Abstract
The iron transport-biosynthesis (ITB) operon in Vibrio anguillarum includes four genes for ferric siderophore transport, fatD, -C, -B, and -A, and two genes for siderophore biosynthesis, angR and angT. This cluster plays an important role in the virulence mechanisms of this bacterium. Despite being part of the same polycistronic mRNA, the relative levels of transcription for the fat portion and for the whole ITB message differ profoundly, the levels of the fat transcript being about 17-fold higher. Using S1 nuclease mapping, lacZ transcriptional fusions, and in vitro studies, we were able to show that the differential gene expression within the ITB operon is due to termination of transcription between the fatA and angR genes, although a few transcripts proceeded beyond the termination site to the end of this operon. This termination process requires a 427-nucleotide antisense RNA that spans the intergenic region and acts as a novel transcriptional terminator.
Small RNAs, some of them antisense, have been found to regulate a wide variety of genes, either in a positive or negative fashion, although some RNAs can act both positively and negatively at different loci. Most work has been carried out in Escherichia coli, although with the explosion in the knowledge on bacterial genomes, small RNAs appear to be ubiquitous in bacteria (3, 17, 28, 30, 32). Functions that have been ascribed to some of these RNAs fall into one or more of the following processes: control of plasmid DNA replication (27), blocking of the ribosome binding site (2), freeing of the ribosome binding site (20), or targeted degradation of mRNA (21). In certain plasmids, such as pT181, harbored by some gram-positive bacteria, small RNAs have been associated with transcription attenuation by binding and folding the target RNA so that a Rho-independent terminator structure forms (7).
We have previously identified two small RNAs in the fish pathogen Vibrio anguillarum: RNAα, which acts on the iron transport gene expression, and RNAβ (11, 22, 23). These two RNAs are transcribed as countertranscripts at different loci within the iron uptake-biosynthesis (ITB [iron transport-biosynthesis]) operon of the virulence plasmid pJM1 (15). This operon encodes the proteins FatD, -C, -B, and -A, which are involved in the transport of the ferric siderophore across the membranes, and AngR and -T, which are both active in the synthesis of the siderophore anguibactin and thus responsible for the ability of this bacterium to cause septicemia in salmonid fishes (26).
Although transcribed as one operon, the level of fatDCBA mRNA is higher than the level of the full-length mRNA (fatDCBA angRT), thus causing differential expression of the genes encoding the ferric siderophore transport proteins as compared to the genes encoding the proteins involved in the siderophore biosynthesis. In this work, we report that the differential expression within this operon is mediated by RNAβ and discuss its biological significance.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Vibrio anguillarum strains were grown at 25°C in Trypticase soy broth or Trypticase soy agar (Difco) supplemented with 1% (wt/vol) NaCl and antibiotics, where appropriate. The concentrations of antibiotics (Sigma) were as follows: chloramphenicol, 30 μg/ml (E. coli) and 10 μg/ml (V. anguillarum); kanamycin, 50 μg/ml (E. coli) and 200 μg/ml (V. anguillarum); and ampicillin, 100 μg/ml (E. coli) and 500 μg/ml (V. anguillarum). For β-galactosidase experiments and RNA isolation, V. anguillarum was grown in M9 minimal medium supplemented with 1% (wt/vol) NaCl, 0.2% Casamino Acids (Difco), and 0.5% glucose (CM9) and the appropriate antibiotics to an optical density at 600 nm of 0.4 (mid-log phase). To achieve various iron levels, the CM9 minimal medium was supplemented with either ferric ammonium citrate for iron-rich conditions or ethylenediamine-di-(o-hydroxyphenylacetic) acid (EDDA) for iron-limiting conditions, as indicated in the figure legends. E. coli strain MM294 harboring the pRK2073 vector was used as a helper strain for conjugation into V. anguillarum (26), and E. coli strain HB101 was used for cloning.
General methods.
Conjugation to V. anguillarum, β-galactosidase assays, plasmid DNA purifications, and transformation were performed as previously described (6, 8, 12, 26, 29).
Construction of plasmids.
For the constitutive expression of RNAβ, plasmid pSC45 was constructed by cloning a 430-bp SnaBI-AflIII fragment of pJM1 DNA encoding RNAβ without its natural promoter into pACYC184 (9) between HindIII and EcoRV sites downstream of the promoter for the tetracycline resistance gene. Both insert and vector had been blunted using the Klenow fragment of DNA polymerase I. For the transcriptional lacZ fusion construct, the HindIII-PstI fragment containing the 3′ end of the fatA gene, the intergenic region, and the 5′ end of angR was cloned into the broad-host-range vector pKT231 (4) between the BamHI and BglII sites downstream of the kanamycin resistance (Kmr) gene promoter. A BstEII-SalI fragment containing this promoter and the downstream fatA-angR region were cloned into the SmaI-SalI sites of pTL61T (19), upstream of the lacZ gene, to create the transcriptional fusion. Site-directed mutagenesis was carried out using the Quickchange site-directed mutagenesis kit (Stratagene) according to the manufacturer's recommendations and the following primers (base changes underlined): for the −10 mutation, 5′-TTACGTATCAGGAGTTCGATTACGAAAACATTTCG-3′ and the complementary primer for pMS125; for the loop I change, 5′-CTACCATCGCCAATCAATTACACATTTGGCGTCTTGCGGT-3′ and the complementary primers for pSC45-L1C and pMS125-L1C; for the loop II change, 5′-GTCAATTTTGCCCTTGACCAAAGTGGTTAAGGGCATCGTC-3′ and the complementary primers for pSC45-L2C and pMS125-L2C; for the loop III change, 5′-TTCCCCTTCCCCCCATTTCCGAATTTATGAGAATTTAGAA-3′ and the complementary primer for pSC45-L3C; and for the loop IV change, 5′-AGAATGAATCAAAATGAACTAGGGTTCGCTTTCCCTGAGA-3′ and the complementary primer for pSC45-L4C. All mutants were sequenced prior to conjugation to verify that the only bases affected were the ones induced by site-directed mutagenesis. The construct pSC45-L1C/L2C was made by back-to-back mutagenesis of pSC45, first for loop I and then for loop II.
