Abstract
We previously reported that SadB, a protein of unknown function, is required for an early step in biofilm formation by the opportunistic pathogen Pseudomonas aeruginosa. Here we report that a mutation in sadB also results in increased swarming compared to the wild-type strain. Our data are consistent with a model in which SadB inversely regulates biofilm formation and swarming motility via its ability both to modulate flagellar reversals in a viscosity-dependent fashion and to influence the production of the Pel exopolysaccharide. We also show that SadB is required to properly modulate flagellar reversal rates via chemotaxis cluster IV (CheIV cluster). Mutational analyses of two components of the CheIV cluster, the methyl-accepting chemotaxis protein PilJ and the PilJ demethylase ChpB, support a model wherein this chemotaxis cluster participates in the inverse regulation of biofilm formation and swarming motility. Epistasis analysis indicates that SadB functions upstream of the CheIV cluster. We propose that P. aeruginosa utilizes a SadB-dependent, chemotaxis-like regulatory pathway to inversely regulate two key surface behaviors, biofilm formation and swarming motility.
Pseudomonas aeruginosa is an important model organism for the study of gram-negative biofilm development, yet little is known about the molecular mechanisms underlying the initial events leading to the surface interactions that characterize the early steps in bacterial biofilm formation. Microscopic observations (23, 26, 40, 51) and genetic analyses (2) revealed two sequential events that lead to stable surface interactions. First, a bacterial cell pole contacts the surface in a process referred to as reversible attachment. This is a relatively unstable interaction, as reversibly attached bacteria can readily return to a planktonic existence. The second event is a transition from the polar association to one that is mediated by the long axis of the cell body, referred to as irreversible attachment. In P. aeruginosa, the only mutation known to block the transition from reversible to irreversible attachment is in the sadB gene (2).
Another key aspect of biofilm formation by P. aeruginosa is the production of an extracellular matrix. In pseudomonads, this matrix is thought to be comprised of exopolysaccharides (EPS), DNA, and protein (19). The biofilm matrix has typically been credited with structuring the mature biofilm (4). Studies have identified the pel and psl loci as two sets of genes predicted to be involved in the production of the polysaccharide component of the matrix required for biofilm maturation by P. aeruginosa on abiotic surfaces, although only the pel gene cluster is found in P. aeruginosa strain PA14 (7, 8, 15, 27), the focus of study in this report. Interestingly, recent studies suggest that the pel locus also plays a role in early biofilm formation. A pel mutant of P. aeruginosa PAK shows a strong attachment defect in a strain lacking type IV pili (48) and P. aeruginosa PAO1 with a mutation in the psl locus has a block in biofilm initiation (24).
Swarming motility, another surface behavior of P. aeruginosa, allows this microbe to move across surfaces (11). Swarming motility requires both a functional flagellum and the production of the surface-wetting agent rhamnolipid surfactant, but the mechanism by which P. aeruginosa propels itself across the surface has not been explored. Swarming motility can be distinguished from swimming motility in that swarming is required to move across a hydrated, viscous semisolid surface, while swimming allows movement through a relatively low-viscosity liquid environment. We have shown that sadB is also involved in modulating swarming motility in response to rhamnolipid surfactants (3).
Here we show that SadB participates in the inverse regulation of biofilm formation and swarming motility and requires chemotaxis cluster IV to mediate these effects. Our data are consistent with a model in which SadB inversely regulates these behaviors via its ability both to modulate flagellar reversals in a viscosity-dependent fashion and to influence the production of the Pel polysaccharide. We propose that P. aeruginosa inversely regulates biofilm formation and swarming motility upon transitioning to a surface lifestyle.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, media, and chemicals.
Bacterial strains, plasmids and primers used in this study are shown in Table 1. P. aeruginosa PA14 was cultured on lysogeny broth (LB) medium (1) solidified with 1.5% agar. M63 minimal salts (35) medium supplemented with MgSO4 (1 mM), glucose (0.2%), and (where indicated) Casamino Acids (CAA, 0.5%) was used for static biofilm assays. Swarm (0.5% and 0.55% agar) and swim (0.3% agar) plates consisted of M8 minimal medium (20) supplemented with MgSO4 (1 mM), glucose (0.2%), and CAA (0.5%). Twitch plates consisted of LB medium solidified with 1.0% agar. Unless noted otherwise, antibiotics were used at the following concentrations for P. aeruginosa: carbenicillin, 500 μg/ml; tetracycline, 150 μg/ml, gentamicin, 100 μg/ml. All enzymes used for DNA manipulation were purchased from Invitrogen (Carlsbad, CA).
TABLE 1.
