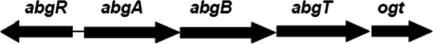Abstract
Escherichia coli AbgT was first identified as a structural protein enabling the growth of p-aminobenzoate auxotrophs on exogenous p-aminobenzoyl-glutamate (M. J. Hussein, J. M. Green, and B. P. Nichols, J. Bacteriol. 180:6260-6268, 1998). The abg region includes abgA, abgB, abgT, and ogt; these genes may be regulated by AbgR, a divergently transcribed LysR-type protein. Wild-type cells transformed with a high-copy-number plasmid encoding abgT demonstrate saturable uptake of p-aminobenzoyl-glutamate (KT = 123 μM); control cells expressing vector demonstrate negligible uptake. The addition of metabolic poisons inhibited uptake of p-aminobenzoyl-glutamate, consistent with this process requiring energy. p-Aminobenzoyl-glutamate taken in by cells expressing large amounts of AbgT alone is not rapidly metabolized to a form that is trapped in the cell, as the addition of nonradioactive p-aminobenzoyl-glutamate to these cells results in a rapid loss of intracellular label. The addition of nonradioactive p-aminobenzoate has no effect. The abgA, abgB, and abgAB genes were cloned into the medium-copy-number plasmid pACYC184; p-aminobenzoate auxotrophs transformed with the clone encoding abgAB demonstrated enhanced ability to grow on low levels of p-aminobenzoyl-glutamate. When transformed with complementary plasmids encoding high-copy levels of abgT and medium-copy levels of abgAB, p-aminobenzoate auxotrophs grew on 50 nM p-aminobenzoyl-glutamate. Our data are consistent with a model of p-aminobenzoyl-glutamate utilization in which AbgT catalyzes transport of p-aminobenzoyl-glutamate, followed by cleavage to p-aminobenzoate by a protein composed of subunits encoded by abgA and abgB. While endogenous expression of these genes is very low under the conditions in which we performed our experiments, these genes may be induced by AbgR bound to an unknown molecule. The true physiological role of this region may be related to some molecule similar to p-aminobenzoyl-glutamate, such as a dipeptide.
Folic acid, in its reduced forms, is needed for the biosynthesis of DNA, RNA, amino acids, and other important cellular components (reviewed in reference 15). Essential in humans, this vitamin is synthesized de novo in many microorganisms (5, 27). Microorganisms differ vastly in the ability to take in folate (reviewed in references 6 and 24). Bacteria that can make their own folate typically cannot utilize exogenous folate, explaining the widespread success of the sulfonamide antibiotics that target the folate biosynthetic pathway. Those that cannot synthesize folate, such as Lactobacillus casei, require exogenous folate (25, 26, 28). Other bacteria that transport folates include Lactobacillus salivarius (10), Streptococcus faecium, formerly Streptococcus faecalis (16), and Pediococcus cerevisiae (11-13). In contrast, Escherichia coli was observed early on to be unable to grow on exogenous folate (17). While the enzymes and associated genes involved in the folate biosynthetic pathway have been identified and at least partially characterized (5), the genes or enzymes involved in catabolism remain largely unidentified and uncharacterized, though there is evidence that E. coli can catabolize folate to form p-aminobenzoyl-glutamate and a pterin (23, 34). Though E. coli has not been known to take in folate, this bacterium does take in p-aminobenzoate; p-aminobenzoate excreted from wild-type cells can support the growth of strains unable to synthesize this compound (8).
E. coli contains a gene for a transport protein, abgT, that when expressed at elevated levels in p-aminobenzoate auxotrophs, confers the ability to grow on nanomolar quantities of the folate catabolite p-aminobenzoyl-glutamate (9). The gene lies within the abg region of the E. coli genome (named for its growth on p-aminobenzoyl glutamate [Fig. 1]). This region of E. coli was identified from a search for mutants able to grow on folic acid to overcome p-aminobenzoate auxotrophy. Further work revealed that the bacteria were actually growing on p-aminobenzoyl-glutamate, a contaminant of the folic acid. The abg region consists of four genes arranged in a potential operon, including abgA, abgB, and abgT; the next gene downstream, ogt, which codes for O6-methylguanine-DNA methyltransferase, a DNA repair protein, may also be associated with this region (Fig. 2). Analysis of the gene products revealed that AbgA and AbgB have homology to one another and to aminoacyl aminohydrolases and that AbgT has homology to transport proteins. Divergently oriented from abgABT is abgR; the AbgR protein has homology to LysR-type regulatory proteins. The original mutations mapped to the intergenic region between abgR and abgA and resulted in increased expression of the downstream genes. Strains containing interruptions in abgA, abgB, or abgT were unable to grow on p-aminobenzoyl-glutamate. When transformed with a plasmid that overexpresses AbgT, however, all strains were able to grow on p-aminobenzoyl-glutamate. Thus, AbgT was found to be necessary and sufficient for growth on low concentrations of p-aminobenzoyl-glutamate.