RNA isolation and analysis.
RNA was isolated from mid-log cultures of V. anguillarum grown in CM9, with various iron concentrations achieved by supplementing CM9 with ferric ammonium citrate or the iron chelator EDDA (see figure legends for concentrations), using the RNAwiz solution (Ambion) according to the manufacturer's recommendations. For RNase protection assays and Northern blot analysis, the riboprobes were made using the T3 and T7 promoters from pBluescript SK(+) (Stratagene) and the in vitro transcription system (Ambion) in the presence of [α-32P]UTP. Plasmid pSC59, containing the fatA-angR intergenic region as an AflIII-ClaI fragment cloned in pBluescript SK(+) (Stratagene), was used to generate the riboprobe for the fatA-angR region (by linearization of the plasmid with BamHI and transcription from the T7 promoter) and the riboprobe for RNAβ (by linearization with HindIII and transcription from the T3 promoter). Hybridization of the 50-μg total RNA with the riboprobe and subsequent treatment with RNases and urea-polyacrylamide gel electrophoresis (PAGE) (6%) were described previously (11, 14). Primer extension experiments were carried out with the synthetic primer 5′-GAACATCCCTTCGCTTTCCCTG-3′ as follows. Primer end labeling was accomplished using T4-polynucleotide kinase (Life Technologies, Inc.) in the presence of [γ-32P]ATP and hybridized to 50 μg of total RNA, followed by reverse transcription (avian myeloblastosis virus reverse transcriptase [RT]; Promega). DNA sequencing to determine the location of the start site and urea-PAGE (6%) were performed as described previously (8, 14, 25). The S1 mapping analysis to detect the 3′ end of RNAβ was performed with a riboprobe made using the plasmid pSC58 as a template, which carries the AflIII-NheI 171-bp fragment of the ITB operon cloned in pBluescript SK(+) (Stratagene). Following the linearization of the plasmid with SalI, in vitro transcription from the T3 promoter in the presence of [α-32P]UTP generated a 245-nucleotide (nt) product. The hybridization of the total RNA (10 μg) to the riboprobe (42°C) was carried out overnight prior to the digestion with the S1 nuclease. The subsequent urea-PAGE (6%) was performed as described previously (8).
For the S1 mapping of the mRNA in the intergenic region between fatA and angR, we used a PCR product with primers 11905 5′-ATATATTGATGCACAAAACAC-3′ and 11906 5′-GATAATAACTGAGTAAGCTGC-3′. For the 5′-end mapping, primer 11906 was labeled using [γ-32P]ATP and T4-polynucleotide kinase (Life Technologies, Inc.) prior to the PCR. For the 3′-end-labeled probe, the PCR fragment was digested with NheI and the overhang was filled in using [ α-32P]CTP and the Klenow fragment of DNA polymerase I. Hybridizations were performed as described above.
Northern blot analysis was performed according to standard protocols (24). Twenty micrograms of total RNA was loaded on a polyacrylamide gel and transferred to uncharged nylon (Amersham) using the transblot apparatus from Bio-Rad. RNA was fixed under UV from 1 min at 1,200 kJ. The riboprobe was generated from pSC59 as described above.
Quantitative real-time PCR was performed using an ABI PRISM 7000 sequence detection system and SYBR green master mix (both from Applied Biosystems) according to the manufacturer's recommendations. To generate the cDNA, 1 μg of total RNA was reverse transcribed using Moloney murine leukemia virus RT from Ambion according to manufacturer's recommendations. Prior to reverse transcription, total RNA was made DNA free using TURBO DNA-free (Ambion) according to the manufacturer's recommendation. As a control, parallel samples were run in which RT was omitted from the reaction mixture. PCRs were performed in triplicate in a 25-μl volume, with the following cycle parameters: 95°C for 10 min for enzyme activation followed by 95°C for 45 s and 55°C for 1 min for 45 cycles. A melting plot was performed to ensure specific amplification. Data analysis was performed using the comparative cycle threshold method (Applied Biosystems) to determine relative expression levels. Values for the expression of fatA and angR were normalized to the levels of aroC, a chromosomal gene that is not regulated by iron or any of the other regulators identified in V. anguillarum (10). The primers used for the real-time RT-PCR analysis are as follows (5′ to 3′): aroC, TGATTCGATCGGTGCAAAGA and AAGCATGTGCAATATCCGCAT; fatA, AACAAACGGAAAAACGGCAG and TCATATCGAGCTTTAAGCCGG; and angR, GCTCAGTGACGACTTCTACC and CTTGCGGACTCAAGATCAAC.
For the in vitro transcription, we used a PCR product from the transcriptional fusion construct containing the Kmr promoter, 3′ end of fatA, and 5′ end of angR, without the lacZ gene. The resulting 1,154-bp product was incubated for 30 min with E. coli RNA polymerase saturated with σ70 (Epicenter) according to the manufacturer's recommendation and nucleoside triphosphates (NTPs), of which ATP was used as α-32P label, with and without RNAβ as indicated in the figure and figure legend. Mouse liver RNA (5 μg) was used to control for specificity of RNAβ. Reactions were cleaned over G-25 Sepharose spin columns (Amersham) and after precipitation resuspended in gel loading buffer II (Ambion). The samples were then separated by 6% urea-PAGE and visualized on Kodak X-Omat film.