Strains, plasmids and primers used in this studya
| Strain, plasmid, or primer | Relevant characteristics or sequence | Source or reference |
|---|---|---|
| P. aeruginosa PA14 | Wild type | 37 |
| SMC705 | PA14 sadB::Tn5B21; Tcr | 2 |
| SMC257 | PA14 flgK::Tn5B30; Tcr | 32 |
| SMC252 | PA14 pilB::Tn5B30; Tcr | 32 |
| PA14 ΔpelA | 7 | |
| SMC478 | PA14 pilG::Tn5B21; Tcr | This study |
| SMC2992 | PA14 ΔpilJ | This study |
| SMC2990 | PA14 ΔchpB | This study |
| SMC3257 | PA14 ΔchpB pelA::pMQ89 | This study |
| SMC2958 | PA14 PAO178::pKO3PAO178 | This study |
| SMC2962 | PA14 wspA::pKO3wspA | This study |
| SMC2949 | PA14 cheR1::pKO3cheR1 | This study |
| Plasmids | ||
| pUCP18 | Cloning vector, Cbr Apr | 41 |
| pNC5 (pSadB+) | sadB in pUCP18; Cbr Apr | 2 |
| pMQ90 | Cloning vector; Cbr Apr | 42 |
| pChpB+ | chpB gene cloned in pMQ90 | This study |
| pPilJ+ | pilJ gene cloned in pMQ90 | This study |
| pMQ30 | Suicide vector; Gmr SacB+URA3 CEN6/ARSH4 | 42 |
| pMQ30ΔpilJ | pilJ KO construct | This study |
| pMQ30ΔchpB | chpB KO construct | This study |
| pKO3 | Suicide vector; Tcr | 30 |
| pKO3-PAO178 | PAO178 KO construct | This study |
| pKO3-wspA | wspA KO construct | This study |
| pKO3-cheR1 | cheR1 KO construct | This study |
| Primers | ||
| pilJKO1 | 5′-CAGTCACGACGTTGTAAAACGACGGCCAGTGCCAAGCTTGCACCATGGCTCGTATTTTGATTGTTG-3′ | |
| pilJKO2 | 5′-CAGTTTGAAGCCGGATACCGAGCCCGCGAAAAGATTGCCTG-3′ | |
| pilJKO4 | 5′-TCACTCATTAGGCACCCCAGGCTTTACACTTTATGCTTCCCACTCCATTCATGTGCCTGAG-3′ | |
| chpBKO1 | 5′-CCCAGTCACGACGTTGTAAAACGACGGCCAGTGCCAAGCTTGGTGCTCGAGTCCGGTTTCTCC-3′ | |
| chpBKO2 | 5′-CCAGATGCTTGACCAATGCCTCCAACGAGGTGTCGGCGATCAC-3′ | |
| chpBKO3 | 5′-GTGATCGCCGACACCTCGTTGGAGGCATTGGTCAAGCATCTGG-3′ | |
| chpBKO4 | 5′-CATTAGGCACCCCAGGCTTTACACTTTATGCTTCCGGCTCGTGAACTCGACGGAGGAAAGATCC-3′ | |
| pilJ5′ | 5′-GGCGGAATTCAAGGAGCCAAATATGAAGAAAATCAACGC-3′ | |
| pilJ3′ | 5′-GGCGAAGCTTCCTCGTCCAAGGTGTCGAC-3′ | |
| chpB5′ | 5′-GGCGGAATTCCGGCCAGCATGAGTGAGC-3′ | |
| chpB3′ | 5′-GGCGAAGCTTGTGTTCCGGCTCATGCCATC-3′ | |
| PAO178KO1 | 5′-GCGGGGATCCCTCCAGGTGTTCTTCG-3′ | |
| PAO178KO2 | 5′-GCGGGAATTCGGTGATCACCAGTTCG-3′ | |
| wspAKO1 | 5′-GCGGCTGCAGGAACTGGACTGTTCG-3′ | |
| wspAKO2 | 5′-GGCGAAGCTTCGTGACGAATTCGG-3′ | |
| cheR1KO1 | 5′-GCGGGGATCCGGATTTCGAGTTGTTCAGG-3′ | |
| cheR1KO2 | 5′-GCGGGAATTCCGAAATAGCGTTGCAGAC-3′ |
Ap, ampicillin; Cb, carbenicillin; Gm, gentamicin; Tc, tetracycline; KO, knockout.
Molecular techniques.
The DNA flanking the transposon carried by the pilG::Tn5B21 mutant was mapped to the pilG locus using the published P. aeruginosa PAO1 genome (45) and arbitrarily primed PCR, as previously described (34). In-frame deletions of pilJ and chpB were generated using pMQ30 (42), constructed as reported elsewhere (13), and the resolved integrants were confirmed by PCR. Single-crossover insertion of plasmid pKO3 (30) by homologous recombination was used to disrupt the PAO178, wspA, and cheR1 genes. The constructs were made using the primers in Table 1 and confirmed by PCR. Plasmids for chpB and pilJ complementation were constructed in pMQ90 by amplifying chpB and pilJ by PCR using the primers listed in Table 1.