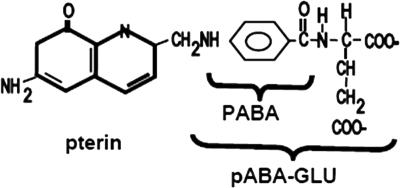
FIG. 1.
Structures of folic acid, p-aminobenzoate (PABA), and p-aminobenzoyl-glutamate (pABA-GLU).
FIG. 2.
abg region of the chromosome.
Sequence analyses have placed AbgT as a member of the ion transporter superfamily; in fact, it provides the name to its subfamily, the p-aminobenzoyl-glutamate transporter family (19). Twenty bacterial homologues have been identified; all were predicted to have 12 transmembrane segments and 26 fully conserved residues, and all members were identified in bacteria, not in archaea or eukaryotes. Only one other member of the family, Neisseria gonorrhea MtrF, has been characterized; this protein has a role in antimicrobial (hydrophobic detergent) resistance (32).
The focus of this study was to further characterize the roles of abgA, abgB, and abgT in the transport and utilization of p-aminobenzoyl-glutamate in E. coli.
MATERIALS AND METHODS
Materials.
[3H]Folic acid diammonium salt (3′,5′,7,9-3H) (1 mCi/ml; 64 Ci/mmol) was obtained from Moravek Biochemicals (Brea, CA). Ampicillin sodium salt, chloramphenicol, DEAE cellulose, ammonium bicarbonate, folic acid, N-(p-aminobenzoyl)-glutamic acid, and p-aminobenzoic acid were obtained from Sigma (St. Louis, MO).
Strains and plasmids.
Wild-type E. coli MG1655, BN1103 (pabA rpsL704), and BN1140 (abg-1 zda-3061::Tn10 pabA1 rpsL704) were obtained from the laboratory of Brian Nichols, University of Illinois at Chicago (9). Plasmids used included pUC18 and pJMG128, a pUC18-derived high-copy-number plasmid encoding abgT; this plasmid was constructed to be identical to pMJH128 (9). Plasmids constructed in this study are described below.
Microbiological and molecular methods.
Bacteria were maintained in either rich medium (Luria broth; Fisher, Pittsburgh, PA) or Vogel-Bonner glucose (0.4%) minimal medium (33). This solution is henceforth referred to simply as “minimal medium.” Ampicillin and chloramphenicol were used at 100 μg ml−1 and 50 μg ml−1, respectively, for maintenance of plasmids (1, 29). E. coli chromosomal DNA was prepared as described previously (1). Small-scale plasmid preparations were done with QIAprep kits (QIAGEN, Valencia, CA). Restriction endonuclease digestions and ligation reactions were performed in accordance with the manufacturer's recommendations (New England Biolabs, Inc., Ipswitch, MA). Transformations were performed with calcium chloride as described previously (1). Agarose gel electrophoresis was performed as described previously (1).
PCR amplification and subsequent cloning of abgA, abgB, and abgAB.
Using the published nucleotide sequences for abgA and abgB (2), we designed primers for cloning into the medium-copy-number plasmid pACYC184 (3). Each primer was designed so that a BamHI site was at the 5′ end and a HindIII site was at the 3′ end; four additional nucleotides were added at the 5′ end to ease restriction. For cloning of abgA, the following primers were used: 5′-GATCAAGCTTATGGAGTCTTTGAATCAATTT-3′ (abgA-up) and 5′-GATCGGATCCTCAGATACCTCGCGTCCAG-3′ (abgA-down); this encompassed the abgA genomic fragment from 1402589 to 1401279(ASAP [4]). For cloning of abgB, the following primers were used: 5′-GATCAAGCTTATGCAGGAAATCTATCGTTTT-3′ (abgB-up) and 5′-GATCGGATCCTTATTTTAAAGGTGACGGTG-3′ (abgB-down); this encompassed the abgB genomic fragment from 1401279 to 1399834. For cloning of abgAB, primers abgA-up and abgB-down were used; this encompassed the abgAB genomic fragment from 1402589 to 1399834. PCR amplification was performed with Taq DNA polymerase according to the manufacturer's instructions (U.S.B. Corporation, Cleveland, OH). PCR conditions were as follows: 94°C for 3 min and 30 cycles of denaturing at 94°C for 0.5 min, annealing at 66°C for 1 min, and extending at 72°C for 2.5 min (PTC-200 Peltier Thermal Cycler; MJ Research, Waltham, MA). PCR products were purified with the QIAquick PCR purification kit from QIAGEN (Valencia, CA). Agarose gel electrophoresis was used to confirm that products were the correct predicted size. Purified PCR products of abgA, abgB, and abgAB were digested with BamHI and HindIII and ligated into pACYC184, which was similarly restricted, by using T4 DNA ligase according to the manufacturer's instructions (New England Biolabs, Ipswitch, MA). Ligation was performed overnight at 15°C. Ligation mixtures were transformed into BN1103 (pabA) and selected for growth on LB plates containing chloramphenicol (50 μg/ml). Plasmid DNA was purified with QIAGEN QIAquick miniprep kits. Candidates were confirmed by restriction mapping and sequence analysis (Molecular Cloning Laboratories, South San Francisco, CA). Those plasmids encoding abgA, abgB, and abgAB were named pLJA, pLJB, and pLJAB, respectively.
