Abstract
The Hha/YmoA family of nucleoid-associated proteins is involved in gene regulation in enterobacteria. In Salmonella enterica serovar Typhimurium, virulence genes required for intracellular growth are induced following host cell invasion but the proteins responsible for repressing these genes prior to host cell entry have not been fully identified. We demonstrate here that Hha is the major repressor responsible for silencing virulence genes carried in Salmonella pathogenicity island 2 prior to bacteria sensing an intracellular environmental cue.
Virulence gene regulation in bacterial pathogens is a highly coordinated process involving extracellular sensors and transcription factors that are activated in response to specific environmental cues (4). In Salmonella enterica serovar Typhimurium, the virulence factors required for intracellular growth are encoded on a large pathogenicity island called Salmonella pathogenicity island 2 (SPI-2), which encodes a type III secretion system and a two-component regulatory system called SsrA-SsrB that activates this type III system in the intracellular environment (2, 5). SsrA is a sensor kinase that phosphorylates the cognate response regulator, SsrB, leading to the induction of SPI-2 gene expression. Because standard LB broth is noninducing for SPI-2 gene expression, we (6) and others (10) previously established an in vitro-acidified culture medium (LPM) with micromolar concentrations of phosphate and magnesium that induces robust SsrB-dependent gene expression, the assembly of the type III secretion system, and the secretion of virulence effectors at pH 5.8. This growth medium is suitable for the production of SPI-2-secreted proteins from cultures ranging in scale from a few milliliters (6) to 40-liter fermentors (our unpublished data).
The specific environmental context required for SPI-2 activation implied the existence of a repressing system to silence intracellular virulence genes in SPI-2 in the absence of an activating environmental signal. Because SPI-2 lacks an obvious negative regulator by sequence similarity to other repressors, we hypothesized that SPI-2 integrates with ancestral negative regulatory proteins to achieve appropriate genetic control. We found one repressor, YdgT, which affected the expression of SPI-2 genes and contributed to the contextual activation of virulence in animals (7). However, other unidentified repressors were likely involved because ydgT null mutants still repressed SPI-2 genes in LB medium. YdgT is a member of the Hha/YmoA family of small nucleoid-like proteins involved in gene regulation (8, 13, 16, 20). YmoA modulates the expression of Yersinia enterocolitica virulence factors, including Yop proteins, the YadA adhesin, and invasin (8, 11). Hha was originally shown to increase the cytoplasmic expression of hemolysin in Escherichia coli (18) and has also been shown to negatively regulate invasion genes in Salmonella (12) and virulence genes in the locus of enterocyte effacement in enterohemorrhagic E. coli (22). Heretofore, there has been no report on the role of Hha in SPI-2 gene expression. Here, we identify Hha as the major repressor that silences SPI-2 virulence genes prior to bacteria sensing an activating environmental cue consisting of low Mg2+, low PO43−, and acidification. These data add important insight into the understanding of intracellular virulence gene regulation in Salmonella and make accessible new culture conditions and a genetic probe for examining this essential virulence determinant in Salmonella serovar Typhimurium.
Because the ydgT mutants created in our earlier work (7) still repressed intracellular virulence genes in SPI-2 when grown in LB medium, we hypothesized the existence of another repressor protein that contributed to SPI-2 gene regulation. We focused our attention on the small nucleoid-like protein Hha because of its amino acid similarity to YdgT and YmoA (7). To begin, we deleted hha from wild-type Salmonella serovar Typhimurium strain SL1344 and from a ΔydgT strain to create single and double mutants, respectively (Table 1). Chromosomal deletions of hha were created using the λ Red recombination method as described by Datsenko and Wanner (9). The primers for PCRs were designed to contain 5′ end complementarity to the hha gene and 3′ end complementarity to the FRT (Flp recognition target)-flanked antibiotic resistance cassettes of plasmids pKD3 and pKD4. The primer sequences used were as follows: hhaF, 5′-GCG TGT TCT CTA AAA AGT AAT GTA GCG TGA TTA ACG GTG TAG GCT GGA GCT GCT TCG-3′; and hhaR, 5′-CTT GTT AAA AAT TAT TAC AAT CAT AGG TAG AAT TTA TGT CTG ATA AAC CAT ATG AAT ATC CTC CTT A-3′. The resulting PCR products were purified, electroporated into Salmonella strains, and subjected to selection as described previously (9).