The microarray experiments are part of a separate study, and results will be published elsewhere. In short, the tiled microarray of the pJM1 plasmid was designed by Nimblegen (www.nimblegen.com). For each strand, a 24-mer probe was made that overlaps the previous probe by 12 nucleotides. For each strand, a total of 5,403 probes encompassing the total 65,009 nucleotides of the pJM1 plasmid were added on a chip hybridized with 10 μg total RNA isolated from wild-type V. anguillarum strain 775 grown under either iron-rich (4 μg/ml) or iron-limiting (2 μM EDDA) conditions using RNAwiz (Ambion) as described above. Each RNA sample was extracted from equal amounts of cells of three independent cultures, to account for the biological error. The intensity for each probe was measured using the 5-μm scanning platform at Nimblegen. The data were analyzed using the Excel program (Microsoft).
RESULTS
Identification of antisense RNA species encoded in the fatA-angR intergenic region.
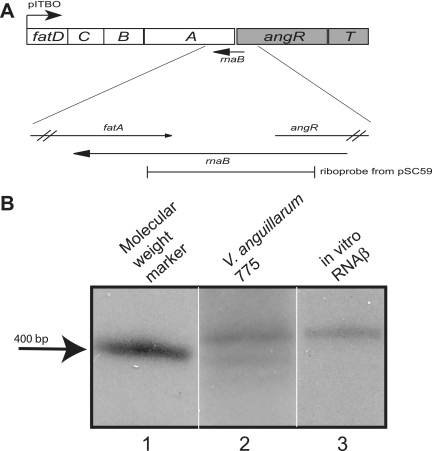
From previous studies on the angR gene, we had some indication about the existence of an antisense RNA transcribed opposite of the 5′ end of the angR gene (22). We therefore performed a Northern blot analysis to determine the full length of this antisense RNA. We used a riboprobe generated from pSC59 (Fig. 1A and see Materials and Methods). In the Northern blot, we could detect two bands—one slightly larger than 400 nt and one slightly smaller (Fig. 1B, lane 2). When in vitro-transcribed RNAβ was used in the same Northern blot analysis, only one band that is slightly larger than 400 nucleotides can be identified (Fig. 1B, lane 3). This analysis revealed that two species of RNA were found in the region complementary to angR when we used RNA extracted from wild-type V. anguillarum cells grown under iron-limiting conditions (same conditions as those used previously by Salinas and Crosa [22]). Since we cannot rule out the relevance of either of these species, we designated both as RNAβ independently of their size.
FIG. 1.
Genetic organization of the iron transport and biosynthesis operon on the pJM1 plasmid and Northern blot analysis of RNAβ. Panel A shows the operon organization of the ITB operon. The location of the probe derived from pSC59 is also shown. (B) Northern blot analysis. Lane 1, molecular weight marker; lane 2, RNA from V. anguillarum 775 grown in CM9 minimal medium; lane 3, in vitro-synthesized RNAβ. The lanes are from the same gel, but shown are different exposure times for lanes 1 and 3, since at the exposure time of lane 2, these were saturating the film.
RNAβ is complementary to the 5′ end of angR and the 3′ end of fatA.
To determine the exact size and start and end points of RNAβ, we mapped the 5′ end by primer extension and the 3′ end by S1 mapping analyses using RNA samples obtained from cultures grown at various iron concentrations. The primer extension (Fig. 2A) was performed as described in Materials and Methods and shows that a predominant band of 110 nt was detected in the presence of 3 μM EDDA (lane 1) or iron concentrations that range from 0 to 8 μg/ml ferric ammonium citrate (lanes 2 to 5). Based on the size of the band, the transcription start site (+1) of RNAβ was mapped within the angR open reading frame at 119 nt downstream from the predicted angR translational start (Fig. 2C). Under conditions of iron limitation (3 μM EDDA; lane 1) and in cells grown in high iron concentration (lane 5), the levels of RNAβ were decreased. In even higher iron concentrations (16 μg/ml ferric ammonium citrate, lane 6), we did not find the 110-nt band and only detected the smaller products also seen in lane 5. These smaller species could be the product of degradation of the larger, 110-nt, species. It should be noted that cells grown in minimal medium without the addition of the iron chelator EDDA are already nutritionally limited with respect to the availability of iron (13).
FIG. 2.
(A and B) Mapping of the 5′ end and 3′ end of RNAβ. (A) Primer extension analyses was carried out to determine the 5′ end of RNAβ. The products that extended from the 32P-labeled primer (highlighted in gray in panel C) were analyzed in parallel with a known sequence marker (lanes GACT) by urea-PAGE (6%). Panel B shows the results of S1 mapping analysis, which was performed with a 245-nt 32P-labeled riboprobe containing the 171-nt AflIII-NheI DNA fragment from the fatA-angR intergenic region (see panel C). The riboprobe without S1 nuclease treatment is shown in lanes U, alongside a known sequence marker (lanes TCAG). In both panels, the supplements to the medium were as follows: lane 1, 3 μM EDDA; lane 2, none; and lanes 3 to 6, ferric ammonium citrate at concentrations of 2 μg/ml (lane 3), 4 μg/ml (lane 4), 8 μg/ml (lane 5), and 16 μg/ml (lane 6). Lanes marked with E are empty. (C) The nucleotide sequence of the region encompassing the fatA 3′ end and angR 5′ end is shown. The 5′ end (+1) and both 3′ ends of RNAβ and the putative −10 promoter sequence for RNAβ are indicated. Boxes contain the nucleotides of the loops of the stem-loop structures shown in panel D. (D) The putative stem-loop structures were generated using the computer program FoldRNA, GCG (University of Wisconsin). The complementary stem-loop structures (I-I*, II-II*, III-III*, and IV-IV*) are darkly shaded. Position 1 in the sense RNA structure corresponds to the first nucleotide in the AflIII site shown in the sequence in panel C.
