Abstract
We have constructed defined deletions in the hmuO gene from Corynebacterium diphtheriae and Corynebacterium ulcerans and show that the C. ulcerans hmuO mutation results in a significant reduction in hemoglobin-iron utilization, whereas in C. diphtheriae strains, deletion of hmuO caused no or only partial reduction in the utilization of heme as an iron source. We also show that expression from the C. ulcerans hmuO promoter exhibits minimal regulation by iron and heme whereas transcription from the C. diphtheriae hmuO promoter shows both significant iron repression and heme-dependent activation. These findings indicate that variability in HmuO function and expression exists among Corynebacterium species.
Iron is an essential nutrient for almost all bacteria, and the ability to acquire sufficient amounts of this element is important for many virulent organisms to cause disease in a mammalian host (6, 7). Corynebacterium diphtheriae, the etiological agent of diphtheria, uses various compounds as iron sources, including heme and hemoglobin (10, 18, 21, 22). Corynebacterium proteins involved in the uptake and utilization of heme as an iron source include an ABC-type heme transporter encoded by the hmuTUV genes (10, 22) and HmuO, a heme oxygenase that has been shown in vitro to oxidatively cleave the heme macrocycle, which results in the release of heme-bound iron (18, 31). In a previous study, it was shown that point mutations in the hmuO gene in the HC1 strain of C. diphtheriae C7 resulted in a diminished ability to use heme-iron (9, 18). This earlier study utilized a qualitative assay that measured the ability of C. diphtheriae HC1 mutants to use heme compounds as a sole iron source and used chemical mutagenesis to create the mutations (18). Due to the limitations of the method used to identify and screen for the heme utilization mutants in this earlier study, wild-type C. diphtheriae C7 could not be used for the isolation of hmuO mutations (18). Moreover, while the C. diphtheriae HC1 parent strain was useful for the identification of hmuO mutants, it was not an ideal background for the analysis of these mutants, since the strain is severely defective in iron uptake and likely contains multiple mutations in genes involved in iron transport (13, 16). Chemical mutagenesis was also used to obtain heme-iron utilization mutants of Corynebacterium ulcerans 712 (CU712), and two distinct groups of mutants were identified: one group was complemented by the C. diphtheriae hmuTUV transport genes, while the second group was complemented by the cloned hmuO gene (10, 18). The specific mutations in these C. ulcerans heme utilization mutants were not, however, specifically identified or further characterized.
Although C. diphtheriae HmuO was the first bacterial heme oxygenase to be identified, heme oxygenases that share sequence or structural similarities to HmuO have been found in both gram-negative and gram-positive organisms (17, 27, 33). A novel group of proteins that possess a heme-degrading activity, but have no amino acid sequence similarity to HmuO, have recently been described for several bacterial species, including Staphylococcus aureus (25), Bacillus anthracis (26), and Bradyrhizobium japonicum (15). Moreover, some of the genes that encode these novel heme degradation enzymes were shown to complement the defect in a C. ulcerans heme utilization mutant (15, 25), which suggests that these genes are associated with the utilization of heme as an iron source.
Expression of the C. diphtheriae hmuO gene has been previously examined, and its transcription was shown to be under a dual mode of regulation, which involves iron repression, mediated by DtxR (5, 19, 20), and heme-dependent activation that requires the ChrS-ChrA and HrrS-HrrA two-component signal transduction systems (3, 4). Previous studies showed that a DtxR binding site overlaps the hmuO promoter region and also demonstrated that a region upstream of the hmuO promoter is critical for the heme-dependent activation (20).
In this study, we characterized a defined deletion in the hmuO gene in the wild-type strain C. diphtheriae C7(−) (2) and also in Russian clinical isolates 1737 and 1716 (12, 14, 18). Additionally, the hmuO gene from CU712 was cloned and a hmuO deletion mutant of CU712 was constructed and characterized. We also show that iron and heme regulation of the C. ulcerans hmuO promoter is significantly different from that observed previously with the C. diphtheriae hmuO promoter.