Synthesis of tritiated p-aminobenzoyl-glutamate.
Radiolabeled p-aminobenzoyl-glutamate is unavailable commercially, so it was necessary to synthesize it via oxidative cleavage by a method adapted from Maruyama et al. (14). Folic acid (8 to 12 mg) was dissolved in 4.6 ml of 0.1 M ammonium bicarbonate, pH 9; radioactive folate (200 to 300 μCi) was then added. Oxidation was initiated by the addition of 0.5 ml of 2% KMnO4 and was terminated after 2 min with 0.5 ml of 30% H2O2. Further purification was performed at 4°C. Following centrifugation, the supernatant was applied to a DEAE cellulose column (2.0 cm by 45 cm) and eluted with a linear gradient of water to 1 M NH4HCO3 (500 ml each). Fractions (10 min; 10 to 14 ml) were collected with a HeliFrac fraction collector (Amersham Biosciences, Piscataway, NJ) and analyzed for radioactivity and absorbance at 273 nm by using the molar extinction coefficient for p-aminobenzoyl-glutamate (15,400−1 at 273 nm in 0.1 m NaOH) (14). The absorbance spectrum for radioactive p-aminobenzoyl-glutamate synthesized as described was identical to that of commercial p-aminobenzoyl-glutamate. In addition, we analyzed our product by thin-layer chromatography according to published protocols and found that elution was consistent with its identification as p-aminobenzoyl-glutamate (18).
Transport assays.
Cells were inoculated from a single colony and grown, with shaking, for 16 h at 37°C. Cell growth was monitored by measuring the absorbance at 600 nm. Cells were washed twice with ice-cold minimal medium. Washed cells were resuspended at 1/25 of the original volume in 1× minimal medium, including any antibiotics needed to maintain plasmid selection, and preincubated for 15 min at 37°C. Uptake was initiated by the addition of the labeled compound (also in 1× minimal medium), which had been preincubated at 37°C for 5 min prior to the initiation of transport. At various time points, 100 to 400 μl was applied directly to a prewetted filter and washed immediately with 3- to 10-ml aliquots of ice-cold minimal medium; 0.2-μm Millipore nitrocellulose filters (GSWP02500) were used with a Hoeffer filter apparatus (model FH 225, serial no. 91-25V1003; Hoeffer Scientific Instruments, San Francisco, CA). Filters were placed in scintillation fluid (Scintisafe Econo 2; Fisher Chemicals, Pittsburgh, PA) and counted with a Beckman 6500 scintillation counter. For each transport experiment, aliquots of the reaction mixture (identical to those applied to the filter) were placed into a preweighed microfuge vial, centrifuged briefly to pellet the cells, desiccated to dryness, and then weighed again. The difference was taken to be the dry weight of the cells and was used in subsequent data analysis.
Data analysis.
Statistical analyses were performed and graphs were made with GraphPad Prism 4 (GraphPad Software, Inc., San Diego, CA).
RESULTS
Direct measurement of p-aminobenzoyl-glutamate uptake.