To locate the 3′ end of RNAβ, S1-mapping analysis was performed with a 171-nt riboprobe (see Fig. 2C and Materials and Methods). S1 nuclease digestion of the RNAβ-riboprobe hybrid yielded a major band of 166 nt only seen when iron was added to the medium at concentrations ranging from 2 to 16 μg/ml ferric ammonium citrate (Fig. 2B, lanes 3 to 6). We also detected a smaller band of about 94 nt, possibly the smaller species of RNAβ detected by Northern blot analysis (Fig. 1B).
Based on the size of the major band (166 nt), we were able to locate the 3′ end of the major species of RNAβ within the fatA open reading frame, 222 nt upstream of the fatA translational stop (Fig. 2C). However, the 3′ end of RNAβ could not be detected under iron-limiting conditions (lanes 1 and 2). The accessibility to the probe may have been obscured by the interaction with the ITB operon transcript under iron-limiting conditions. The results from the primer extension, S1 mapping, and Northern blot analysis suggest that there are at least two species of RNAβ according to their size. The first species is the full-length RNAβ of 427 nt that is found under low iron concentrations (0 to 4 μg/ml ferric ammonium citrate) and is possibly also present in smaller amounts under iron-limiting conditions (3 μM EDDA). The second species is smaller (about 355 nt) and contains only the 5′ end of the 427-nt RNAβ but is shorter at the 3′ end. As part of a different study, we have performed a tiled microarray analysis of the complete sequence of the pJM1 plasmid. This microarray shows that the expression levels of RNAβ are similar under iron-rich (4 μg/ml ferric ammonium citrate) and iron-limiting (3 μM EDDA) conditions. The average expression levels for RNAβ of two independent tiled microarrays, including the error for each condition and the change between the two conditions, are as follows: for ferric ammonium citrate and EDDA, the expression values were 2,392 ± 64 and 2,172 ± 52, respectively, representing a 1.10-fold change. The complete analysis of this tiled microarray will be published elsewhere. In addition, when the rnaB promoter was fused to a promoterless lacZ gene, similar activities were measured under iron-depleted and iron-replete conditions (data not shown). It should be noted that the promoter activities under both conditions are very low. Based on the Northern blot analysis, primer extension, and S1 mapping, microarray analysis, and β-galactosidase studies, we believe that RNAβ is expressed constitutively under the various iron concentrations, with possible rapid degradation in higher iron concentrations (>8 μg/ml ferric ammonium citrate).
In silico analysis of the secondary structure of full-length RNAβ and prediction of the sense mRNA in its complementary region were performed with the GCG Wisconsin package program FoldRNA (Fig. 2D). The sequences of both RNAs predict several stem-loop structures, of which four have sequence complementarities in the loops between the sense and antisense RNA. We designated these structures I, II, III, and IV in the sense RNA and I*, II*, III* and IV* in RNAβ. The predicted stem-loops I*, II*, III*, and IV* are present in both the full-length and the shorter RNAβ.
Differential gene expression within the ITB operon.
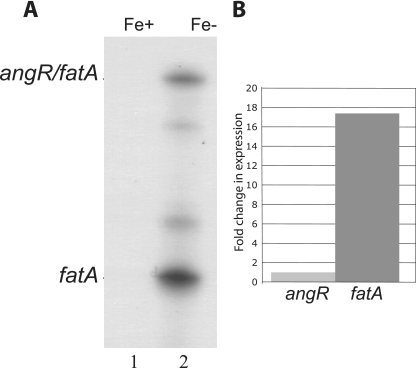
An RNase protection assay was performed to detect the ITB operon mRNA in the fatA-angR intergenic region by using a riboprobe made from plasmid pSC59 (Fig. 1A). This probe was hybridized with total RNA extracted from wild-type V. anguillarum 775 cultures that had been grown under iron-limiting as well as iron-rich conditions (see Materials and Methods and figure legend). Instead of just one protected band, as expected for a polycistronic mRNA encompassing this region (22), we found two major protected bands under iron-limiting conditions (Fig. 3A, lane 2). The band marked angR/fatA corresponds to the fully protected probe that includes both the 3′ end of fatA and the 5′ end of angR, while the lower, more abundant band marked fatA corresponds to a fraction of the whole probe. These two major mRNA bands and additional minor bands appeared only under iron-limiting conditions, when the ITB operon promoter is active (8). We believe that the smaller band (fatA) corresponds to only the mRNA for the fatA and upstream genes and results from either termination or processing of the longer message that includes angR and angT. Either of these situations would result in a reduction of the angR-specific mRNA as seen in Fig. 3A. We believe that the additional minor bands either result from degradation of the protected probe or are termination/processing intermediaries. To determine the ratio between the full-length mRNA and the truncated mRNA, we performed real-time quantitative PCR (Materials and Methods) using primer sets for fatA (near the 3′ end) and angR (near the 5′ end) and used the aroC gene as a control to normalize the expression levels. We determined that the mRNA ratio between fatA and angR is 17.4:1 (Fig. 3B). During the reverse transcription reaction, prior to the real-time quantitative PCR, the primer for the angR gene could reverse transcribe through the intergenic region and fatA. In this case, the expression value for the fatA gene is overestimated. Therefore, in calculating the ratio between fatA and angR, we subtracted the expression level of the angR gene from that of the fatA gene. Since not every primer is reverse transcribed to the fatA gene, the calculated ratio is at least 17.4:1 but could be higher than we report here. From the microarray experiment described above, we also looked at the ratio between fatA and angR. When we compared the expression levels between fatA and angR, there was indeed more fatA than there is angR. The tendency is the same as that detected by RNase protection assay and real-time quantitative PCR, but the ratio is lower. This could be due to the fact that AngR is a nonribosomal peptide synthetase and thus carries domains that are similar to other nonribosomal peptide synthetases in the bacterium. Therefore, the expression level of angR is detected as being higher than it possibly actually is, and this will lead to a lower fatA/angR ratio.
FIG. 3.