Analysis of the CU712 hmuO gene and construction of hmuO deletion mutants.
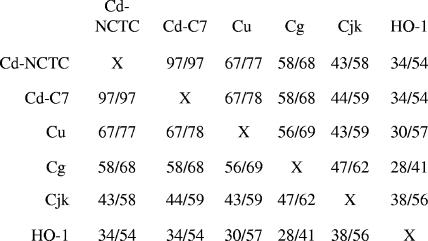
The genome sequence of C. ulcerans has not been determined; therefore, two oligonucleotide primers derived from C. diphtheriae sequences, primer hm1a (5′-GGCGGATCCCGACAAGTAGTAGATGAGGTGTTTTGGGGC-3′) and primer thrC-1 (5′-CTGCATTGCCATGTCTTTAAATGC-3′), were used to amplify a 4-kb DNA fragment from CU712 that contained the hmuO gene, and this amplicon was subsequently cloned into pCRBluntII-TOPO vector (Invitrogen). The nucleotide sequence of the C. ulcerans hmuO gene was determined, and the predicted product (24.0 kDa) was found to share 67% identity with and 77% similarity to HmuO from C. diphtheriae. A comparison of the amino acid sequences among HmuO proteins from various Corynebacterium species and the human heme oxygenase HO-1 is shown in Fig. 1. A BLAST search (1) of the recently completed genome sequence of C. efficiens, a species related to C. glutamicum, failed to identify a hmuO ortholog.
FIG. 1.
Amino acid comparison between various Corynebacterium HmuO proteins and the human heme oxygenase HO-1. The numbers separated by a slash represent percent identity/similarity. Cd-NCTC, C. diphtheriae genome strain NCTC 13129 (8); Cd-C7, C7(−) strain; Cu, C. ulcerans 712; Cg, Corynebacterium glutamicum; Cjk, Corynebacterium jeikeium.
C. ulcerans strain CU29, a chemically derived mutant of CU712, was previously reported to have a reduced ability to use heme as an iron source, and it was shown that this defect is complemented by the presence of the cloned C. diphtheriae hmuO gene (18). The hmuO allele from CU29 was cloned and sequenced (as described above for the wild-type hmuO gene) and shown to contain a single point mutation that results in the amino acid change at position 86 from D to N, which is likely responsible for the heme-iron utilization defect observed in this mutant (data not shown). Examination of the recently solved crystal structure of C. diphtheriae HmuO reveals that D86 is located on the protein surface and is associated with a region involved in HmuO enzymatic activity (11).
The hmuO deletion mutant in C. diphtheriae C7 (C7 hmuOΔ) was previously described (4), and construction of an hmuO deletion in Russian clinical isolates 1737 (1737 hmuOΔ) and 1716 (1716 hmuOΔ) was done following the same procedure as described for C7. A hmuO deletion mutant was also constructed using CU712 (CU712 hmuOΔ) by allelic replacement as previously reported (4, 29). CU712 hmuOΔ contains a 305-bp internal deletion of hmuO and is predicted to generate an in-frame fusion between 84 amino acids at the N terminus and 26 residues from the C terminus. The mutation was confirmed by PCR and DNA sequence analysis (not shown). Regions essential for the catalytic activity of HmuO are deleted in both the C. diphtheriae and C. ulcerans hmuO deletion mutants.
Hemoglobin utilization assays.