Quantitative reverse transcription PCR (qRT-PCR) was performed as previously reported (21). Bacteria were harvested either from static planktonic cultures incubated for 8 h (typical biofilm assay growth conditions) or on agar plates used for Congo red assays (see below) but lacking the dyes and solidified with 1.0% agar.
Biofilm assays.
Ninety-six-well microtiter plate assays and microscopic analysis were performed as previously described (29, 33). To quantify reversible versus irreversible attachment, overnight cultures were normalized by optical density at 600 nm and diluted 1:100 in M63 supplemented with glucose, CAA, and MgSO4. A 500-μl aliquot of this suspension was inoculated into the wells of a 24-well plate (in duplicate) and incubated for 1 h at 37°C. The medium was removed and gently replaced, and images were captured at two frames/second for 30 s. Images were converted to QuickTime files for analysis.
Determining swim reversals.
Swim reversal rate measures the frequency at which a swimming cell changes its direction. Overnight LB-grown cultures were diluted 1:100 into fresh M63 medium supplemented with glucose. Ficoll was added at 3% for low-viscosity (swimming) conditions; this added Ficoll slowed the swimming cells sufficiently to facilitate monitoring of reversal rates. High-viscosity conditions, mimicking conditions of swarming motility based on our previous studies (47), were achieved by adding Ficoll to 15%. Subcultured bacteria were incubated at 37°C for 1 to 2 h, and then 500-μl aliquots were added to the wells of 24-well plates. Phase-contrast, time-lapse images were acquired every 0.3 s at a ×1,400 magnification using the OpenLab software package. The time-lapse images were converted to QuickTime movies for subsequent analysis. QuickTime movies were advanced frame by frame, and individual cells were monitored for the number of times they reversed swim direction while within the field of view. Approximately 25 cells were counted in each of six wells (∼125 cells total) to determine reversal rates, which are expressed as reversals per cell.
Swarm assays.
Plate preparation, inoculation, and incubation were performed as previously described (47). To determine the percentage of swarm plate surface coverage by a given bacterial swarm, an image of the swarm plate was captured using a Nikon 990 digital camera (Nikon, Melville, NY) and false colored using Photoshop software (Adobe, Mountain View, CA) to provide ample contrast between the bacterial swarm and the agar surface so that the pixels could be counted using Kodak 1D Image Analysis software (Kodak, Rochester, NY). The number of pixels that comprised the swarm was expressed as a percentage of the number of pixels that comprised an image of the surface of the plate. Alternatively, the captured image was imported into PowerPoint (Microsoft), where the outline of the bacterial swarm was traced in order to distinguish it from the agar surface. ImageJ software (National Institutes of Health) was used to calculate both the area within the swarm and that of the plate surface for comparison.
Polysaccharide assays.
Congo red (CR) assays were performed as reported elsewhere, except that the base medium used was M63 medium supplemented with glucose, MgSO4, and CAA at the concentrations used for biofilm assays (7, 8). Scanning electron microscopy (SEM) was performed as reported elsewhere (16), except that glutaraldehyde was used at 2.5% and bacteria were grown on the plastic substrate for 38 h in minimal medium plus glucose and CAA.
RESULTS
Relationship of SadB to biofilm formation and swarming.
We postulated that upon encountering a surface, P. aeruginosa would likely coregulate its surface-associated behaviors, including biofilm formation and swarming. The fact that SadB appeared to be required for both biofilm formation (2) and swarming motility (3) suggested that this protein might be involved in coregulating these processes; therefore, we pursued sadB both as a genetic link between these phenomena and to gain greater insight into how P. aeruginosa regulates its surface behaviors.
As part of the published characterization of SadB, we demonstrated that SadB protein levels were elevated under conditions that promote robust biofilm formation. These data suggested that there might be a correlation between the levels of SadB and the extent of biofilm formation. To test this hypothesis, we took advantage of the observation that expressing sadB in multicopy from a plasmid (pSadB+) resulted in elevated cytoplasmic levels of the SadB protein (2). We examined the effects of overexpressing SadB from the pSadB+ plasmid on biofilm formation in glucose minimal medium, a medium that does not promote robust biofilm formation by P. aeruginosa PA14. In a 96-well biofilm assay, biofilm formation was enhanced ∼3-fold at 4 h in the wild type (WT) overexpressing SadB (WT/pSadB+) in comparison to the vector control strain (WT/pUCP18) (Fig. 1A). The WT strain overexpressing SadB also showed a 2.5-fold increase in the number of bacteria attached to the surface in comparison to the vector control at this early time point, as determined by phase-contrast microscopy (Fig. 1B).
