttM rapidly hydrolyzed GBL to GHB (peak at 5.49 min in Fig. S1A and Table S1 in the supplemental material), while an extract lacking this protein did not (Table S2 in the supplemental material). Similarly, an extract containing AttK efficiently converted SSA to SA (peak at 5.79 min in Fig. S1C and Table S2 in the supplemental material), while a control extract did not (Table S2 in the supplemental material). The latter reaction required NAD+, although NADP+ could substitute at low efficiency (data not shown). An extract containing AttL converted GHB to SSA (peak at 4.80 min in Fig. S1B and Table S2 in the supplemental material). However, only ca. 3% of the substrate was converted even after prolonged incubation. This was expected, since this reaction is thermodynamically unfavorable (15). When GHB was added to an extract containing both AttK and AttL, it was efficiently converted to SA (peak at 5.80 min in Fig. S1D and Table S2 in the supplemental material).
We did similar in vitro assays for 3-oxooctanoyl-homoserine lactone and octanoyl-homoserine lactone. AttM hydrolyzed the ring of these compounds, as demonstrated earlier (22), forming the corresponding N-acylhomoserines (data not shown). However, extracts containing AttL, AttK, or both proteins did not further metabolize either compound.
GBL metabolism in vivo requires attM and attL, whereas attK is functionally redundant.
We wanted to determine whether the attKLM operon was solely responsible for GBL metabolism in vivo. To test this, we constructed two strains containing a nonpolar deletion of either attK or attL. We also disrupted attM by Campbell integration mutagenesis. We added GBL (1 mM) to early-log-phase cultures of these three strains plus the parental strain NTL4 (which lacks a Ti plasmid and therefore has just one known AHL lactonase). Samples were collected at various time intervals and analyzed by gas chromatography-MS (GC/MS). Among the metabolites in the degradation pathway of GBL, GHB was detected as a discrete peak by GC/MS. SSA and SA were not detected or were detected in trace amounts due to their rapid further catabolism. In the wild-type strain, GBL was rapidly degraded, and GHB appeared transiently and was not detected at later time points (Table S3 in the supplemental material). In contrast, in the attM mutant (strain YC1), GBL was not converted to other metabolites and gradually decreased over the incubation period, possibly due to its volatility (Table S3 in the supplemental material). The attL mutant (strain YC5) converted GBL to GHB, which was not further metabolized (Table S3 in the supplemental material). This indicates that AttL is the sole enzyme catalyzing the reaction from GHB to SSA.
The attK mutant (strain YC6) gave somewhat more unexpected results. We had anticipated that GBL and GHB would be depleted in this mutant and that SSA would accumulate. Although GBL and GHB were rapidly depleted, SSA accumulated only transiently and to very low levels (Table S3 in the supplemental material). There could be two reasons for the poor accumulation SSA. First, the conversion of GHB to SSA is thermodynamically unfavorable (15), so the equilibrium favors the reactant. Second, A. tumefaciens could express a second enzyme that converts GHB to SSA. The depletion of GHB supports the second hypothesis. Analysis of the A. tumefaciens genome reveals several genes whose products closely resemble AttK. Of these, the most similar are atu3403, atu3498, atu4247, and atu4762 (whose translation products are 62, 41, 61, and 51% identical to AttK, respectively). Starting with the attK mutant YC6, we disrupted each of these four genes by Campbell integration mutagenesis, and the degradation of GBL in all four double mutants was monitored by GC/MS analysis. As expected from thermodynamic considerations, SSA did not accumulate to significant levels in the supernatant of any strain (Table S3 in the supplemental material). In one of the double mutants (strain HC154, which is defective for attK and atu3498), GHB accumulated to higher levels than it did in with YC6 (the attK single mutant) or any of the other double mutants. Keeping in mind that (for thermodynamic reasons) a block in SSA metabolism causes accumulation of GHB rather than SSA, we believe that the attK atu3498 double mutant is blocked for SSA metabolism. The atu3498 gene was found to encode an SSA dehydrogenase in a separate study (16). The other AttK homologs may use other substrates or might simply not be expressed under the conditions of this experiment.
Induction of the attKLM operon by GBL, GHB, and SSA.