To assess the effect of the deletion of hmuO in Corynebacterium strains, a quantitative growth assay was developed to determine the capacity of wild-type and mutant bacteria to use hemoglobin as a sole iron source. Briefly, Corynebacterium strains were grown overnight (20 to 22 h at 37°C) in heart infusion broth (Becton Dickinson, Sparks, MD) with 0.2% Tween 80 (Aldrich, Milwaukee, WI) (HIBTW) and then inoculated to an optical density at 600 nm (OD600) of 0.2 into fresh HIBTW that contained 12 μg/ml of the iron chelator ethylenediamine di(o-hydroxyphenylacetic acid) (EDDA; Sigma, St. Louis, MO). Strains were grown for several hours at 37°C until log phase, at which time bacteria were recovered by centrifugation, resuspended in mPGT medium, a low-iron semidefined minimal medium (28), and then inoculated at an OD600 of 0.03 into fresh mPGT medium that contained various supplements as indicated. After 20 to 22 h of growth at 37°C, the OD600 of the cultures was determined. All strains grew well in the presence of FeSO4 (+Fe; mPGT containing 1 μM FeSO4), and all were inhibited for growth under low-iron conditions (−Fe; mPGT containing 3.6 μg/ml EDDA) (Table 1). All Corynebacterium strains grew to an OD600 of 0.1 or less in mPGT medium that contained no added supplements (data not shown). EDDA was added to mPGT medium (−Fe) at 3.6 μg/ml to chelate trace iron that may have contaminated or subsequently dissociated from the heme and hemoglobin. Wild-type Corynebacterium strains were examined for their ability to use hemoglobin as an iron source over a range of concentrations. Wild-type CU712 and C. diphtheriae C7(−) utilized hemoglobin as an iron source at levels as low as 5 μg/ml, and hemoglobin levels higher than 50 μg/ml did not further enhance growth of any of the strains (Table 1 and data not shown). Heme also stimulated growth of both C. diphtheriae and CU712; however, as previously reported (4), heme was shown to be toxic to C. diphtheriae, but not to C. ulcerans, at concentrations higher than 5 μM (data not shown).
TABLE 1.
Growth stimulation of Corynebacterium strains by iron and hemoglobin
| Strain/plasmid | Growth (OD600) with various iron sourcesa
|
|||||
|---|---|---|---|---|---|---|
| +Feb | −Fec | −Fe with hemoglobin (μg/ml)
|
||||
| Hbf (50) | Hb (25) | Hb (10) | Hb (5) | |||
| CU712 wild type | 4.5 | <0.1 | 4.8 | 2.8 | 0.5 | 0.3 |
| CU712 hmuOΔ | 4.0 | <0.1 | 0.14 | 0.10 | ndd | nd |
| CU712 hmuOΔ/pCUhmuO | 3.8 | <0.1 | 4.2 | 2.5 | nd | nd |
| CU712 hmuOΔ/p37hmuO | 4.2 | <0.1 | 4.1 | 2.2 | nd | nd |
| CU712 hmuOΔ/p1841 | 3.8 | <0.1 | 0.13 | 0.08 | nd | nd |
| C7(−) wild type | 2.1 | <0.1 | 4.3 | 2.7 | 0.7 | 0.4 |
| C7 hmuOΔ | 2.2 | <0.1 | 1.1 | 0.7 | nd | nd |
| C7 hmuOΔ/pCM2.6 (vector) | 5.7e | <0.1 | 1.9 | 0.9 | nd | nd |
| C7 hmuOΔ/pCD293 (hmuO+) | 5.9 | <0.1 | 5.5 | 2.6 | nd | nd |
| C7 hmuOΔ-1841Δ | 2.0 | <0.1 | 0.9 | 0.5 | nd | nd |
| 1737 wild type | 6.4 | <0.1 | 5.8 | 3.5 | nd | nd |
| 1737 hmuOΔ | 6.8 | <0.1 | 6.6 | 3.3 | nd | nd |
OD600 was determined for each culture after growth for 20 to 22 h in mPGT medium. Values shown are results from a representative experiment. All experiments were repeated at least three times, and the results of each assay varied by less than 20% from the mean.
+Fe medium is mPGT with 1 μM FeSO4.
−Fe medium is mPGT with 3.6 μg/ml EDDA.
nd, not done.
C7 strains carrying pCM2.6 plasmids grow to higher densities than strains without plasmids.
Hb, hemoglobin.