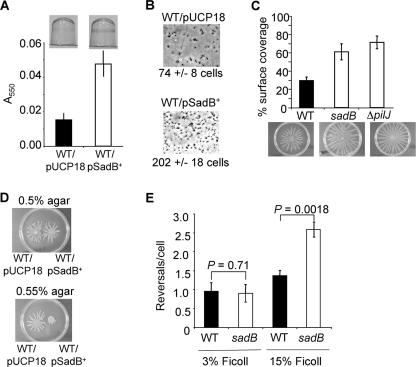
FIG. 1.
Surface-associated behaviors influenced by SadB levels. (A) Biofilm formation phenotypes under static conditions. (Top) Image of crystal violet (CV)-stained biofilms formed by the wild type carrying the vector control (WT/pUCP18) and the wild type overexpressing SadB (WT/pSadB+). Cells were grown in minimal medium containing (0.2%) glucose for 4 h at 37°C before staining with CV. (Bottom) Quantification of CV-stained wells. (B) Phase-contrast images of WT/pUCP18 and WT/pSadB+ attached to the surface of a 24-well plate after incubation at 37°C for 1 h. For each strain, images were recorded at a magnification of ×1,400 over 10 fields of view, and the average number of surface-associated cells for each strain is indicated below the image. (C) The graph shows the average plate surface coverage that results from swarms produced by the WT, sadB, and ΔpilJ strains. Below are representative images of WT, sadB, and ΔpilJ strains after 16 h of incubation at 37°C. The percent surface coverage of the WT is significantly less than that of the sadB mutant (P = 0.0029) and the ΔpilJ mutant (P = 0.000018). (D) Aliquots of the wild type carrying the vector control (WT/pUCP18) and the wild type overexpressing SadB (WT/pSadB+) were spotted on 0.5% and 0.55% swarm agar plates and incubated for 16 h at 37°C. (E) Reversal rates of WT and sadB mutant cells under low-viscosity (3% Ficoll) and high-viscosity (15% Ficoll) conditions.
The sadB mutant also shows a hyperswarming phenotype compared to the WT, resulting in an ∼2-fold increase in surface coverage for the sadB mutant (Fig. 1C). In contrast, overexpression of SadB leads to suppression of swarming motility on 0.55% agar but not on 0.5% agar, suggesting a viscosity-dependent role for SadB in swarm suppression (Fig. 1D). Together, these data suggest an inverse relationship between swarming motility and biofilm formation mediated, at least in part, by SadB.
A sadB mutant has a viscosity-dependent defect in flagellar reversals.
Because both swarming motility and biofilm formation are dependent on a functional flagellum, we assayed the sadB mutant for phenotypes related to motility, in particular, swim speed and flagellar reversal rate. Directly measuring the speed of swimming under low-viscosity conditions (using 3% Ficoll [47]) revealed no difference between the WT (55.3 ± 1.83 μm/s) and the sadB mutant (53.38 ± 1.52 μm/s; P = 0.37), a result consistent with our previous findings (2). Because of the effects of a sadB mutation on swarming motility, we also measured swimming speed under high-viscosity conditions (15% Ficoll), which we have shown previously is a condition analogous to that encountered by the cells when swarming (47). Under high-viscosity conditions, a small but significant difference in swimming speed was measured between the WT (10.03 ± 0.59 μm/s) and the sadB mutant (12.29 ± 0.40 μm/s; P = 0.003), an increase of ∼23% for the sadB mutant. It is formally possible that this small increase in swimming motility can also contribute to the enhanced swarming of the sadB mutant; however, there are no other data to support this conclusion.
Another component of controlling flagellar motility is regulating the rate of flagellar reversals; therefore, we also assessed the flagellar reversal rate of the WT and the sadB mutant (Fig. 1E). The rate of reversals under low-viscosity conditions (3% Ficoll) for the sadB mutant is equal to that of WT. Consistent with a role for SadB in the control of flagellar-mediated reversals, the sadB mutant displayed a >2-fold increase in flagellar reversals compared to WT cells in 15% Ficoll. Also under high-viscosity conditions (15% Ficoll), overexpression of SadB from plasmid pNC5 (pSadB+) in the WT reduced reversals per cell to 0.52 ± 0.26 compared to 1.36 ± 0.04 for the WT carrying the pUCP18 vector control (P = 0.035). This two- to threefold change in reversal rates is on par with the magnitude of change in reversal rate observed for Escherichia coli in the presence of attractants and/or for mutants in the Che machinery (31, 36).
SadB-mediated effects on biofilm formation and swarming require components of chemotaxis cluster IV.
Based on the viscosity-dependent alteration of flagellar reversals in the sadB mutant and the known role of the chemotaxis system of E. coli in the regulation of flagellar reversals (25), we hypothesized that one of the five chemotaxis-like clusters of P. aeruginosa (6) might serve as a link between SadB and flagellar function. We found that a Tn5 insertion in pilG, a component of chemotaxis cluster IV (cheIV) of P. aeruginosa, resulted in a SadB-like swarming phenotype, but mutations in none of the other clusters yielded similar phenotypes (Table 2).