TABLE 1.
Strains used in this study
| Strain | Genotype or description | Source or reference(s) |
|---|---|---|
| E. coli | ||
| BKC1-14 | DH5α supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 | Our collection |
| BKC3-59 | SM10 λpir thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpir(pPsseA::tnpR::lacZ), Apr | 6 |
| Salmonella enterica serovar Typhimurium | ||
| BKC1-1 | SL1344, wild type, Smr | 23 |
| BKC3-25 | SL1344, PsseA::pPsseA::tnpR::lacZ | 6 |
| BKC5-49 | SL1344 ΔydgT | 7 |
| BKC9-49 | SL1344 hha::Kan | This study |
| BKC9-60 | SL1344 ΔydgT hha::Kan | This study |
| BKC1-55 | SL1344 ssrB::Kan | 14 |
| BKC5-56 | SL1344 ssrB::Kan ΔydgT | 7 |
| BKC9-50 | SL1344 ssrB::Kan hha::cat | This study |
| BKC5-57 | SL1344 ΔydgT PsseA::pPsseA::tnpR::lacZ | 7 |
| BKC9-73 | SL1344 hha::Kan PsseA::pPsseA::tnpR::lacZ | This study |
| BKC9-75 | SL1344 ΔydgT hha::Kan PsseA::pPsseA::tnpR::lacZ | This study |
| BKC6-10 | SL1344 ushA::cat, Cmr | 3, 7 |
| BKC10-50 | SL1344(phha), Apr | This study |
| BKC10-51 | SL1344 Δhha(phha) | This study |
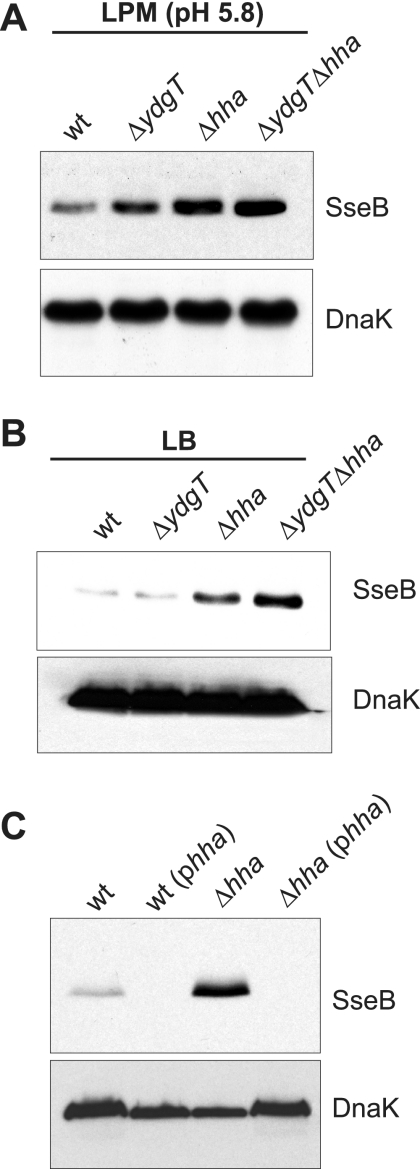
Using established culture conditions for the induction of SPI-2 gene expression (6, 7), we examined the expression of a prototypical SPI-2 gene, sseB, which is controlled by the SsrB-dependent effector/secretory operon promoter in SPI-2 (5). The growth of wild-type Salmonella in LPM (pH 5.8) induced robust expression of SseB as shown previously (6), with a slight increase in SseB levels in a ydgT mutant background as expected. Under these inducing conditions, strains that were deleted for hha or double mutants lacking both ydgT and hha showed greater accumulation of SseB in bacterial lysates than wild-type Salmonella (Fig. 1A).
FIG. 1.
Deletion of hha increases SseB protein levels. Bacterial cultures (wild type [wt] and ΔydgT, ΔhhaT, and ΔydgT Δhha mutants) were grown under SPI-2-inducing conditions in LPM (pH 5.8) (A) and under SPI-2-repressing conditions in LB broth (B) as described in the text. Bacterial cell lysates were probed by Western blot analysis for the SPI-2-encoded protein, SseB (top panels), and a control protein (intracellular DnaK) (bottom panels). (C) Complementation of hha deletion. Wild-type Salmonella serovar Typhimurium and the Δhha mutant were transformed with phha, carrying the full-length wild-type hha strain from an IPTG-inducible promoter. Strains were grown in LB and probed by Western blot analysis for SseB as described in the text. The data shown are representative of experiments performed three times.