CU712 hmuOΔ, the C. ulcerans hmuO deletion mutant, showed minimal growth in the presence of hemoglobin, and CU29, the hmuO point mutant, showed results similar to those seen with CU712 hmuOΔ, which indicates that these strains are strongly defective in their ability to use hemoglobin as an iron source (Table 1 and data not shown). To confirm that HmuO was essential for C. ulcerans to utilize hemoglobin as an iron source, pCUhmuO, a pKN2.6-based vector (23) that contains a copy of the C. ulcerans hmuO gene, was introduced into CU712 hmuOΔ and shown to complement the heme-iron utilization defect observed in this mutant (Table 1). Similarly, the cloned C. diphtheriae hmuO gene on plasmid pCD293 (18) was also able to restore heme utilization to CU712 hmuOΔ (data not shown). All complementing clones used in this study were derived from low-copy-number (1 to 2 copies per cell), pNG2-based, Corynebacterium-Escherichia coli shuttle vectors (23).
The C. diphtheriae C7(−) hmuO deletion mutant C7 hmuOΔ showed a marked decrease relative to the wild-type strain in its ability to use hemoglobin as an iron source for growth; however, this defect was not as severe as that observed for CU712 hmuOΔ, as C7 hmuOΔ could maintain growth in the presence of 25 to 50 μg/ml hemoglobin (Table 1). The hemoglobin utilization defect in C7 hmuOΔ was complemented by the introduction of the cloned C. diphtheriae hmuO gene on plasmid pCD293 (Table 1), which confirms that a defect in HmuO is responsible for the diminished ability of C7 hmuOΔ to use hemoglobin as an iron source. C7 hmuOΔ was also complemented by the presence of the cloned CU712 hmuO gene on plasmid pCUhmuO (data not shown).
Surprisingly, deletion of the hmuO gene in the Russian clinical isolate 1737 hmuOΔ had no effect on its ability to use hemoglobin as an iron source, since growth of the mutant was comparable to that of the wild-type strain (Table 1). The HmuO protein from 1737 differs from C7 HmuO at four residues; however, these differences in sequence do not appear to impair HmuO function, since the cloned 1737 hmuO gene on pKN2.6 (p37hmuO) complemented the hemoglobin-iron utilization defect in CU712 hmuOΔ (Table 1). To further investigate HmuO function in C. diphtheriae strains, an hmuO deletion was constructed in another Russian clinical isolate, 1716 (1716 hmuOΔ), and characterization of this strain indicated a hemoglobin-iron utilization defect similar to that observed in the C7 strain (data not shown). Previous studies have indicated that C. diphtheriae strains exhibit significant genetic diversity in their iron utilization systems (12, 13), and the findings reported here suggest that C. diphtheriae strains C7 and 1716 require HmuO and an additional factor(s) to acquire heme-iron, while in strain 1737, HmuO is not essential for the utilization of heme as an iron source. PCR analysis using three different primer pairs, whose sequences correspond to internal regions of the hmuO gene that are conserved between strains C7, 1737, and CU712, failed to identify a PCR product when 1737 hmuOΔ chromosomal DNA was used as a template for PCR (M. P. Schmitt, unpublished data). These findings suggest that a second or similar copy of the hmuO gene is not encoded on the chromosome of 1737.
Analysis of dip1841.