TABLE 2.
SadB-dependent biofilm and swarming phenotypes of Che cluster mutants
| Chemotaxis cluster | Mutation | Swarming phenotype | SadB biofilm stimulationc,d | SadB swarm repressionc,e |
|---|---|---|---|---|
| ΔmotABa | WT | N | N | |
| I | ΔmotCD | Nonswarming | N | ND |
| II | PAO178::pKO3PAO178 | Decreased | Y | Y |
| III | wspA::pKO3wspA | WT | Y | Y |
| IV | pilG::Tn5B21 | Hyperswarmerb | N | M |
| IV | ΔpilJ | Hyperswarmerb | N | N |
| IV | ΔchpB | Nonswarming | ND | ND |
| V | cheR1::pKO3cheR1 | Decreased | Y | Y |
These genes are not in clusters CheI to CheV.
As determined by a lack of avoidance of other bacterial swarms, as reported elsewhere (3).
N, no; Y, yes; M, moderate; ND, not determined.
Determined by testing whether SadB expressed from a plasmid stimulated biofilm formation (see Fig. 1A).
Determined by testing whether SadB expressed from a plasmid repressed swarming motility on 0.55% agar (see Fig. 1D).
To confirm a role for the CheIV cluster in SadB-dependent effects on biofilm formation and swarming, in-frame deletions in two genes of the cheIV cluster (Fig. 2A), pilJ, encoding a predicted methyl-accepting chemotaxis protein (MCP), and chpB, encoding a predicted MCP demethylase, were constructed. We chose to mutate these genes based on previous work in E. coli—loss of the MCP should block signaling through this chemotaxis-like system, while mutating the demethylase should result in hypermethylation of the MCP and thus presumably result in higher basal receptor activity (25). Therefore, we predicted that mutations in pilJ and chpB should have opposite effects on biofilm formation and swarming.
FIG. 2.
The ΔpilJ mutant is biofilm defective and a hyperswarmer. (A) CheIV chemotaxis cluster. pil genes are shown in light gray and chp genes in dark gray. Black arrows represent open reading frames, and gene names are given below the arrows. (B) Biofilm formation by the WT, ΔpilJ mutant, and complemented strains at 8 h. Cells were grown at 37°C for 8 h in minimal medium containing glucose and CAA. (C) Aliquots of the ΔpilJ mutant carrying the vector control (ΔpilJ/pMQ90) or a construct expressing the pilJ gene in trans (pPilJ+) were spotted on 0.5% swarm agar plates and incubated at 37°C for 16 h. (D) Aliquots of the ΔpilJ mutant carrying the vector control (ΔpilJ/pUCP18) or a construct overexpressing SadB (pSadB+) were spotted on 0.5% and 0.55% swarm agar plates and incubated at 37°C for 16 h. (E) The extent of swarming, expressed as percent surface coverage, is shown on 0.5% agar for the strains indicated. *, statistically significant decrease in swarming compared to the other strains (P < 0.001).
The ΔpilJ mutant is defective for biofilm formation (Fig. 2B) and displays a hyperswarming phenotype (Fig. 1C), and providing pilJ on a plasmid complements these phenotypes (Fig. 2B and C). The ΔpilJ mutant is also defective for twitching motility (Table 3). Furthermore, overexpressing SadB in the ΔpilJ mutant neither stimulates biofilm formation (data not shown) nor suppresses swarming motility (Fig. 2D and E), suggesting that PilJ is genetically downstream of SadB. Consistent with this hypothesis, expressing PilJ from a high-copy-number plasmid suppresses the swarming of the sadB mutant (Fig. 2E). In contrast to the ΔpilJ mutant, the ΔchpB mutant cannot swarm (Fig. 3A) but forms a more robust biofilm than the WT (Fig. 3B).
TABLE 3.
Swimming, twitching and swarming phenotypese
| Strain genotype | Zone (mm)
|
Swarming (% coverage)c | |
|---|---|---|---|
| Swima | Twitchb | ||
| WT | 28.9 ± 1.9 | 14.3 ± 1 | 30 |
| pilG::Tn5B21 | 29.5 ± 0.7 | 0 | ND |
| ΔpilJ | 37.0 ± 1.0 | 0 | 72 |
| ΔchpB | 22.0 ± 1.0 | 13.3 ± 0.4 | <10 |
| flgK::Tn5B30 | 0 | ND | 0 |
| pilB::Tn5B21 | ND | 0 | ND |
| sadB | NDd | NDd | 63 |
The diameter of the circular zone of motility formed on 0.3% agar plates after 24 h incubation at 37°C, as reported previously (32).
The diameter of the circular zone of motility formed on 1.5% agar after 24 h incubation at 37°C, followed by 24 h at 25°C, as reported previously (32).
Swarming is presented as the percentage of the agar plate covered, as reported previously (3).