Previous studies showed that GBL, GHB, and SSA induced the expression of the attKLM promoter (1, 16). However, the strain used in that study was a attKLM+ strain, and the added compounds could therefore be interconverted, complicating the analysis. We constructed an attM-lacZ fusion at the native locus on the pAtC58 plasmid by Campbell integration (strain YC1). This fusion also disrupted attM and therefore blocked GBL metabolism (see above). We tested the attM inducing activity of more than 20 various kinds of AHLs (all added at a final concentration of 1 mM). None of them induced attM-lacZ fusion (data not shown). A smaller set of AHLs were found not to induce this operon in two previous studies (1, 22). Likewise, neither homoserine lactone nor homoserine induced the fusion. The fact that none of these compounds induced the fusion, combined with fact that AttL and AttK do not play a detectable role in the metabolism of these compounds, suggests that this operon did not evolve by selection for AHL degradation.
We then tested the attM-lacZ fusion strain for induction by GBL, GHB, SSA, and SA over a wide range of concentrations (Fig. S2A in the supplemental material). GBL, GHB, and SSA induced the fusion, while SA did not. However, induction by GHB and SSA was much stronger than by GBL (Fig. S2A in the supplemental material). Since the strain used in this experiment is an attM mutant, the added GBL is not readily metabolized (see Table S3 in the supplemental material), whereas exogenous GHB and SSA are metabolized. Furthermore, the slight induction observed with GBL could have resulted from spontaneous ring cleavage of this compound, yielding GHB. This suggests that GBL may have little or no inducer activity, and the earlier findings (1) that GBL is an inducer should be reevaluated with strains that are blocked in GBL metabolism.
Although GHB and SSA appeared to be equally strong inducers in a previous study (1), it is possible that in that study GHB was rapidly converted to SSA and that only SSA was the direct inducer. To test this, we introduced a low-copy plasmid (pYC174) carrying an attK-lacZ fusion into an attL mutant strain (YC5) and into a wild-type control (strain NTL4). GHB significantly induced expression of the fusion in both strains (see Fig. 2B), indicating that this compound is a strong inducer. The fact that GHB appeared to be more effective in the wild-type strain than in an attL mutant suggests that its conversion to SSA may enhance its effectiveness. This hypothesis received additional support using electrophoretic mobility shift assays (EMSA) described below.
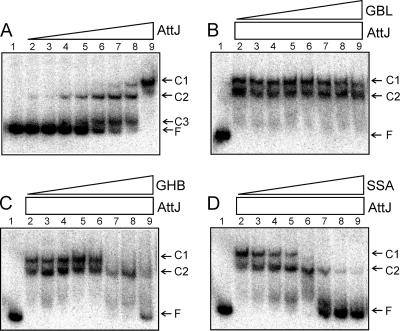
FIG. 2.
(A) EMSA using His-AttJ and radiolabeled attKLM promoter DNA. The positions of the three shifted protein-DNA complexes (C1, C2, and C3) and free DNA (F) are marked. Lane 1 represents free DNA with no added protein. AttJ protein was provided at concentrations of 0.3, 1, 3, 10, 30, 100, 300, and 1,000 nM (lanes 2 to 9). In panels B to D, AttJ was provided at a final concentration of 1,000 nM, and GBL (C), GHB (D), or SSA (E) was provided at concentrations of 10 μM, 30 μM, 100 μM, 300 μM, 1 mM, 3 mM, 10 mM, and 30 mM (lanes 2 to 9, respectively). His6-AttJ was overproduced by using strain BL21(DH3)/pYC160. After lysis and clarification of a 500-ml culture, the soluble fraction was diluted 10-fold with 50 mM phosphate buffer (pH 7.4; 100 mM NaCl, 10% glycerol) and added to a TALON cobalt affinity column (10-ml bed volume) (BD Biosciences). Bound proteins were washed with 40 ml of buffer A (50 mM phosphate buffer [pH 7.4], 300 mM NaCl, 10% glycerol) and eluted using 20 ml of buffer B (buffer A supplemented with 200 mM imidazole). Fractions containing His-AttJ proteins were pooled and dialyzed against 500 ml of phosphate buffer overnight at 4°C. A 217-nucleotide DNA fragment containing the attKLM promoter was amplified by PCR using the oligonucleotides PattKLM-F1 and PattKLM-R1. The resulting PCR products were digested with EcoRI and end labeled using [α-32P]dATP. Binding reactions and size fractionation of the samples were performed according to a previously published protocol (24).