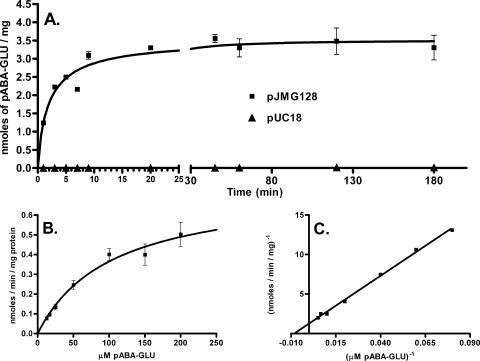
While growth experiments and sequence data were consistent with a role for AbgT as a transport protein, we decided to directly measure the uptake of p-aminobenzoyl-glutamate. For transport experiments, we compared uptake over time in wild-type cells (MG1655) transformed with either pJMG128, encoding abgT on a high-copy-number plasmid, or pUC18, the parent vector of pJMG128 (Fig. 3A). Cells transformed with pJMG128 demonstrated significant transport of p-aminobenzoyl-glutamate, in contrast to cells expressing the vector, which demonstrated very low levels of uptake (0.0009 nmol/min/mg). We also measured the uptake of radioactive p-aminobenzoate in MG1655 cells transformed with pJMG128 versus MG1655 transformed with pUC18 and found no difference (data not shown). In order to determine the KT value for p-aminobenzoyl-glutamate for the AbgT transporter, we measured transport at various concentrations of p-aminobenzoyl-glutamate (Fig. 3B). The KT was found to be 123 μM (Fig. 3C).
FIG. 3.
Transport of radioactive p-aminobenzoyl-glutamate. (A) Time course of p-aminobenzoyl-glutamate uptake by MG1655 cells transformed with pJMG128 (a pUC18-derived plasmid expressing AbgT) and the parent vector pUC18. Assays were performed as described in Materials and Methods. Briefly, cells were grown overnight, washed, and resuspended in minimal medium. After preincubation, the transport assay (final volume, 6 ml) was initiated by the addition of [3H]p-aminobenzoyl-glutamate to a final concentration of 50 μM. Duplicate samples (0.1 ml) were taken at the times shown following initiation. Samples were filtered, immediately washed with ice-cold minimal medium, and counted. Data were analyzed with GraphPad Prism 4; error bars represent standard errors. (B) Effect of p-aminobenzoyl-glutamate concentration on the initial uptake of [3H]p-aminobenzoyl-glutamate by MG1655 transformed with pJMG128. Assays were performed as described in Materials and Methods. After preincubation, the transport assay (final volume, 6 ml) was initiated by the addition of [3H]p-aminobenzoyl-glutamate; final concentrations used were 12.5 μM, 16.7 μM, 25 μM, 50 μM, 100 μM, 150 μM, and 200 μM. For each concentration, a reaction was initiated and duplicate samples (0.4 ml) were taken within the first few minutes following initiation. Samples were filtered, immediately washed with ice-cold minimal medium, and counted. For each concentration, linear regression analysis was performed, and the slope of the line was measured and taken as the initial velocity. Data were analyzed with GraphPad Prism 4; error bars represent standard errors. (C) Lineweaver-Burk plot of the data from panel B.
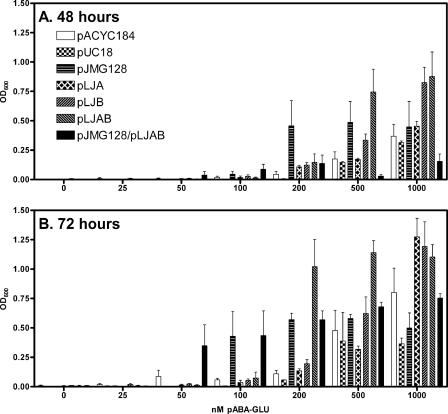
AbgT, the transporter encoded by abgT, is located in a putative operon with two open reading frames that have homology to lyases, abgA and abgB. We hypothesized that that these proteins, either separately or together as dissimilar subunits of a single protein, might catalyze the cleavage of p-aminobenzoyl-glutamate to form p-aminobenzoate. We used PCR to clone abgA, abgB, and abgAB together, into the medium-copy-number plasmid pACYC184. We measured the ability of p-aminobenzoate auxotrophs (BN1103) transformed with these plasmids to grow on various levels of p-aminobenzoyl-glutamate on solid minimal medium; by measuring the average colony size we found that only cells transformed with abgAB showed enhanced growth on 100 nM p-aminobenzoyl-glutamate (data not shown). The background of very tiny colonies was likely due to p-aminobenzoate contamination of the commercial p-aminobenzoyl-glutamate. We performed a similar experiment with liquid culture and monitored growth by measuring the optical density (Fig. 4). The data were consistent in showing that only abgAB together enabled growth on low levels of p-aminobenzoyl-glutamate, supporting the idea that the products of abgA and abgB comprise subunits of an enzyme that cleaves p-aminobenzoyl-glutamate, thereby enabling p-aminobenzoate auxotrophs to grow.
FIG. 4.