Analysis of the expression levels of the fatA-angR intergenic region mRNA. Panel A shows the RNase protection experiment carried out with the total RNA preparations isolated from strain 775 grown under iron-rich (lane 1 [2 μg/ml ferric ammonium citrate]) and low-iron (lane 2 [2 μM EDDA]) conditions. Panel B shows the real-time quantitative PCR results in change (fold) in expression.
We speculate that antisense RNAβ could play a role in the differential gene expression that occurs at the fatA-angR intergenic region because it is expressed when the ITB operon mRNA is transcribed and has sequence complementarities; thus, both RNAs could have a physical interaction.
Antisense RNAβ is necessary for the differential gene expression.
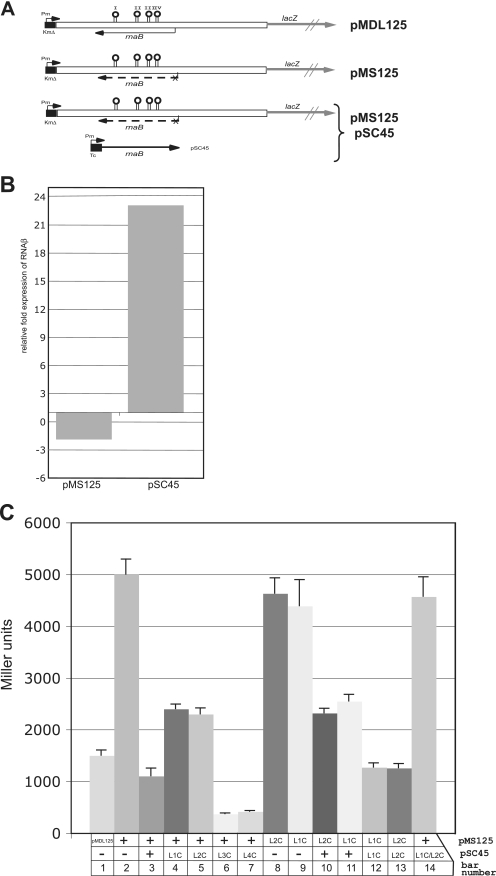
Our attempts to generate RNAβ-deficient strains by mutating the RNAβ promoter in the 65-kb pJM1 plasmid were unsuccessful. Therefore, to address the possible involvement of RNAβ in the differential gene expression within the ITB operon, we generated a transcriptional fusion of the 3′ end of fatA, the intergenic region, and the 5′-end of angR (open bars in Fig. 4A) with the promoterless lacZ gene in the plasmid pTL61T (19), generating plasmid pMDL125. In this construct, the transcription of the sense mRNA is driven by a Kmr gene promoter (Fig. 4A). The vector pTL61T contains an RNase III processing site between the cloning site and the lacZ gene, which ensures independent translation of lacZ irrespective of what sequences are fused upstream (19). Plasmid pMDL125, in which RNAβ is transcribed from its own promoter, was subjected to site-directed mutagenesis to mutate the predicted −10 region of the rnaB promoter from ATTTGA to GTTCGA to abolish or reduce transcription of the antisense RNAβ, generating a new plasmid, pMS125.
FIG. 4.
β-Galactosidase assay of transcriptional fusions of the intergenic region. (A) Schematic representation of the lacZ transcriptional fusions used in this study with the position of the sense mRNA stem-loops and the location of the rnaB gene. (B) Expression levels of RNAβ in pMS125 and pSC45 compared to pMDL125 as measured by real-time quantitative PCR. (C) Measurements of transcription termination using lacZ fusions. Shown are the average β-galactosidase activities (including the error) in Miller units of five independent experiments performed in triplicate for each strain. Under each bar is indicated what plasmids are present in V. anguillarum. L1C indicates that the bases in loop I have been changed to their complementary base, L2C indicates the complementary bases in loop II, and so on. + indicates that the plasmid is present as listed on the side, and − indicates that this particular plasmid is not present.
The −10 and −35 sequences identified in Fig. 2C are in good agreement with those of other promoters identified in V. anguillarum (8, 14) and do not follow the E. coli consensus. These mutations in the −10 region did not affect the amino acid sequence encoded on the remaining portion of the angR gene in the complementary strand. The effect of the −10 mutations on the expression of RNAβ was tested by real-time quantitative PCR to determine the reduction (fold) of the RNAβ promoter activity in pMS125 compared to pMDL125. The levels of RNAβ were normalized to the expression level of the ampicillin gene located on the same plasmid. The results in Fig. 4B show that there is only a twofold reduction in the expression levels that is probably background since the rnaB promoter is very weak. Also shown in Fig. 4B is the increase (fold) in the levels of RNAβ expressed from the Tcr gene promoter in pSC45 compared to that from pMDL125. This construct overexpresses RNAβ by 23-fold. In this experiment, the transcription levels were normalized to the aroC gene located on the chromosome.