The above data suggested that Hha might be involved in the repression of genes carried in SPI-2 under certain environmental conditions. As a more stringent test of this hypothesis, we cultured wild-type Salmonella and the repressor mutants in LB broth, a rich medium that is highly repressing for SPI-2 gene expression, and examined bacterial lysates for SseB. As reported in our previous work (7), the loss of ydgT did not lead to increases in SPI-2-encoded proteins compared to the wild type under noninducing conditions, such as growth in LB broth (Fig. 1B). However, deletion of hha or both hha and ydgT led to a 4.3-fold or 5.6-fold increase, respectively, in SseB protein levels as determined by Western blot and densitometry analyses (Fig. 1B), suggesting a derepression of the promoter activity of sseB in the mutants under study. To verify that the specific deletion of hha in our strains was responsible for the derepression of sseB, we cloned the full-length hha gene into pFLAG-CTC (Sigma) as an NdeI/BglII DNA fragment but eliminated the carboxyl-terminal FLAG tag in our cloning strategy by incorporation of the native stop codon from hha. This generated a complementation plasmid with IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible expression of the wild-type hha gene (designated phha). This complementation plasmid was transformed into wild-type and Δhha Salmonella serovar Typhimurium strains, and the strains were grown in LB as described above. The expression of the wild-type hha strain from the complementation plasmid prevented SseB accumulation from the Δhha strain and also eliminated the low basal expression of SseB from wild-type Salmonella serovar Typhimurium (Fig. 1C), indicating that the deletion of hha was responsible for this overexpression phenotype.
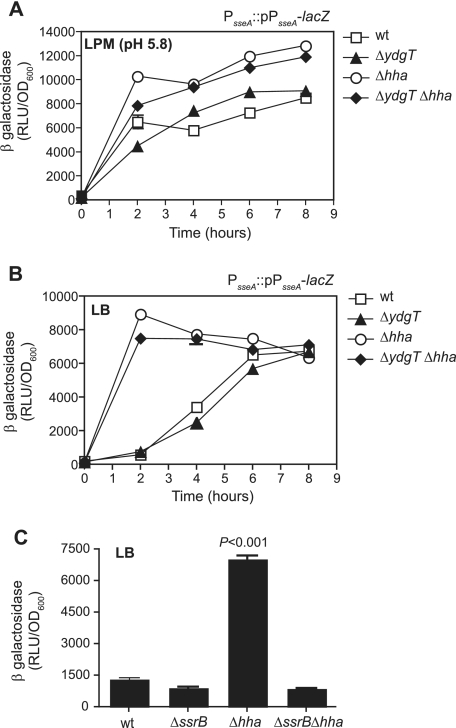
In order to examine the transcriptional activity of the promoter controlling sseB, we next integrated into the chromosome a single-copy transcriptional fusion of the sseAB promoter to lacZ (PsseA::lacZ) by homologous recombination as described previously (6). Reporter strains were constructed in wild-type bacteria and ydgT, hha, and ydgT hha mutants, creating strains designated with PsseA::pPsseA-lacZ nomenclature. Reporter strains were inoculated into either LB or LPM (pH 5.8) to give an A600 of ∼0.05 and then incubated with shaking at 37°C for various times. Samples were removed at regular intervals for the determination of numbers of CFU, A600, and β-galactosidase activity by using a chemiluminescence assay described previously (7). Relative light units of β-galactosidase activity were measured using a top-reading plate luminometer (Molecular Devices, Sunnyvale, CA) after a 60-minute incubation of bacterial lysate and substrate. To facilitate direct comparisons between strains, β-galactosidase activity was normalized to the optical density of the paired culture. Under SPI-2-inducing conditions (LPM at pH 5.8), the transcriptional activity of PsseA was detected from wild-type bacteria and β-galactosidase activity from ydgT mutant bacteria was slightly increased compared to that from wild-type bacteria during the exponential phase of the growth cycle (Fig. 2A). When the experiment was conducted with hha mutants and with ydgT hha double mutants, β-galactosidase activity was significantly higher in both mutant strains at all time points examined (Fig. 2A). Under these conditions, normalized β-galactosidase activity reached significantly greater maximal levels in strains deleted for hha. To further examine the transcriptional activity of SPI-2 under conditions that normally repress SPI-2 gene expression, we repeated the β-galactosidase activity assays using strains grown in LB under SPI-2-repressing conditions. As expected, sseA promoter activity was low in wild-type Salmonella and in ydgT mutants during the exponential phase of growth (Fig. 2B). As cultures plateau into stationary phase, the transcriptional activity of SPI-2 promoters increases, but this expression is not dependent on the acidification of the culture medium and thus represents a different mode of activation, unlike classical SPI-2 gene expression (6). In Δhha and ΔydgT Δhha strains, β-galactosidase activity was 16-fold and 15-fold higher than in the wild-type strain, respectively, at 2 h after subculture (Fig. 2B) and remained