Growth of p-aminobenzoate auxotrophs (BN1103) transformed with various plasmids encoding genes of the abg region in liquid medium containing various concentrations of p-aminobenzoyl-glutamate. (A) Comparison of the growth of BN1103 transformed with pACYC184, pLJA, pLJB, pLJAB, pUC18, pJG128, and pLJAB/pJG128 in minimal medium containing various concentrations of p-aminobenzoyl-glutamate at 48 h after inoculation. BN1103 (pabA) cells transformed with pACYC184, pLJA, pLJB, pLJAB, pUC18, pJG128, and pLJAB/pJG128 were grown overnight in minimal medium containing the appropriate antibiotics for plasmid selection and 100 nM p-aminobenzoate, washed three times with sterile saline, and diluted 105 into 5 ml of minimal medium containing antibiotic and increasing concentrations of p-aminobenzoyl-glutamate. Cultures were incubated at 37°C; the optical density (OD) (λ = 600 nm) of a 1-ml sample was measured every 24 h. The loss in volume due to evaporation was about 2.5% per day (∼8% loss after 72 h). Results are given as means ± standard errors. (B) Readings of the data from panel A at 72 h.
If AbgT enabled uptake of p-aminobenzoyl-glutamate and AbgAB catalyzed subsequent cleavage to form p-aminobenzoate, then p-aminobenzoate auxotrophs expressing both proteins should demonstrate enhanced growth at low concentrations of p-aminobenzoyl-glutamate. To test this, we performed a double transformation of BN1103 cells with both pJMG128 and pLJAB and monitored the ability of cells to grow in liquid minimal medium supplemented with various concentrations of p-aminobenzoyl-glutamate. As shown in Fig. 4, cells expressing both pLJAB and pJMG128 can grow at the lowest concentrations of p-aminobenzoyl-glutamate (50 nM), while cells expressing pJMG128 alone or pLJAB require 100 nM p-aminobenzoyl-glutamate.
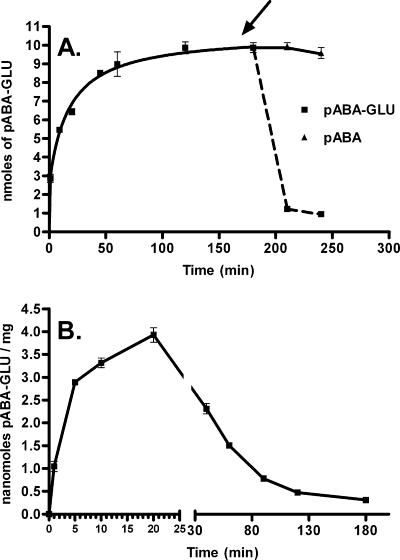
In order to determine whether p-aminobenzoyl-glutamate was metabolized immediately after import, we allowed cells expressing large amounts of abgT (MG1655 transformed with pJMG128) to take in radioactive p-aminobenzoyl-glutamate, taking aliquots with time; at the180-min time point the sample was supplemented with excess nonradioactive p-aminobenzoyl-glutamate or p-aminobenzoate (Fig. 5A). We subsequently measured the effect of these additions on the retention of cellular radioactivity. Uptake increased and reached a plateau; when the cells were subjected to an addition of excess nonradioactive p-aminobenzoyl-glutamate, there was a rapid loss of label. We interpreted this to mean that p-aminobenzoyl-glutamate was taken up in these cells but was not rapidly metabolized to a form that was retained by the cell. In other words, p-aminobenzoyl-glutamate accumulates in the cell. In a parallel sample, cells were challenged with nonradioactive p-aminobenzoate; these cells showed no loss of label. This is consistent with the observation that p-aminobenzoate does not interact with the AbgT transporter and does not exchange with intracellular p-aminobenzoyl-glutamate.
FIG. 5.
Transport and retention of radioactive p-aminobenzoyl-glutamate. (A) Effect of the addition of unlabeled p-aminobenzoyl-glutamate or p-aminobenzoate on cells (MG1655 transformed with pJMG128) that have accumulated radioactive p-aminobenzoyl-glutamate. Assays were performed as described in Materials and Methods. Briefly, cells were grown overnight, washed, and resuspended in minimal medium. After preincubation, the transport assay (final volume, 6 ml) was initiated by the addition of [3H]p-aminobenzoyl-glutamate to a final concentration of 50 μM. Duplicate samples (0.4 ml) were taken at the times shown. After the 180-min sample was taken, the mixture was divided in two equal portions and a nonradioactive sample of p-aminobenzoyl-glutamate (final concentration, 10 mM) or p-aminobenzoate (final concentration, 2 mM) was added. Duplicate samples were then taken of each mixture at 210 and 240 min. Samples were filtered, immediately washed with ice-cold minimal medium, and counted. Data were analyzed with GraphPad Prism 4; error bars represent standard errors. (B) Transport and retention of [3H]p-aminobenzoyl-glutamate by MG1655 transformed with both pJMG128 and pLJAB. Assays were performed as described in Materials and Methods. After preincubation, the transport assay (final volume, 6 ml) was initiated by the addition of [3H]p-aminobenzoyl-glutamate to a final concentration of 50 μM. Duplicate samples (0.1 ml) were taken at the times shown. Samples were filtered, immediately washed with ice-cold minimal medium, and counted. Data were analyzed with GraphPad Prism 4; error bars represent standard errors.