Bacterial proteins with heme-degrading activity that are distinct from previously described heme oxygenases have recently been reported in studies of S. aureus (IsdG and IsdI) (25), B. anthracis (IsdG and IsdI) (26), and B. japonicum (HmuQ and HmuD) (15). In addition, the isdG and isdI genes from S. aureus and hmuQ and hmuD from B. japonicum were shown to complement the heme-iron utilization defect in C. ulcerans CU29 (15, 25). A BLAST search of the recently completed genome sequence of C. diphtheriae NCTC 13129 (8) has identified a putative ortholog of the IsdG/HmuQ proteins (designated dip1841) that is predicted to encode a 101-amino-acid product that has 25% and 28% identity and 46% and 44% similarity to IsdG and HmuQ, respectively. Studies based on the crystal structure of IsdG have identified residues important for enzymatic activity (32), some of which are conserved in Dip1841; moreover, Pfam analysis (http://pfam.wustl.edu/) indicates that Dip1841 is a member of the ABM family of monooxygenases, which also includes HmuQ and IsdG (15). Analysis of the C. diphtheriae genome suggests that dip1841 is not linked to a heme or iron transport locus and is the distal gene in a two-gene operon (dip1842-dip1841). To determine whether Dip1841 can complement an hmuO mutant, both the dip1841 gene alone and the complete dip1841-dip1842 operon were cloned onto plasmid vector pKN2.6 to generate p1841 and p1841-42, respectively, and these constructs were tested for their ability to correct the hemoglobin-iron utilization defect in CU712 hmuOΔ. Neither p1841 nor p1841-42 was able to restore heme-iron utilization to CU712 hmuOΔ or CU29 (Table 1 and not shown). To determine whether Dip1841 has a role in the use of heme as an iron source in C. diphtheriae, a dip1841 nonpolar in-frame deletion was constructed using wild-type and hmuO mutant strains of C7 (C7-1841Δ and C7 hmuOΔ-1841Δ) and 1737 (1737-1841Δ and 1737 hmuOΔ-1841Δ) using previously described methods (4, 29). The dip1841 mutation was confirmed by PCR and is predicted to encode a product that contains four amino acids from the N terminus of Dip1841 fused in frame to two amino acids at the C terminus. The dip1841 deletion in the wild-type C7 and 1737 strains or in 1737 hmuOΔ did not result in a decreased ability to use hemoglobin as an iron source, and the C7 double mutant, C7 hmuOΔ-1841Δ, exhibited a phenotype similar to that seen in the single hmuO mutant strain C7 hmuOΔ (Table 1 and data not shown). These findings suggest that dip1841 is not involved in the utilization of hemoglobin as an iron source in C. diphtheriae and indicate the need for additional studies to identify the factor(s) involved in the extraction of heme-iron in the 1737 and C7 hmuOΔ strains.
Hemoglobin and iron regulation at the C. ulcerans hmuO promoter.
Previous studies demonstrated that transcription from the C. diphtheriae hmuO promoter was repressed under high-iron conditions (with the repression mediated by DtxR) and activated by the ChrA-ChrS and HrrA-HrrS two-component systems in the presence of a heme source (3, 4, 19, 20). A DtxR binding site overlaps the C. diphtheriae hmuO promoter, and a 50-bp region upstream of the promoter is required for full heme-dependent expression (20); however, specific DNA sequences required for activation have not been identified. A comparison of the hmuO promoter regions between C. diphtheriae and C. ulcerans shows that critical residues in the −10 and −35 regions are relatively well conserved (Fig. 2A); however, the DtxR binding site in C. ulcerans has less homology to the 19-bp consensus binding sequence than that observed in C. diphtheriae. Specifically, C. ulcerans matches 13 of 19 bases whereas C. diphtheriae matches 15 of 19 bases with the consensus DtxR binding site, and 14 of 19 bases of the two species match (Fig. 2A and B). The region upstream of the promoter is poorly conserved between the species except for a 9-bp contiguous sequence located within 20 bp of the −35 region (Fig. 2A).
FIG. 2.
A. Alignment of the nucleotide sequences in the hmuO promoter region between C. ulcerans (CU) and C. diphtheriae C7 (CD). DtxR binding sites are underlined. rbs, putative ribosome binding site; ATG, start codon for hmuO. The arrow indicates the start of transcription in C. diphtheriae (19). B. Alignment of the 19-bp DtxR binding sites from CU712 and C. diphtheriae C7 with the DtxR consensus binding sequence. Residues represented by bold characters indicate the most highly conserved nucleotides.