We reported previously that a sadB mutant has no swimming or twitching defect (2).
ND, not determined.
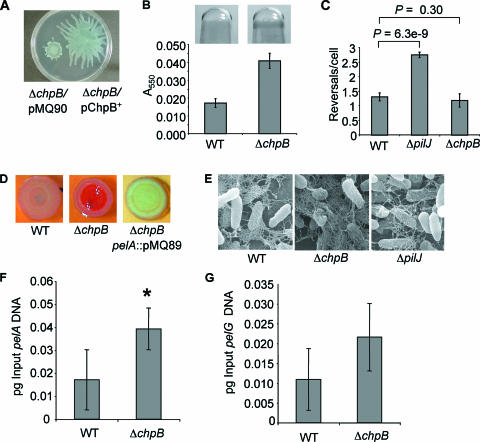
FIG. 3.
The ΔchpB mutant is defective for swarming motility and displays increased biofilm formation and CR binding. (A) Aliquots of the ΔchpB mutant carrying the vector control (ΔchpB/pMQ90) and a construct expressing the chpB gene in trans (pChpB+) were spotted on 0.5% swarm agar plates and incubated at 37°C for 16 h. (B) Biofilm formation by the WT and ΔchpB mutant was assessed after 4 h of growth at 37°C in minimal medium containing glucose. Shown are representative wells (top) and quantification of the biofilm assays (bottom). (C) Reversal rates of the WT and the ΔpilJ and ΔchpB mutants under high-viscosity (15% Ficoll) conditions. (D) CR assays with the WT, the ΔchpB mutant, and the ΔchpB pelA double mutant. Plates were incubated for 24 h at 37°C and an additional 24 h at room temperature. (E) SEM of WT and the ΔchpB and ΔpilJ mutants. Images were prepared with ruthenium red to highlight polysaccharides. (F and G) Quantitative RT-PCR analysis of pelA (F) and pelG (G) gene expression in agar-grown cultures of the indicated strains. *, statistically significant difference from the WT (P < 0.05).
If the CheIV cluster is in the same genetic pathway as SadB, we predict that mutations in ΔpilJ and/or ΔchpB might cause altered flagellar reversal rates. Consistent with the observation that the ΔpilJ mutant and the sadB mutant have similar biofilm and swarming phenotypes, the ΔpilJ mutant also shows an ∼2-fold increase in flagellar reversal rates compared to the WT under high-viscosity conditions; however, the reversal rate of the ΔchpB mutant is not significantly different from that of the WT (Fig. 3C).
Mutating chpB increases production of the pel-encoded matrix.
We investigated whether the ΔchpB mutant is altered for other biofilm-related functions that might explain the hyperbiofilm and nonswarming phenotypes of this mutant. The ΔchpB mutant did not display any defects in swimming or twitching motility (Table 3). The ΔchpB mutant did show increased binding to CR compared to the WT (Fig. 3D, compare left and center panels). CR has been shown to bind the pel-encoded polysaccharide of P. aeruginosa PA14 (7, 8). Consistent with the conclusion that mutating chpB alters production of the Pel polysaccharide, introducing a pelA mutation into the ΔchpB genetic background completely eliminated CR binding (Fig. 3D).
The CR data were confirmed by SEM (Fig. 3E) using methods similar to those reported for the analysis of the Pel polysaccharide (7). The WT produced an amorphous material characteristic of extracellular polysaccharides, and consistent with the CR studies, the ΔchpB mutant produced more of this material (Fig. 3E, compare left and center panels).
To determine whether the ΔchpB mutant affected pel gene transcription, we measured the transcript levels of pelA and pelG using qRT-PCR. We chose to assess the expression of pelA and pelG because both of these genes are predicted to code for enzymes required to produce the glucose-rich polysaccharide component of the P. aeruginosa matrix (7). The WT and the ΔchpB mutant were grown either planktonically under static conditions (identical to conditions used for biofilm assays) or on agar plates under conditions identical to those used for CR assays (minus the dyes). A small (∼2-fold) but significant increase in pelA transcript, but no difference in pelG transcript level, was observed between the WT and ΔchpB mutant grown as a colony (Fig. 3F and G), and no difference was observed when the cultures were grown planktonically (data not shown). These data suggest that mutating chpB has little or no effect on the expression of the pelA and pelG genes.
The mutation in pilJ also results in a small decrease in CR staining and an altered colony morphology, but to a degree identical to that observed for a mutant lacking type IV pili (data not shown), suggesting that loss of PilJ function plays little or no role in EPS production. However, it may not be possible to observe subtle changes in EPS production using the CR assay. Consistent with the CR findings, SEM studies indicated that the WT and ΔpilJ mutant produced similar levels of extracellular material (Fig. 3E, right panel).
Role for sadB in Pel polysaccharide production.