In view of the possible role that AbgAB has in cleaving p-aminobenzoyl-glutamate upon entry into the cell, we decided to use MG1655 cells transformed with both pJMG128 and pLJAB to measure uptake and retention of radioactive p-aminobenzoyl-glutamate over time (Fig. 5B). Interestingly, within the first few minutes the pattern of uptake of p-aminobenzoyl-glutamate for cells expressing both plasmids resembled the pattern for cells expressing just high-copy abgT (pJMG128). Then, instead of reaching and maintaining a plateau as in Fig. 5A, the label remaining in the cell decreased. This is consistent with AbgT catalyzing the uptake of p-aminobenzoyl-glutamate into the cell and accumulating p-aminobenzoyl-glutamate to a concentration enabling AbgAB to catalyze cleavage into p-aminobenzoate and glutamate. p-Aminobenzoate enters cells in a nonsaturable manner consistent with diffusion (30; also data not shown); therefore, once radioactive p-aminobenzoate is generated in the cell by cleavage of radioactive p-aminobenzoyl-glutamate, it can depart the cell.
Metabolic poisons are useful for measuring the dependence of transport on cellular features such as the presence of ATP or a proton gradient. We found that transport of p-aminobenzoyl-glutamate by AbgT was inhibited completely (0% of normal activity) by 100 nM sodium azide (an electron transport inhibitor), was inhibited to 15.1% of normal activity by 100 mM potassium fluoride (an inhibitor of the glycolytic enzyme enolase), and was inhibited to 0.2% of normal activity by 100 μM carbonyl cyanide 3-chlorophenylhydrazone (a protonophore and electron transport inhibitor). These data are consistent with the transport of p-aminobenzoyl-glutamate requiring a source of ATP.
DISCUSSION
In this study, we further characterized the E. coli transport protein AbgT. E. coli AbgT catalyzes the uptake of p-aminobenzoyl-glutamate in a concentration-dependent, saturable manner, and is characterized by a KT of 123 μM. It is interesting to compare the properties of AbgT with known bacterial transporters of vitamins and related compounds. Bacterial folate transport systems typically have KT values in the nanomolar-micromolar range; for folate transporters, L. casei has the lowest KT (16 nM) for folate (7, 25), while S. faecalis is characterized by a KT for its substrate, 5-methyl-tetrahydrofolate, of 2 μM (6). Compared to these values, our KT for p-aminobenzoyl-glutamate is relatively high; it is possible that AbgT may actually serve physiologically as a transporter for some other molecule, perhaps a dipeptide, and that it transports p-aminobenzoyl-glutamate as a secondary activity.
Our data are consistent with a model of p-aminobenzoyl-glutamate utilization in which AbgT catalyzes the uptake of p-aminobenzoyl-glutamate and the gene products of abgA and abgB form a protein that catalyzes the cleavage of p-aminobenzoyl-glutamate to p-aminobenzoate and glutamate. The role of the abg region in E. coli physiology is not known. Gene Brown, whose elegant research delineated the folate biosynthetic pathway in E. coli, first considered the physiological relevance of p-aminobenzoyl-glutamate. Dihydropteroate synthetase catalyzes the reaction between dihydropteridine pyrophosphate and p-aminobenzoate to form dihydropteroate and pyrophosphate (21). Brown dismissed the possibility that p-aminobenzoyl-glutamate was physiologically relevant as a substrate of dihydropteroate synthetase because kinetic measurements revealed that the Km value (p-aminobenzoate) for dihydropteroate synthetase was 2.5 μM while the Ki for p-aminobenzoyl-glutamate as a competitive inhibitor was 1.1 to 1.3 mM. Observations of the enzyme preference were extended by others to a variety of bacteria (27). This, in addition to the observation that no enzyme had been found from any source that catalyzed the synthesis of p-aminobenzoyl-glutamate from p-aminobenzoate and glutamate, led these authors to conclude that p-aminobenzoate, not p-aminobenzoyl-glutamate, was the true physiological substrate.