Localization of the attKLM transcription start site and repressor binding site.
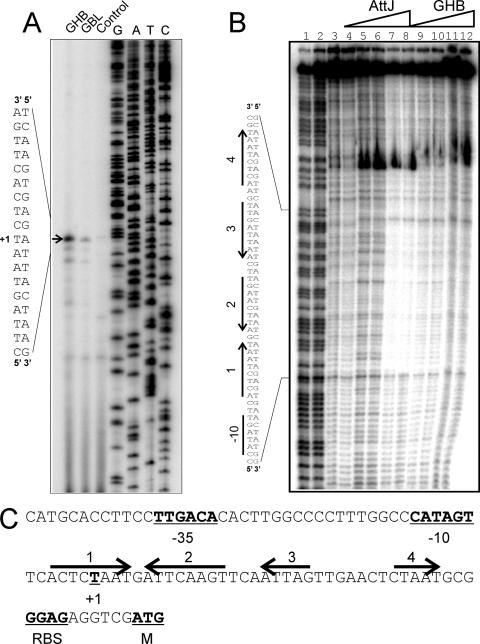
Using primer extension analysis, we identified the 5′ end of a transcript that was weakly induced by GBL and more strongly induced by GHB (marked as +1 in Fig. 1A and C). This site lies directly downstream of sequences similar to consensus −10 and −35 promoter motifs (underlined nucleotides in Fig. 1C). This transcription start site lies well upstream of a possible start site identified by sequence inspection in an earlier study (22).
FIG. 1.
Primer extension and DNase I footprinting assays of the attK promoter. (A) Primer extension assays. Lanes 1 and 2 identify mRNA from cultures incubated with GHB or GBL, respectively, added at a final concentration of 10 mM to strain NTL4. Primer extension assays were performed by using a protocol described previously (3). RNA was prepared by using RNeasy minikits (QIAGEN) from exponential-phase cultures of NTL4 after 2 h of growth in the presence of 100 μM GBL or GHB. The arrow indicates the position of the transcription start. (B) Footprinting assays using purified His6-AttJ and the radiolabeled attK promoter DNA. Lanes 1 and 2 represent G+C ladders; lanes 3 and 4 represent free DNA; in lanes 5 and 6, His-AttJ was added at 1 μM; in lanes 7 and 8, His-AttJ was added at 3 μM; in lanes 9 and 10, His-AttJ was added at 3 μM and GHB (sodium salt) was added at 10 mM; and in lanes 11 and 12, His-AttJ was added at 3 μM and GHB was added at 30 mM. Sequences protected by AttJ are indicated on the left. DNase I footprinting assays were carried out using a 217-nucleotide PCR product digested with EcoRI and SacII and end labeled at the EcoRI site using [α-32P]dATP and Klenow DNA polymerase. AttJ was added to the DNA fragment and allowed to bind for 30 min in a 50-μl volume. DNase I (0.25 U) was then added to the samples, and the reactions were terminated after 30 s by the addition of 0.25 M EDTA and 5 mg of yeast tRNA/ml. DNA was precipitated using ethanol and size fractionated using 6% polyacrylamide denaturing gels in 1× Tris-borate-EDTA buffer. (C) Sequence analysis of the attKLM promoter. The −35 and −10 RNA polymerase binding site motifs, the transcriptional start, and the predicted ribosome-binding site and translational start of the attK gene are underlined and in boldface. The four arrows represent DNA sequences with a limited dyad symmetry that overlap the region that is protected by His-AttJ in footprinting assays. All gels were analyzed by using a Storm B840 PhosphorImager (Molecular Dynamics).
AttJ was previously shown by EMSA to bind to a fragment containing the attK promoter (22). However, the binding site was not mapped, and the effect of inducing ligands was not tested. To map the binding site for AttJ, we constructed a His6-AttJ fusion, purified it to virtual homogeneity by immobilized metal affinity chromatography, and used this protein and a 217-nucleotide DNA fragment containing the attKLM promoter for EMSA. Since AttJ was titrated in a series of binding reactions, a total of three complexes were detected (Fig. 2A), suggesting that multiple AttJ subunits can bind the operator. The Kd for the binding was estimated to be approximately 2 × 10−7 M. Formation of complex C1 appeared to be highly cooperative, whereas formation of complex C2 and C3 seemed to be noncooperative (Fig. 2A).