To compare expression between the hmuO promoters of C. diphtheriae and C. ulcerans, a C. ulcerans hmuO-lacZ promoter construct (pPO712), which contains a 315-bp DNA sequence from CU712 that extends from the 5′ portion of the hmuO coding region to 60 bp upstream of the −35 sequence, was constructed in the low-copy-number promoter probe vector pCM502 (19). Plasmids pPO712 and pCPO-1, a pCM502-based plasmid that contains a C. diphtheriae hmuO promoter-lacZ fusion (20), were subsequently introduced into various Corynebacterium strains, and promoter activity was determined after overnight growth at 37°C in HIBTW medium containing various supplements. As shown in Table 2, and as reported previously (4, 19, 20), the C. diphtheriae hmuO promoter on pCPO-1 was strongly repressed by iron (+Fe) and showed only minimal expression in low-iron conditions (−Fe); however, in the presence of hemoglobin, expression of hmuO was enhanced more than 30-fold. In contrast, transcription from the CU712 hmuO promoter was weakly repressed by iron, and expression in the presence of hemoglobin was enhanced only twofold; however, overall promoter activity was more robust in C. ulcerans than in C. diphtheriae (Table 2). CU712 hmuOΔ/pPO712 showed significantly enhanced levels of hmuO expression in the presence of hemoglobin relative to wild-type results, and since this mutant showed a reduced ability to degrade heme, excess intracellular heme may function to further activate expression. Expression of pPO712 in C. diphtheriae C7 indicates that the C. ulcerans hmuO promoter is more strongly expressed under all conditions relative to the C. diphtheriae hmuO promoter (pCPO-1), and the lack of hemoglobin-dependent activation from the C. ulcerans hmuO promoter in a C7 mutant that is defective for both ChrA-ChrS and HrrA-HrrS suggests that the C. ulcerans hmuO gene is regulated by similar two-component systems in C. ulcerans. Additional studies will be required to determine whether C. ulcerans harbors these signal transduction systems or other putative activators that may be involved in the expression of hmuO.
TABLE 2.
hmuO promoter activity in Corynebacterium strains
| Strain/plasmide | LacZ activitya in:
|
||
|---|---|---|---|
| +Feb | −Fec | −Fe/Hbd | |
| C7 wt/pCP01 (C7/hmuO-lacZ) | <0.5 | 3.0 (0.7) | 112.0 (3.0) |
| CU712 wt/pPO712 (CU/hmuO-lacZ) | 46.7 (2.3) | 129.7 (11.1) | 256.6 (20.0) |
| CU712 hmuOΔ/pPO712 | 121.3 (15.5) | 196.0 (38.0) | 922.0 (120.0) |
| C7 wt/pPO712 | 10.9 (3.0) | 15.5 (3.4) | 155.3 (24.5) |
| C7chrASΔ-hrrASΔ/ pPO712 | 1.7 (0.3) | 2.6 (0.1) | 2.5 (0.3) |
LacZ activity was determined as described previously (24). Values represent the means (± standard deviations) of the results of three experiments.
+Fe, HIBTW medium.
−Fe, HIBTW medium with 12.5 μg/ml EDDA.
−Fe HIBTW medium with Hb (hemoglobin) at 140 μg/ml.
wt, wild type.
Conclusions.
The results from this study indicate that expression of hmuO in C. ulcerans is only weakly influenced by the presence of iron and heme and that the level of transcription of C. ulcerans hmuO is relatively high under all conditions examined, which is in contrast to what was observed with C. diphtheriae, for which significant levels of hmuO transcription are only observed in the presence of a heme source (Table 2 and reference 4). These observations suggest that in C. ulcerans, HmuO function is needed regardless of the iron or heme levels, whereas in C. diphtheriae, HmuO activity is primarily required in the presence of an external heme source. One possible explanation for the overall higher hmuO expression levels in C. ulcerans is that CU712 may have higher endogenous intracellular heme levels than C. diphtheriae. This observation would be consistent with what is observed in CU712 hmuOΔ, where the absence of HmuO results in higher levels of hmuO expression, which could be caused by higher intracellular heme levels.