We predicted that the sadB mutant might also be altered for polysaccharide production. Given the lack of biofilm formation and increased swarming of the sadB mutant strain, phenotypes opposite those of the ΔchpB mutant, we predicted that the sadB mutant would bind less rather than more CR, and this is what we observed (Fig. 4A). Furthermore, SEM analysis revealed that the sadB mutant produced noticeably less extracellular matrix material than the WT (Fig. 4B). The ΔpelA mutant, defective in production of the Pel polysaccharide, served as a control in this study.
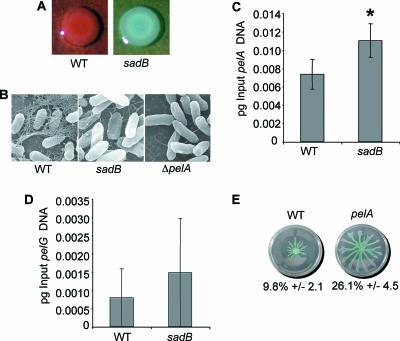
FIG. 4.
A sadB mutant displays decreased Pel polysaccharide production but no decrease in pel gene expression. (A) CR binding assays with the WT and the sadB mutant. Plates were incubated for 24 h at 37°C and an additional 24 h at room temperature. (B) SEM of the indicated strains. Images were prepared with ruthenium red to highlight polysaccharides. (C and D) Quantitative RT-PCR analysis of pelA (C) and pelG (D) gene expression in agar-grown cultures of the indicated strains. *, statistically significant difference from the WT (P < 0.05). (E) Swarming phenotype of the WT and the ΔpelA mutant. Shown are a representative swarm plate for each strain and quantification of plate coverage for each strain (five plates).
We determined whether the sadB mutant affected pel gene transcription. A small (∼2-fold) but significant increase was observed for the pelA or pelG transcript level in the sadB mutant versus the WT grown under colony growth conditions (Fig. 4C and D), and no difference in transcript levels was observed under planktonic conditions (data not shown), indicating that the decrease in CR staining in the sadB mutant cannot be explained by decreased transcription of the pel locus.
Role for the pel locus in swarming and early biofilm formation.
Given the apparent relationship between swarming and CR binding described above, we also determined whether a mutation in pelA might impact swarming motility. A strain mutated in the pelA gene showed a ∼2.5-fold increase in swarming motility compared to the WT strain (Fig. 4E). A mutation in the pelA gene does not impact swimming or twitching motility (data not shown).
Several mutations described above impact both biofilm formation and motility, and furthermore, sadB is known to impact biofilm formation at the transition from reversible to irreversible attachment. If the pel-encoded matrix also plays a role at this early step in biofilm formation, we would predict that a strain defective for matrix production would also have a defect in irreversible attachment. Consistent with this prediction, in a static assay, we observed a statistically significant decrease in irreversible attachment of the ΔpelA mutant (86.1% ± 3.2%) compared to the WT (94.7% ± 4.2%; P = 0.0000583). The decrease in irreversible attachment of the pelA strain is not as striking as that observed for the sadB mutant (67.8% ± 8.1%; P = 0.0001417).
DISCUSSION
Here we show that SadB, originally identified as required for early biofilm formation, is also a negative effector of swarming motility, a result consistent with our previous findings (3). We also showed previously that RpoN and FleR, known regulators of flagellum and rhamnolipid production in P. aeruginosa (14, 38, 46), also regulate SadB levels (2), suggesting that SadB is coregulated with other functions required for swarming and biofilm formation. How does SadB contribute to both biofilm formation and swarming behaviors? A model summarizing the findings from this study is shown in Fig. 5. While we have yet to identify the biochemical function of the SadB protein, our results implicate this protein in two pathways that impact swarming motility and biofilm formation.
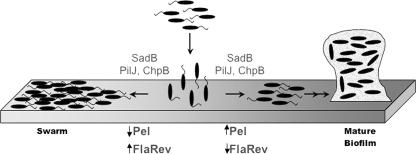
FIG. 5.
Model for inverse control of biofilm formation and swarming motility. Planktonic bacteria (top) initially interact with the surface, likely via reversible polar attachment, although which end of the cell interacts with the abiotic surface is unclear. A pathway that includes SadB and components of the CheIV chemotaxis cluster (PilJ and ChpB) controls the decision to initiate biofilm formation or move via swarming. Biofilm formation is associated with an increase in the production of Pel-dependent polysaccharide (Pel) and a decrease in flagellar reversal rate (FlaRev). Swarming is associated with decreased production of polysaccharide and an increased rate of flagellar reversals. The substratum is shown as a gradient of gray to represent the fact that variations in surface properties (viscosity, wetness, etc.) might also impact the biofilm-versus-swarming decision.