We added various amounts of GBL, GHB, and SSA to DNA-binding reactions to see whether these compounds affected AttJ binding affinity. GBL had virtually no effect on AttJ binding except possibly at the highest concentration (lane 9 in Fig. 2B), supporting the idea that this compound is not a significant inducer of the attKLM operon. In contrast, both GHB and SSA caused dissociation of AttJ from the promoter (Fig. 2C and D). SSA was effective in a similar EMSA in a previous study (16). Of these, SSA was more effective than GHB at low concentrations. These results matched results from in vivo assays (described above), which again suggest that SSA is the stronger inducer than GBL or GHB. The fact that purified AttJ was released by these compounds also indicates that it is the direct receptor of these chemical signals.
To identify AttJ binding sites, we carried out DNase I footprinting experiments. The binding site extends from nucleotides −11 to +34 with respect to the transcription start site (Fig. 1B), a position well suited for a repressor binding site (11). This 45-nucleotide region includes two pairs of repeated sequences (arrows 1, 2, 3, and 4 in Fig. 1B and C). Of these, sites 1 and 2 form one pair of inverted repeats, and sites 3 and 4 form a second pair. When GHB was added at 10 or 30 mM, it partially released AttJ from the DNA (Fig. 1B, right lanes).
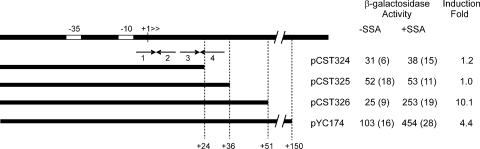
We also used a genetic approach to identify a cis-acting site required for regulation. We made three promoter resections from the 3′ end of the regulatory region and made transcriptional fusions between the resected promoter fragments and lacZ. His6-AttJ was overexpressed by fusion to a Ptac promoter. A promoter fragment containing 51 nucleotides downstream of the transcription start site showed a similar response to SSA as a fragment containing 150 nucleotides (Fig. 3). In contrast, fragments containing 24 or 36 nucleotides downstream of the start site showed no response to SSA (Fig. 3). Evidently, some sequence between +35 and +51 is needed for repression. These data are in close agreement with the DNase I footprinting data described above.
FIG. 3.
Resections of the attK promoter. Plasmids pCST324, pCST325, pCST326, and pYC174 contain fusions of the attK promoter fragments indicated. They were introduced into A. tumefaciens strain YC6(pHis6-AttJ) and assayed for β-galactosidase after overnight incubation with or without 1.5 mM SSA. β-Galactosidase activity is expressed in Miller units (10); values in parentheses represent standard deviations.
Arabidopsis and squash seedlings release GHB-like inducer of attKLM.
A proteomic analysis showed that AttM and AttK are preferentially expressed when bacteria are incubated with tomato root sections (14). We tested exudates of four different nonwounded seedlings (Arabidopsis, squash, tomato, and tobacco) for induction of the attKLM operon. Two seedling exudates (from Arabidopsis and squash) caused a mild induction of the attKLM-lacZ fusion (Fig. S3A in the supplemental material).
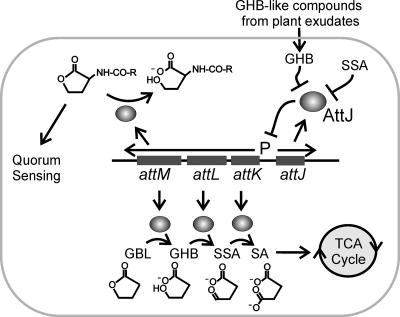
We used MS to test whether these seedling exudates contained known attKLM inducers. None of the four exudates contained detectable levels of GBL or SSA. However, GHB or a similar compound was readily detectable in exudates of Arabidopsis, squash, and tomato seedlings. Figure S3B and C show the retention time (5.530 min) and the mass spectrum of the GHB-like compound from Arabidopsis seedling exudate, which are similar to those of the reagent GHB (Fig. S3D and 3E in the supplemental material). This compound could also be γ-amino butyric acid (GABA), which closely resembles GHB in mass spectrum and which is known to induce the attKLM operon (4). This finding could have significant ecological implications for cell-cell communication by this bacterium. The release of GHB or related compounds into the rhizosphere could in principle block AHL-mediated signaling by inducing the transcription of the AttM lactonase (Fig. 4).