significantly higher throughout log phase. Remarkably, the level of β-galactosidase activity in log-phase hha and hha ydgT mutant strains in noninducing growth medium was greater than the level of activity observed for wild-type Salmonella grown in SPI-2-inducing medium. Next, to examine whether the increase in sseAB promoter activity from Δhha strains required the response regulator, SsrB, we deleted hha from a strain lacking ssrB to generate an ssrB hha double mutant. We integrated the chromosomal PsseA::pPsseA-lacZ reporter into this strain and examined the β-galactosidase activity of the SPI-2 promoter as described above. The deletion of ssrB eliminated the increase in β-galactosidase activity from the hha strain (Fig. 2C), indicating that SsrB was required for this transcriptional activity. As expected, the β-galactosidase activities of both wild-type and ssrB mutant Salmonella serovar Typhimurium strains were low (Fig. 2C). Together, these experiments demonstrated that the deletion of hha results in the overproduction of SPI-2-encoded SseB protein and the transcriptional activation of the sseAB promoter under classically noninducing conditions where wild-type Salmonella represses this expression. This increase in promoter activity requires SsrB since an ssrB hha double mutant shows transcriptional activity similar to those of wild-type bacteria and an ssrB mutant under these conditions. Importantly, the identification of Hha as the major repressor of SPI-2 gene expression makes available facile culture conditions under which to examine the SPI-2 regulon and identify potentially new genes that coregulate with this virulence system.
FIG. 2.
Transcriptional activity of the SPI-2 effector operon promoter (PsseA) is derepressed upon deletion of hha and requires SsrB for activity. The β-galactosidase activity of a chromosomal PsseA::lacZ reporter was examined in the indicated strains during growth under (A) SPI-2-inducing conditions (LPM at pH 5.8) and (B) noninducing conditions (LB). The data are means and standard deviations (SD) from triplicate determinations from two independent experiments. (C) The β-galactosidase activity of a chromosomal PsseA::lacZ reporter was examined in wild-type Salmonella (wt), in mutants lacking the response regulator, ssrB, or the repressor, hha, and in a double mutant lacking both ssrB and hha. The data are the means and SD from triplicate determinations from two experiments. P was <0.001 for the Δhha strain compared to the wild type. RLU, relative light units; OD600, optical density at 600 nm.
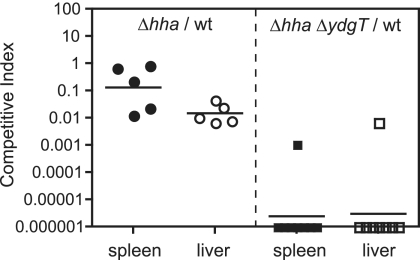
Previously, we reported that Salmonella serovar Typhimurium YdgT was involved in the contextual control of virulence factor expression during murine typhoid (7). To examine the in vivo phenotype of our hha and hha ydgT mutant strains, we performed competitive infections of mice (1) by using a wild-type strain (SL1344 ushA::cat) (7) and each of the mutants generated in this work. Groups of C57BL/6 mice (n = 5 to 10) were infected by the oral route with ∼107 CFU of a mixed inoculum of wild-type Salmonella and the mutants indicated. All animal procedures were approved by the McMaster University Animal Care Committee and were in accordance with guidelines set by the Canadian Council on Animal Care. Competitive indices (CI) for the spleen and liver of infected mice were calculated at 72 h postinfection by using the following equation: CI = (mutant/wild type)output/(mutant/wild type)input. The mean CI for the spleen and liver for the hha mutant were 0.125 (P = 0.02) and 0.0151 (P < 0.0001), respectively, indicating that the hha mutant was significantly attenuated compared to wild-type bacteria (Fig. 3). This level of attenuation was similar to that observed previously for single ydgT mutants at the same time point (7). However, the deletion of both ydgT and hha reduced the virulence of the double mutant by 6 orders of magnitude, producing mean CI values for the spleen and liver of 2.63 × 10−6 (P < 0.0001) and 3.16 × 10−6 (P < 0.0001), respectively. These data support our previously articulated notion (7) that the negative regulation of virulence genes is a key evolutionary strategy that is essential for pathogen virulence. However, further research of the extent to which SPI-2 deregulation contributes to this attenuation is required.