While Brown's studies have stood the test of time, one possibility that they failed to consider was that p-aminobenzoyl-glutamate may be a natural product of folate catabolism, not a biosynthetic intermediate. In nature, E. coli has adapted itself rather well to a variety of environmental stresses that it may face in the lower gastrointestinal tract of animals. The major end product of folate catabolism in humans is p-aminobenzoyl-glutamate and its derivative acetamido-aminobenzoyl-glutamate; these catabolic end products in humans are present in both urine and fecal matter.
A report from Quinlivan et al. has provided some insights into E. coli folate metabolism under stress (20). Using high-performance liquid chromatography analytic techniques, these authors investigated the mechanism of the antibacterial drug trimethoprim. Drug effectiveness has long been attributed to inhibition of dihydrofolate reductase, accumulation of dihydrofolate derivatives, and starvation of the cell for reduced folates. Trimethoprim treatment caused a depletion in total cellular folates and a progressive conversion of tetrahydrofolate derivatives (representing 100% of the cellular folates in untreated cells) to folic acid (45%) and p-aminobenzoyl-glutamate (53%). The authors concluded that trimethoprim acts to increase bacterial catabolism of reduced folic acid cofactors; they postulated that this may be catalyzed by an as-yet-unidentified enzyme or perhaps by chemical cleavage, possibly owing to changes in the cellular redox state and decreased intracellular pH levels.
It is interesting to compare the abg region to the aae operon in E. coli, which shares several features (31). The aae operon (for “aromatic carboxylic acid efflux”) is comprised of three genes, aaeX, aaeA, and aaeB, that are transcribed divergently from aaeR. AaeR, like AbgR, is similar to LysR transcriptional regulators (22). AaeR binds to specific aromatic acids, including p-hydroxybenzoic acid, to induce expression of aaeA and aaeB by more than 10-fold; these proteins form subunits of an efflux protein that exports p-hydroxybenzoic acid. Various aromatic carboxylic acids induce this operon, which has a very low basal (uninduced) level of activity.
Our studies indicate that in wild-type cells growing aerobically, chromosomal expression of abgT is very low. It is likely that AbgR, the putative regulator of the abg region, binds to some as-yet-unknown molecule to stimulate expression. abgT may comprise an operon with abgA, abgB, and ogt (Fig. 2). It is possible that E. coli has the ability to utilize p-aminobenzoyl-glutamate in a “salvage” or recycling pathway, whereby part of the folate structure is scavenged. The abg region of the chromosome remains one whose role in the microorganism remains poorly understood. We are continuing our studies of the genes of the abg region in hopes of obtaining more clues into the molecular physiology of E. coli as it relates to folate catabolism.
Acknowledgments
This work was supported in part by funds from Midwestern University and by grant R15 GM071009, from the National Institutes of Health, to J.M.G.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Short protocols in molecular biology, 4th ed. John Wiley & Sons, Inc., New York, NY.
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glasner, J. D., P. Liss, G. Plunkett III, A. Darling, T. Prasad, M. Rusch, A. Byrnes, M. Gilson, B. Biehl, F. R. Blattner, and N. T. Perna. 2003. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 31:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green, J. M., B. P. Nichols, and R. G. Matthews. 1996. Folate biosynthesis, reduction, and polyglutamylation, p. 665-673. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. I. ASM Press, Washington, DC. [Google Scholar]
- 6.Henderson, G. B. 1986. Transport of folate compounds into cells, p. 207-250. In S. J. Benkovic (ed.), Folates and pterins, vol. III. John Wiley and Sons, Inc., New York, NY. [Google Scholar]
- 7.Henderson, G. B., and F. M. Huennekens. 1974. Transport of folate compounds into Lactobacillus casei. Arch. Biochem. Biophys. 164:722-728. [DOI] [PubMed] [Google Scholar]
- 8.Huang, M., and J. Pittard. 1967. Genetic analysis of mutant strains of Escherichia coli requiring p-aminobenzoic acid for growth. J. Bacteriol. 93:1938-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussein, M. J., J. M. Green, and B. P. Nichols. 1998. Characterization of mutations that allow p-aminobenzoyl-glutamate utilization by Escherichia coli. J. Bacteriol. 180:6260-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar, H. P., J. M. Tsuji, and G. B. Henderson. 1987. Folate transport in Lactobacillus salivarius. Characterization of the transport mechanism and purification and properties of the binding component. J. Biol. Chem. 262:7171-7179. [PubMed] [Google Scholar]