The plasmids pMDL125 and pMS125 were conjugated to a V. anguillarum strain from which the pJM1 plasmid had been cured to avoid ambiguous results due to the presence of wild-type mRNAs of fatA, angR, and RNAβ from the pJM1 plasmid. Bars 1 and 2 in Fig. 4C show a comparison of the β-galactosidase activity for each of these two plasmids. It is clear that the β-galactosidase activity was increased by about fivefold when the mutant plasmid was used (pMS125) compared with the wild type (pMDL125), indicating that RNAβ is involved in this phenomenon (i.e., transcription termination is relieved or processing is abolished when RNAβ is either not present or present at very low levels). Furthermore, we could restore the mutant phenotype of the V. anguillarum strain carrying pMS125 to the wild-type levels by supplying RNAβ in trans from plasmid pSC45 (Fig. 4C, bar 3), where RNAβ is transcribed from the Tcr gene promoter (Fig. 4A). Additionally, when pMDL125 was complemented with pSC45, even lower levels of β-galactosidase were detected (not shown). To identify the features of RNAβ that are involved in the reduction of the full-length mRNA, we mutated the predicted loops I through IV (Fig. 2C and D and 4A) to their complementary bases in the plasmid pSC45, which constitutively expresses RNAβ. These constructs (pSC45L1C, pSC45L2C, pSC45L3C, and pSC45L4C) were used in β-galactosidase experiments together with pMS125. The mutants are described in Materials and Methods, and the results of the β-galactosidase experiments with these mutants are shown in Fig. 4C, bars 4 to 7. Only the base changes in loop I and II have an effect on RNAβ activity, as seen by the increase in the β-galactosidase activity (Fig. 4C, bars 4 and 5). Although the levels of β-galactosidase activity in the strains harboring the mutation in loop III or IV (Fig. 4C, bars 6 and 7) are lower that that of the strain harboring the wild-type constructs and could potentially be of interest, we only focused on loops I and II. We first mutated loops I and II in the transcriptional fusion pMS125 and used these constructs, pMS125L1C and pMS125L2C, in β-galactosidase experiments: neither sequence change has a cis effect on the β-galactosidase activity (Fig. 4C, compare bars 8 and 9 to bars 1 and 2). When these mutants are complemented with wild-type RNAβ, the activity is reduced (Fig. 4C, bars 10 and 11), but not to the same extent as when the sequences are both complementary as in the wild-type constructs (Fig. 4C, bar 3), indicating that loops I and II are both involved in the decrease in full-length mRNA. When the corresponding loop is also mutated to the complementary bases: i.e., base changes in both the transcriptional fusion construct and the plasmid expressing RNAβ, the expression of β-galactosidase is decreased to wild-type levels, indicating that both loops are involved in the action of RNAβ and suggesting an interaction between RNAβ and the mRNA (Fig. 4C, bars 12 and 13). When both loops I and II are mutated in pSC45, the effect of each single mutation is amplified (Fig. 4C, bar 14) and the β-galactosidase levels are increased to the level of the rnaB −10 mutant strain.
Transcription termination of the polycistronic mRNA is the cause for the differential gene expression within the ITB operon.
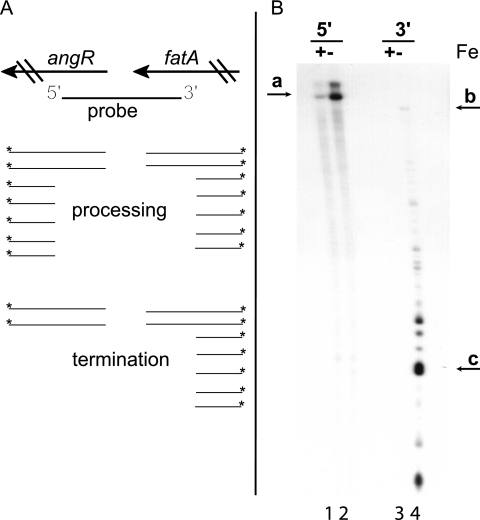
The RNase protection experiment and the quantitative PCR results presented in Fig. 3 show a dramatic decrease in the levels of the full-length ITB mRNA as compared to the shorter species up to the end of the fatA gene. The results from the lacZ fusion experiments show a necessity for RNAβ in the decreased levels of full-length mRNA. However, the analyses done so far cannot resolve whether the differential gene expression observed is due to termination of transcription or processing of the full-length mRNA followed by rapid degradation of the angRT mRNA. In order to discriminate between these two possibilities, both 5′-end S1 mapping and 3′-end S1 mapping were performed as described in Materials and Methods. Figure 5A shows a scheme of the expected banding patterns if either of the processes is responsible for the reduced transcription between fatA and angR. If processing is occurring, even with rapid degradation of the processed RNA, one would expect the appearance of low-molecular-weight bands corresponding to protection of the labeled probe by transcripts representing both the 3′ and 5′ ends of the processed RNA, in addition to a band corresponding to the full-length transcript resulting from protection by the unprocessed mRNAs. In the case of termination, the 5′-end-labeled probe should detect only the mRNA species that have not been terminated, while the 3′-end probe should detect a band corresponding to the unterminated mRNA and smaller distinct bands representing the 3′ end of the fatA gene up to the termination site. The results (Fig. 5B) are consistent with a termination mechanism. Analysis using the 5′-end-labeled probe detected only the full-length transcript (band a), with no detection of smaller processed bands. Using the 3′-end-labeled probe, several smaller, yet distinct, bands were detected along with a small amount of the full-length mRNA (band b). The band labeled c (Fig. 5B, lane 4) is the predominant terminated species and maps to a stem-loop within the fat-angR intergenic region that we have previously identified (1). Comparing the results obtained from the S1 mapping (Fig. 5) with the sequence (Fig. 2C) of the mRNA, the major transcription termination site maps to stem-loop II. Because the S1 mapping was not sensitive enough to detect single nucleotides, we can only map the band (Fig. 5B, band c) to the region encompassing stem-loop 2. Although the results are consistent with transcription termination, we cannot rule out processing from this experiment alone because, although unlikely, the degradation could proceed extremely rapidly and degradation intermediates could therefore be missed. As described in Materials and Methods, the 5′-end-labeled probe was made by PCR using a labeled primer, while the 3′-end-labeled probe was generated using the same PCR product as the 5′-end-labeled probe (without a labeled primer), but cleaved with NheI and subsequently filled in with the Klenow fragment of DNA polymerase I and a radioactively labeled nucleotide. The resulting probe is therefore a few bases shorter.
FIG. 5.
3′-end and 5′-end S1 mapping of the fatA-angR intergenic region. (A) Scheme of the expected banding patterns in the S1-treated samples if the difference in expression is the result of processing of a large mRNA including fatA-angR or transcription termination at a site between fatA and angR (see text for details). Asterisks indicate the position of the radioactive label. Please note that the order of the genes is depicted opposite from those for the other figures in this study. Panel B shows the actual results of the 5′- and 3′-end S1 mapping experiments using a 32P-labeled DNA probe and total RNA obtained from V. anguillarum 775 grown under iron-rich (lanes 1 and 3 [2 μg/ml ferric ammonium citrate]) and low-iron (lanes 2 and 4 [2 μM EDDA]) conditions.