FIG. 4.
Model for the role of plant-released GHB during A. tumefaciens colonization. In this model, GHB induces transcription of the attKLM operon by inactivating the AttJ repressor. The AttK and AttL proteins metabolize GHB to SA, which enters the tricarboxylic acid cycle. AttM can convert GBL to GHB. AttM can also hydrolyze 3-oxooctanoyl-homoserine lactone, forming N-acylhomoserine, thereby blocking quorum sensing.
Supplementary Material
Acknowledgments
We thank Anja Brencic for preparing plant exudates and members of Winans laboratory for helpful discussions and critical evaluations of the manuscript.
This study was supported by a grant from the National Institute of General Medical Sciences (GM42983).
Footnotes
Published ahead of print on 16 February 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Carlier, A., R. Chevrot, Y. Dessaux, and D. Faure. 2004. The assimilation of gamma-butyrolactone in Agrobacterium tumefaciens C58 interferes with the accumulation of the N-acyl-homoserine lactone signal. Mol. Plant-Microbe. Interact. 17:951-957. [DOI] [PubMed] [Google Scholar]
- 2.Carlier, A., S. Uroz, B. Smadja, R. Fray, X. Latour, Y. Dessaux, and D. Faure. 2003. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactonase activity. Appl. Environ. Microbiol. 69:4989-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chai, Y., and S. C. Winans. 2005. A small antisense RNA downregulates expression of an essential replicase protein of an Agrobacterium tumefaciens Ti plasmid. Mol. Microbiol. 56:1574-1585. [DOI] [PubMed] [Google Scholar]
- 4.Chevrot, R., R. Rosen, E. Haudecoeur, A. Cirou, B. J. Shelp, E. Ron, and D. Faure. 2006. GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 103:7460-7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun, C. K., E. A. Ozer, M. J. Welsh, J. Zabner, and E. P. Greenberg. 2004. Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc. Natl. Acad. Sci. USA 101:3587-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong, Y.-H., L.-H. Wang, J.-L. Xu, H.-B. Zhang, X.-F. Zhang, and L.-H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 7.Dong, Y.-H., J.-L. Xu, X.-Z. Li, and L.-H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 9.Gralla, J., and J. Collado-Vides. 1996. Organization and function of transcription regulatory elements, p. 1232-1245. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology Press, Washington, DC.
- 10.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 11.Park, S.-Y., S. J. Lee, T.-K. Oh, J.-W. Oh, B.-T. Koo, D.-Y. Yum, and J.-K. Lee. 2003. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 149:1541-1550. [DOI] [PubMed] [Google Scholar]
- 12.Parsek, M. R., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 96:4360-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin, Y., Z. Q. Luo, A. J. Smyth, P. Gao, S. Beck von Bodman, and S. K. Farrand. 2000. Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. EMBO J. 19:5212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosen, R., A. G. Matthysse, D. Becher, D. Biran, T. Yura, M. Hecker, and E. Z. Ron. 2003. Proteome analysis of plant-induced proteins of Agrobacterium tumefaciens. FEMS Microbiol. Ecol. 44:355-360. [DOI] [PubMed] [Google Scholar]
- 15.Segel, I. H. 1976. Biochemical calculations, 2nd ed. John Wiley & Sons, Inc., New York, NY.
- 16.Wang, C., H. B. Zhang, L. H. Wang, and L. H. Zhang. 2006. Succinic semialdehyde couples stress response to quorum-sensing signal decay in Agrobacterium tumefaciens. Mol. Microbiol. 62:45-56. [DOI] [PubMed] [Google Scholar]
- 17.Wang, Y. J., and J. R. Leadbetter. 2005. Rapid acyl-homoserine lactone quorum signal biodegradation in diverse soils. Appl. Environ. Microbiol. 71:1291-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 19.Welch, M., D. E. Todd, N. A. Whitehead, S. J. McGowan, B. W. Bycroft, and G. P. C. Salmond. 2000. N-acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 19:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 21.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M.-J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z.-Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J.-F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, H.-B., L.-H. Wang, and L.-H. Zhang. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, L.-H. 2003. Quorum quenching and proactive host defense. Trends Plant Sci. 8:238-244. [DOI] [PubMed] [Google Scholar]
- 24.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA 96:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu, J., and S. C. Winans. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. USA 98:1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.