FIG. 3.
Competitive infections in mice. Mice were infected with a mixed inoculum of wild-type Salmonella serovar Typhimurium (wt) and either an hha mutant (left) or an hha ydgT double mutant (right). The CI for the hha strain and ydgT hha strain were determined for the spleen and liver at 72 h postinfection. Each data point represents one animal, and horizontal bars indicate geometric means. CI for hha/wt, P = 0.021 (spleen) and P = 0.001 (liver); CI for ydgT hha/wt, P < 0.0001 for spleen and liver.
A previous report describing the role of Hha in the transcriptional regulation of hilA, a positive activator of virulence genes in SPI-1 (12), examined bacterial invasion into the ileal mucosae of anesthetized mice. Our in vivo data extend this previous observation by demonstrating a prominent role for hha in the virulence of systemic murine typhoid and by confirming the importance of ydgT for systemic colonization (7), as demonstrated by the severe virulence defect of the ydgT hha double mutant. The major virulence defect of the hha ydgT double mutant strain, significantly more attenuated than either the hha or ydgT single mutant, suggests a potential negative synergistic interaction between YdgT and Hha that warrants additional work to determine the exact nature of this genetic interaction. We are also currently engaged in studies to determine the relationship between the SsrB transcriptional activator and the repressors identified in our work because the transcriptional reporter data indicate that SsrB is required for the overexpression phenotype of hha mutants. It was recently reported that H-NS, another nucleoid-associated protein in gram-negative bacteria, could silence genes acquired by horizontal gene transfer (15, 17). Given that H-NS can form heterodimeric complexes with both Hha and YdgT (19, 21), it is possible that H-NS is also involved in the repression phenotype we observe. However, since the deletion of hns is lethal in Salmonella serovar Typhimurium unless additional mutations in the alternative sigma factor gene, rpoS, or the virulence regulator, phoP, are also introduced (17), uncovering the potential role of H-NS in this repressive activity will require further study.
In addition to identifying a major regulatory node controlling intracellular virulence of Salmonella, this study draws attention to highly accessible culture conditions and genetic mutants in which to examine SPI-2 regulation and the expanded regulon coexpressed with the SPI-2 type III secretion system. In addition, the hha mutant and hha ydgT double mutant we describe are novel genetic probes of an essential virulence pathway in Salmonella, facilitating a detailed examination of genetic interactions between virulence gene repressors and transcriptional activators that collectively control the virulence behavior of this pathogen. We believe that the seemingly diametric actions of activators and repressors sculpt the virulence gene program during the evolution of bacterial pathogenesis, which is then fine-tuned by using multiple regulatory inputs for the optimal colonization of host niches.
These data support the notion that both positive regulators and repressors of virulence genes are crucial for the control of bacterial pathogenesis in animals. In the case of Salmonella serovar Typhimurium, the integration of a two-component regulatory system acquired by horizontal gene transfer (SsrA-SsrB) with ancestral repressors such as YdgT and Hha is a salient feature controlling the virulent behavior of the pathogen during the infection of host animals. Thus, such regulators would seem to be befitting targets for new anti-infectives that upset the highly coordinated expression of virulence traits in Salmonella and likely other pathogens. Realizing the therapeutic potential of targeting virulence gene regulators stands to bridge a widening innovation gap in anti-infective targets and to revolutionize our approach to fighting pathogens in human and animal medicine.