In vitro termination by RNAβ.
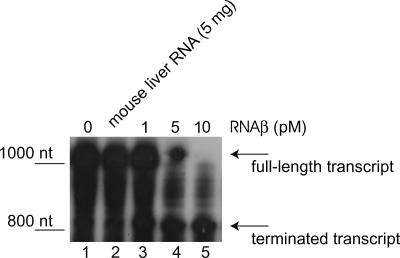
While the above-mentioned experiment hints at transcription termination, processing cannot be ruled out completely. To prove that transcription termination is what is happening, we reconstructed the system in vitro using as a template a linear PCR product of 1,154 nt consisting of the Kmr gene promoter followed by the 3′ end of fatA, the intergenic region, and the 5′ end of angR. The in vitro system also included E. coli RNA polymerase saturated with σ70 and NTPs, of which ATP was labeled with 32P at the α position. The PCR construct carries the whole rnaB gene but not the promoter; therefore, only the fatA/angR mRNA can be transcribed in the reaction. Reaction mixtures were incubated with and without in vitro-synthesized RNAβ for 30 min at 37°C. Reaction mixtures were cleaned over Sepharose G-25 spin columns and loaded on 6% urea-PAGE gel. The results in Fig. 6 show that when no RNAβ is added (lane 1) or if mouse liver RNA (lane 2) is added to the reaction mixture, mainly the full-length transcript is detected. When RNAβ is added in increasing amounts, the full-length transcript disappears and a smaller band appears (lanes 3 to 5). From the experiments shown in Fig. 6, it is clear that termination is induced by RNAβ and no other factors are necessary. However, it is possible that in vivo, where transcription factors might be more limited, other additional factors are required to enhance the probability of interactions between the interplaying molecules.
FIG. 6.
In vitro termination assay for RNAβ. We used E. coli RNA polymerase saturated with σ70, [α-32P]ATP and no addition (lane 1), 5 μg (20 pM) mouse liver RNA (lane 2), 1 pM RNAβ (lane 3), 5 pM RNAβ (lane 4), and 10 pM RNAβ (lane 5).
DISCUSSION
The ferric anguibactin transporter proteins, FatD, FatC, FatB, and FatA, and two of the proteins involved in anguibactin biosynthesis, AngR and AngT, are encoded as part of an operon (ITB) that originates upstream of the fatD gene (8). These genes are an important component of the virulence repertoire of V. anguillarum (1). Transcription of this operon is negatively regulated by the Fur protein and positively regulated by AngR and the trans-acting factor, TAF (8, 18, 31). On the complementary strand to this operon, there are two genes, rnaA and rnaB, that encode countertranscripts that we named antisense RNAα and RNAβ, respectively (11, 31). In this work, we investigated the role of RNAβ in the complex regulatory network of the ITB operon. We showed that the abundance of the transcript corresponding to the full-length ITB operon is 17-fold lower than the shorter message that spans up to the end of fatA. We showed that this reduction is caused by transcription termination between angR and fatA, with a major termination event occurring at stem-loop II. This stem-loop does not fall into any of the present classifications for transcription terminators (i.e., no Rho-independent stem-loop structure and no Rho binding site upstream of this stem-loop), suggesting a novel mechanism for the termination event within this operon. Because this is the first report of such a mechanism, we cannot identify a “termination” motif. Perhaps when this type of transcription termination is identified in other organisms, a motif may become apparent. Because of the location of the rnaB gene and the complementary nature of the loops in both RNAs, we investigated the potential of RNAβ as an effector of transcription termination within this operon. In this work, we also showed that RNAβ is a 427-nt-long antisense RNA that can be found as a full-length molecule and as a shorter species that contains the same 5′ end but differs at the 3′ end. The promoter for RNAβ is somewhat independent of the iron concentration (i.e., it is active at the high and low iron concentrations used in this study). We do not believe that RNAβ is translated, like RNAIII in Staphylococcus aureus (5), but translation of a predicted peptide of up to 25 amino acids cannot be ruled out at present.
Using lacZ transcriptional fusions of the fatA-angR intergenic region, we were able to show that the transcription termination event, mainly at stem-loop II, relies on the presence of RNAβ. We further demonstrated that loops I and II of RNAβ are directly involved and necessary for maximum termination to maintain the balance of transport versus biosynthesis gene transcripts. Mutation in either loop I or II changes the termination phenotype, and the complementary mutation restores the phenotype, indicating that loops I and II actually exist. Mutations in loops III or IV do not seem to eliminate or reduce termination, but they could simply not exist in RNAβ in the cell. It seems from the in vitro experiments that RNAβ does not need any other factor for termination to occur; however, it is possible that in vivo, where transcription factors might be more diluted, additional factors are required to enhance the interactions between the interplaying molecules.
This novel mechanism for transcription termination by an antisense RNA without any other element, like Rho-independent terminator structures or Rho binding sites, is not only uncommon, but it also results in a higher level of the transport gene mRNA compared to that for the two genes encoding the siderophore biosynthetic proteins, AngR and AngT. Therefore, although all of the components for transport and biosynthesis are controlled by modulating the activity of a single promoter (22), the antisense RNA-mediated transcription termination mechanism results in a fine-tuning of the relative contributions of transport and biosynthesis genes: a few molecules of the biosynthetic enzymes could produce enough siderophore, but an excess in the level of the transport proteins benefits the cell in its chance encounter with ferric siderophore molecules in the extracellular environment. Other studies done by Yarnell and Roberts (33) in vitro have reported that formation of RNA stem-loop structures leads to destabilization of the RNA polymerase-template-transcript elongation complex and consequently to a termination event. In that study (33), addition of oligoribonucleotides that hybridize to the nascent RNA strand would also lead to destabilization of the RNA polymerase-template-transcript elongation complex and subsequent termination. Comparing their result with the results reported here, it is possible that what was seen in vitro by Yarnell and Roberts (33) happens in vivo with RNAβ in V. anguillarum in a much more efficient manner, since only a few molecules of RNAβ are sufficient.