In higher organisms, heme oxygenases are involved in both the acquisition of heme-iron and heme homeostasis (30). It is not known whether the Corynebacterium HmuO proteins are involved in heme homeostasis or other heme-associated metabolic activities. It is clear, however, from the findings in this study that HmuO is involved in the acquisition of heme-iron in C. ulcerans and in some C. diphtheriae strains. It is possible that in C. ulcerans, HmuO is involved in heme homeostasis or other related heme metabolic activities in both the presence and absence of environmental heme sources, while in C. diphtheriae, proteins other than HmuO may perform these functions in the absence of externally supplemented heme. This hypothesis may explain why an hmuO mutation in C. ulcerans fully abolishes heme utilization whereas a C. diphtheriae hmuO mutant is only partially defective in heme-iron acquisition (or has no defect, as seen with strain 1737). This reasoning assumes that in C. diphtheriae there are one or more factors in addition to HmuO that are involved in heme-iron utilization and possibly required for additional heme-associated activities, while it predicts that in C. ulcerans, HmuO is the only protein involved in the extraction of iron from heme. Whether there are additional proteins involved in heme homeostasis in C. ulcerans remains to be determined. Clearly, further studies are required to fully understand the mechanism and factors involved in the acquisition of iron from heme in pathogenic Corynebacterium species.
Nucleotide sequence accession number.
The nucleotide sequence of a 777-bp region containing the C. ulcerans hmuO gene and flanking sequences has been assigned GenBank accession number EF186829.
Acknowledgments
We thank Karen Meysick and Lori Bibb for helpful comments on the manuscript.
Footnotes
Published ahead of print on 23 February 2007.
REFERENCES
- 1.Altschul, S. F., G. Warren, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barksdale, W. L., and A. M. Pappenheimer, Jr. 1954. Phage-host relationships in nontoxigenic and toxigenic diphtheria bacilli. J. Bacteriol. 67:220-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibb, L. A., C. A. Kunkle, and M. P. Schmitt. The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect. Immun., in press IAI01821-06. [DOI] [PMC free article] [PubMed]
- 4.Bibb, L. A., N. D. King, C. A. Kunkle, and M. P. Schmitt. 2005. Analysis of a heme-dependent signal transduction system in Corynebacterium diphtheriae: deletion of the chrAS genes results in heme sensitivity and diminished heme-dependent activation of the hmuO promoter. Infect. Immun. 73:7406-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, J. M., O. N. Manish, and J. R. Murphy. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 87:5968-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, V. 2005. Bacterial iron transport related to virulence. Contrib. Microbiol. 12:210-233. [DOI] [PubMed] [Google Scholar]
- 7.Brown, J. S., and D. W. Holden. 2002. Iron acquisition by gram-positive bacterial pathogens. Microbes Infect. 4:1149-1156. [DOI] [PubMed] [Google Scholar]
- 8.Cerdeño-Tárraga, A. M., A. Efstratiou, L. G. Dover, M. T. Holden, M. Pallen, S. D. Bentley, G. S. Besra, C. Churcher, K. D. James, A. De Zoysa, T. Chillingworth, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, M. A. Quail, E. Rabbinowitsch, K. M. Rutherford, N. R. Thomson, L. Unwin, S. Whitehead, B. G. Barrell, and J. Parkhill. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 31:6516-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cryz, S. J., L. M. Russell, and R. K. Holmes. 1983. Regulation of toxinogenesis in Corynebacterium diphtheriae: mutations in the bacterial genome that alter the effects of iron on toxin production. J. Bacteriol. 154:245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drazek, E. S., C. A. Hammack, and M. P. Schmitt. 2000. Corynebacterium diphtheriae genes required for acquisition of iron from hemin and hemoglobin are homologous to ABC hemin transporters. Mol. Microbiol. 36:68-84. [DOI] [PubMed] [Google Scholar]