Acknowledgments
We thank Carl de Boer for his technical assistance in creating the bacterial mutants used in this work.
This work was supported by a grant to B.K.C. from the Public Health Agency of Canada. Infrastructure support for the Coombes laboratory was provided by the Public Health Agency of Canada and McMaster University.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Beuzón, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. [DOI] [PubMed] [Google Scholar]
- 2.Bijlsma, J. J., and E. A. Groisman. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57:85-96. [DOI] [PubMed] [Google Scholar]
- 3.Brown, N. F., B. A. Vallance, B. K. Coombes, Y. Valdez, B. A. Coburn, and B. B. Finlay. 2005. Salmonella pathogenicity island 2 is expressed prior to penetrating the intestine. PLoS Pathogens 1:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calva, E., and R. Oropeza. 2006. Two-component signal transduction systems, environmental signals, and virulence. Microb. Ecol. 51:166-176. [DOI] [PubMed] [Google Scholar]
- 5.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 6.Coombes, B. K., N. F. Brown, Y. Valdez, J. H. Brumell, and B. B. Finlay. 2004. Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J. Biol. Chem. 279:49804-49815. [DOI] [PubMed] [Google Scholar]
- 7.Coombes, B. K., M. E. Wickham, M. J. Lowden, N. F. Brown, and B. B. Finlay. 2005. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc. Natl. Acad. Sci. USA 102:17460-17465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis, G. R., C. Sluiters, I. Delor, D. Geib, K. Kaniga, C. Lambert de Rouvroit, M. P. Sory, J. C. Vanooteghem, and T. Michiels. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5:1023-1034. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 11.Ellison, D. W., B. Young, K. Nelson, and V. L. Miller. 2003. YmoA negatively regulates expression of invasin from Yersinia enterocolitica. J. Bacteriol. 185:7153-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juárez, A., J. M. Nieto, A. Prenafeta, E. Miquelay, C. Balsalobre, M. Carrascal, and C. Madrid. 2000. Interaction of the nucleoid-associated proteins Hha and H-NS to modulate expression of the hemolysin operon in Escherichia coli. Adv. Exp. Med. Biol. 485:127-131. [DOI] [PubMed] [Google Scholar]
- 14.Knodler, L. A., J. Celli, W. D. Hardt, B. A. Vallance, C. Yip, and B. B. Finlay. 2002. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol. Microbiol. 43:1089-1103. [DOI] [PubMed] [Google Scholar]
- 15.Lucchini, S., G. Rowley, M. D. Goldberg, D. Hurd, M. Harrison, and J. C. Hinton. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathogens 2:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madrid, C., J. M. Nieto, and A. Juarez. 2002. Role of the Hha/YmoA family of proteins in the thermoregulation of the expression of virulence factors. Int. J. Med. Microbiol. 291:425-432. [DOI] [PubMed] [Google Scholar]
- 17.Navarre, W. W., S. Porwollik, Y. Wang, M. McClelland, H. Rosen, S. J. Libby, and F. C. Fang. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236-238. [DOI] [PubMed] [Google Scholar]
- 18.Nieto, J. M., M. Carmona, S. Bolland, Y. Jubete, F. de la Cruz, and A. Juarez. 1991. The hha gene modulates haemolysin expression in Escherichia coli. Mol. Microbiol. 5:1285-1293. [DOI] [PubMed] [Google Scholar]
- 19.Nieto, J. M., C. Madrid, E. Miquelay, J. L. Parra, S. Rodríguez, and A. Juárez. 2002. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 184:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nieto, J. M., C. Madrid, A. Prenafeta, E. Miquelay, C. Balsalobre, M. Carrascal, and A. Juarez. 2000. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol. Gen. Genet. 263:349-358. [DOI] [PubMed] [Google Scholar]
- 21.Paytubi, S., C. Madrid, N. Forns, J. M. Nieto, C. Balsalobre, B. E. Uhlin, and A. Juarez. 2004. YdgT, the Hha paralogue in Escherichia coli, forms heteromeric complexes with H-NS and StpA. Mol. Microbiol. 54:251-263. [DOI] [PubMed] [Google Scholar]
- 22.Sharma, V. K., and R. L. Zuerner. 2004. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186:7290-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]