In E. coli, antisense RNAs that are about 100 nt long can function at multiple targets (16). It is thus possible, given the large size of RNAβ and the fact that it is expressed differentially compared to the ITB operon mRNA, that RNAβ could act pleiotropically in the regulation of other genes in the V. anguillarum genome.
Acknowledgments
This work was supported by grant AI19018 from the National Institutes of Health to J.H.C.
We are very grateful to Sunghee Chai for invaluable contributions.
Footnotes
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Actis, L. A., M. E. Tolmasky, D. H. Farrell, and J. H. Crosa. 1988. Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid-mediated iron-transport system. J. Biol. Chem. 263:2853-2860. [PubMed] [Google Scholar]
- 2.Altuvia, S., A. Zhang, L. Argaman, A. Tiwari, and G. Storz. 1998. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 17:6069-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 4.Bagdasarian, M., R. Lurz, B. Ruckert, F. C. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. [DOI] [PubMed] [Google Scholar]
- 5.Balaban, N., and R. P. Novick. 1995. Translation of RNAIII, the Staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3′-end deletion. FEMS Microbiol. Lett. 133:155-161. [DOI] [PubMed] [Google Scholar]
- 6.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brantl, S., and E. G. Wagner. 2000. Antisense RNA-mediated transcriptional attenuation: an in vitro study of plasmid pT181. Mol. Microbiol. 35:1469-1482. [DOI] [PubMed] [Google Scholar]
- 8.Chai, S., T. J. Welch, and J. H. Crosa. 1998. Characterization of the interaction between Fur and the iron transport promoter of the virulence plasmid in Vibrio anguillarum. J. Biol. Chem. 273:33841-33847. [DOI] [PubMed] [Google Scholar]
- 9.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Q., L. A. Actis, M. E. Tolmasky, and J. H. Crosa. 1994. Chromosome-mediated 2,3-dihydroxybenzoic acid is a precursor in the biosynthesis of the plasmid-mediated siderophore anguibactin in Vibrio anguillarum. J. Bacteriol. 176:4226-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Q., and J. H. Crosa. 1996. Antisense RNA, fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J. Biol. Chem. 271:18885-18891. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Q., A. M. Wertheimer, M. E. Tolmasky, and J. H. Crosa. 1996. The AngR protein and the siderophore anguibactin positively regulate the expression of iron-transport genes in Vibrio anguillarum. Mol. Microbiol. 22:127-134. [DOI] [PubMed] [Google Scholar]
- 13.Crosa, J. H., and L. L. Hodges. 1981. Outer membrane proteins induced under conditions of iron limitation in the marine fish pathogen Vibrio anguillarum 775. Infect. Immun. 31:223-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Lorenzo, M., S. Poppelaars, M. Stork, M. Nagasawa, M. E. Tolmasky, and J. H. Crosa. 2004. A nonribosomal peptide synthetase with a novel domain organization is essential for siderophore biosynthesis in Vibrio anguillarum. J. Bacteriol. 186:7327-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Lorenzo, M., M. Stork, M. E. Tolmasky, L. A. Actis, D. Farrell, T. J. Welch, L. M. Crosa, A. M. Wertheimer, Q. Chen, P. Salinas, L. Waldbeser, and J. H. Crosa. 2003. Complete sequence of virulence plasmid pJM1 from the marine fish pathogen Vibrio anguillarum strain 775. J. Bacteriol. 185:5822-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303-328. [DOI] [PubMed] [Google Scholar]
- 17.Hershberg, R., S. Altuvia, and H. Margalit. 2003. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 31:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koster, W. L., L. A. Actis, L. S. Waldbeser, M. E. Tolmasky, and J. H. Crosa. 1991. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J. Biol. Chem. 266:23829-23833. [PubMed] [Google Scholar]
- 19.Linn, T., and R. St. Pierre. 1990. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J. Bacteriol. 172:1077-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masse, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salinas, P. C., and J. H. Crosa. 1995. Regulation of angR, a gene with regulatory and biosynthetic functions in the pJM1 plasmid-mediated iron uptake system of Vibrio anguillarum. Gene 160:17-23. [DOI] [PubMed] [Google Scholar]
- 23.Salinas, P. C., L. S. Waldbeser, and J. H. Crosa. 1993. Regulation of the expression of bacterial iron transport genes: possible role of an antisense RNA as a repressor. Gene 123:33-38. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tolmasky, M. E., L. A. Actis, and J. H. Crosa. 1988. Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel trans activator of siderophore biosynthesis. J. Bacteriol. 170:1913-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomizawa, J. 1990. Control of ColE1 plasmid replication. Intermediates in the binding of RNA I and RNA II. J. Mol. Biol. 212:683-694. [DOI] [PubMed] [Google Scholar]
- 28.Vogel, J., V. Bartels, T. H. Tang, G. Churakov, J. G. Slagter-Jager, A. Huttenhofer, and E. G. Wagner. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31:6435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Gabain, A., J. G. Belasco, J. L. Schottel, A. C. Chang, and S. N. Cohen. 1983. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc. Natl. Acad. Sci. USA 80:653-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wertheimer, A. M., W. Verweij, Q. Chen, L. M. Crosa, M. Nagasawa, M. E. Tolmasky, L. A. Actis, and J. H. Crosa. 1999. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect. Immun. 67:6496-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 101:9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yarnell, W. S., and J. W. Roberts. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611-615. [DOI] [PubMed] [Google Scholar]