- 11.Mandelbaum-Shavit, F., and N. Grossowicz. 1973. Carrier-mediated transport of folate in a mutant of Pediococcus cerevisiae. J. Bacteriol. 114:485-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandelbaum-Shavit, F., and N. Grossowicz. 1975. Pediococcus cerevisiae mutant with altered transport of folates. J. Bacteriol. 123:400-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandelbaum-Shavit, F., and N. Grossowicz. 1970. Transport of folinate and related compounds in Pediococcus cerevisiae. J. Bacteriol. 104:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maruyama, T., T. Shiota, and C. L. Krumdieck. 1978. The oxidative cleavage of folates. A critical study. Anal. Biochem. 84:277-295. [DOI] [PubMed] [Google Scholar]
- 15.Matthews, R. G. 1996. One-carbon metabolism, p. 600-611. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. I. ASM Press, Washington, DC. [Google Scholar]
- 16.McElwee, P. G., and J. M. Scott. 1972. Folate metabolism in Streptococcus faecalis. Biochem. J. 127:901-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nickerson, W. J., and M. Webb. 1956. Effect of folic acid analogues on growth and cell division of nonexacting microorganisms. J. Bacteriol. 71:129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orsomando, G., G. G. Bozzo, R. D. de la Garza, G. J. Basset, E. P. Quinlivan, V. Naponelli, F. Rebeille, S. Ravanel, J. F. Gregory III, and A. D. Hanson. 2006. Evidence for folate-salvage reactions in plants. Plant J. 46:426-435. [DOI] [PubMed] [Google Scholar]
- 19.Prakash, S., G. Cooper, S. Singhi, and M. H. Saier, Jr. 2003. The ion transporter superfamily. Biochim. Biophys. Acta 1618:79-92. [DOI] [PubMed] [Google Scholar]
- 20.Quinlivan, E. P., J. McPartlin, D. G. Weir, and J. Scott. 2000. Mechanism of the antimicrobial drug trimethoprim revisited. FASEB J. 14:2519-2524. [DOI] [PubMed] [Google Scholar]
- 21.Richey, D. P., and G. M. Brown. 1969. The biosynthesis of folic acid. IX. Purification and properties of the enzymes required for the formation of dihydropteroic acid. J. Biol. Chem. 244:1582-1592. [PubMed] [Google Scholar]
- 22.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 23.Scott, J. M. 1984. Catabolism of folates, p. 307-327. In S. J. Benkovic (ed.), Folates and pterins, vol. III. John Wiley and Sons, Inc., New York, NY. [Google Scholar]
- 24.Shane, B. 1986. Pterins and folates in the nutrition of lower organisms, p. 1-29. In S. J. Benkovic (ed.), Folates and pterins, vol. III. John Wiley and Sons, Inc., New York, NY. [Google Scholar]
- 25.Shane, B., and E. L. Stokstad. 1975. Transport and metabolism of folates by bacteria. J. Biol. Chem. 250:2243-2253. [PubMed] [Google Scholar]
- 26.Shane, B., and E. L. Stokstad. 1976. Transport and utilization of methyltetrahydrofolates by Lactobacillus casei. J. Biol. Chem. 251:3405-3410. [PubMed] [Google Scholar]
- 27.Shiota, T. 1984. Biosynthesis of folate from pterin precursors, p. 121-134. In S. J. Benkovic (ed.), Folates and pterins, vol. I. John Wiley and Sons, Inc., New York, NY. [Google Scholar]
- 28.Stokstad, E. L., and J. Koch. 1967. Folic acid metabolism. Physiol. Rev. 47:83-116. [DOI] [PubMed] [Google Scholar]
- 29.Tran, P. V., T. A. Bannor, S. Z. Doktor, and B. P. Nichols. 1990. Chromosomal organization and expression of Escherichia coli pabA. J. Bacteriol. 172:397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran, P. V., and B. P. Nichols. 1991. Expression of Escherichia coli pabA. J. Bacteriol. 173:3680-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Dyk, T. K., L. J. Templeton, K. A. Cantera, P. L. Sharpe, and F. S. Sariaslani. 2004. Characterization of the Escherichia coli AaeAB efflux pump: a metabolic relief valve? J. Bacteriol. 186:7196-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veal, W. L., and W. M. Shafer. 2003. Identification of a cell envelope protein (MtrF) involved in hydrophobic antimicrobial resistance in Neisseria gonorrhoeae. J. Antimicrob. Chemother. 51:27-37. [DOI] [PubMed] [Google Scholar]
- 33.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 34.Wachter, H., A. Hausen, E. Reider, and M. Schweiger. 1980. Pteridine excretion from cells as indicator of cell proliferation. Naturwissenschaften 67:610-611. [DOI] [PubMed] [Google Scholar]