- 11.Hirotsu, S., G. C. Chu, M. Unno, D.-S. Lee, T. Yoshida, S.-Y. Park, Y. Shiro, and M. Ikeda-Saito. 2004. The crystal structure of the ferric and ferrous forms of the heme complex of HmuO, a heme oxygenase of Corynebacterium diphtheriae. J. Biol. Chem. 279:11937-11947. [DOI] [PubMed] [Google Scholar]
- 12.Kunkle, C. A., and M. P. Schmitt. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J. Bacteriol. 185:6826-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunkle, C. A., and M. P. Schmitt. 2005. Analysis of a DtxR-regulated iron transport and siderophore biosynthesis gene cluster in Corynebacterium diphtheriae. J. Bacteriol. 187:422-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popovic, T., S. Y. Kombarova, M. W. Reeves, H. Nakao, I. K. Mazurova, M. Wharton, I. K. Wachsmuth, and J. D. Wenger. 1996. Molecular epidemiology of diphtheria in Russia, 1985-1994. J. Infect. Dis. 174:1064-1072. [DOI] [PubMed] [Google Scholar]
- 15.Puri, S., and M. R. O'Brian. 2006. The hmuQ and hmuD genes from Bradyrhizobium japonicum encode heme-degrading enzymes. J. Bacteriol. 188:6476-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian, Y., J. H. Lee, and R. K. Holmes. 2002. Identification of a DtxR-regulated operon that is essential for siderophore-dependent iron uptake in Corynebacterium diphtheriae. J. Bacteriol. 184:4846-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratliff, M., W. Zhu, R. Deshmukh, A. Wilks, and I. Stojiljkovic. 2001. Homologues of neisserial heme oxygenases in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183:6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt, M. P. 1997. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenase and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitt, M. P. 1997. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect. Immun. 65:4634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt, M. P. 1999. Identification of a two-component signal transduction system from Corynebacterium diphtheriae that activates gene expression in response to the presence of heme and hemoglobin. J. Bacteriol. 181:5330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt, M. P. 2004. Corynebacterium diphtheriae, p. 344-359. In Jorge H. Crosa, Alexandra R. Mey, and Shelley M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 22.Schmitt, M. P., and E. S. Drazek. 2001. Construction and consequences of directed mutations affecting the hemin receptor in pathogenic Corynebacterium species. J. Bacteriol. 183:1476-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 59:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt, M. P., and R. K. Holmes. 1991. Characterization of a defective diphtheria toxin repressor (dtxR) allele and analysis of dtxR transcription in wild-type and mutant strains of Corynebacterium diphtheriae. Infect. Immun. 59:3903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skaar, E. P., A. H. Gaspar, and O. Schneewind. 2004. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279:436-443. [DOI] [PubMed] [Google Scholar]
- 26.Skaar, E. P., A. H. Gaspar, and O. Schneewind. 2006. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J. Bacteriol. 188:1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stojiljkovic, I., and D. Perkins-Balding. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21:281-295. [DOI] [PubMed] [Google Scholar]
- 28.Tai, S.-P. S., A. E. Krafft, P. Nootheti, and R. K. Holmes. 1990. Coordinate regulation of siderophore and diphtheria toxin production by iron in Corynebacterium diphtheriae. Microb. Pathog. 9:267-273. [DOI] [PubMed] [Google Scholar]
- 29.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 30.Wilks, A. 2002. Heme oxygenase: evolution, structure, and mechanism. Antioxid. Redox Signal. 4:603-614. [DOI] [PubMed] [Google Scholar]
- 31.Wilks, A., and M. P. Schmitt. 1998. Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. J. Biol. Chem. 273:837-841. [DOI] [PubMed] [Google Scholar]
- 32.Wu, R., E. P. Skaar, R. Zhang, G. Joachimiak, P. Gornicki, O. Schneewind, and A. Joachimiak. 2005. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J. Biol. Chem. 280:2840-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu, W., D. J. Hunt, A. R. Richardson, and I. Stojiljkovic. 2000. Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J. Bacteriol. 182:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]