First, SadB is involved in mediating flagellar reversals, but only under high-viscosity conditions likely similar to those encountered during either biofilm formation or swarming, but not swimming. A role for the chemotaxis cluster in E. coli in controlling flagellar reversal rates prompted us to investigate the potential involvement of the five chemosensory-like clusters of P. aeruginosa as a mechanism for linking SadB to flagellar function. An in-frame deletion of pilJ, an MCP homolog, rendered the strain biofilm defective and a hyperswarmer and resulted in increased flagellar reversals. The biofilm, swarming and flagellar reversal phenotypes of the ΔpilJ mutant are identical to those of a sadB mutant strain. Epistasis analysis indicates that SadB is genetically upstream of pilJ, consistent with a model wherein sadB exerts its effects on flagellar rotation via the CheIV chemosensory system.
Based on the E. coli paradigm (25), loss of the PilJ MCP should block signaling via this chemosensory system. In contrast, mutating the demethylase gene homolog, chpB, should result in a hypermethylated MCP with higher basal receptor activity. Consistent with these hypotheses, the ΔchpB mutant is defective for swarming but forms a more robust biofilm than the WT, phenotypes opposite those observed for the ΔpilJ mutant.
While mutations in sadB and pilJ resulted in increased flagellar reversals, to our surprise the ΔchpB mutant did not have the predicted decrease in reversals and in fact showed no discernible effect on this behavior. Perhaps the repression of flagellar reversals is accomplished via another pathway. Alternatively, while the ΔchpB mutation alters the basal activity of the MCP, perhaps a second input signal is still required to observe decreased reversals in this mutant background. We do show here that mutations in chpB impact another known biofilm-related factor, namely, the production of the putative pel-encoded matrix. A ΔchpB mutant has a CR-hyperbinding phenotype that is pelA dependent and results in increased matrix production as judged by SEM, but the ΔchpB mutation does not alter pel gene expression, suggesting that this mutation increased production of the Pel polysaccharide by a nontranscriptional mechanism. In contrast to the ΔchpB mutant, the sadB mutant showed decreased CR binding and matrix production, suggesting that SadB positively impacts production of the Pel polysaccharide. Despite the decrease in apparent production of the Pel polysaccharide by the sadB mutant, expression of the pelA and pelG mRNAs is slightly up-regulated in the sadB mutant versus the WT. These data indicate that the reduction of Pel polysaccharide production in a sadB mutant occurs via a nontranscriptional mechanism.
At this point, we do not understand how SadB or components of the CheIV cluster impact EPS production. Given the lack of change in pel gene expression in the sadB and chpB mutants, one obvious explanation for the changes in matrix production in strains mutated in these functions is that Pel production is controlled by a mechanism other than regulation of pel operon gene expression. To date, the only known means of nontranscriptional regulation of EPS production in pseudomonads is thought to be via the nucleotide signaling molecule c-di-GMP (10, 12, 17, 22, 44). However, there are no proteins with known c-di-GMP-related motifs in the CheIV cluster or in the CheI, CheII, or CheV cluster. The WspR protein, a component of the wsp chemosensory system (CheIII cluster) of P. aeruginosa, which plays a role in biofilm formation and EPS production, contains a GGDEF domain, an amino acid motif associated with the synthesis of c-di-GMP from GTP, and has been shown in vitro to catalyze c-di-GMP synthesis (12), but mutations in wspR do not yield SadB-like phenotypes. Furthermore, SadB lacks the EAL, GGDEF, and HD-GYP domains (39, 44) associated with c-di-GMP metabolism, and we have no biochemical evidence that SadB is involved in c-di-GMP metabolism (data not shown), nor does it appear to alter cellular pools of c-di-GMP (J. Hickman, J. Merritt, C. Harwood, and G. O'Toole, unpublished data). Therefore, the mechanism by which SadB and ChpB modulate Pel polysaccharide production remains to be elucidated.
Components of the CheIV cluster, including pilJ and the previously described chpA (49), also play a role in twitching motility, indicating that this putative chemosensory system participates in coordinating all three known surface behaviors of this microbe. We also showed that mutations in pilJ and chpB had no effect on swimming motility, further reinforcing a role for the CheIV cluster specifically in surface-associated behaviors of this microbe.
Our data suggest that SadB and PilJ modulate flagellar reversals under high-viscosity conditions but do not contribute to the control of flagellar reversals under the low-viscosity conditions that favor swimming. We hypothesize that SadB-dependent control of flagellar reversals upon polar, reversible attachment to a solid surface might decrease rotation about the cell pole and thus increase the time of interaction between the bacterium and its substratum, thereby promoting biofilm formation. In contrast, increased reversal rates appear to favor swarming motility. Wolfe and Berg postulated that for E. coli, increasing flagellar reversals might in some circumstances facilitate the ability to move through a semisolid matrix (50). Based on their microscopic observations, they concluded that “cells that do not tumble tend to get trapped in agar” and move less efficiently through this matrix (50). While that study was performed in the context of swimming through 0.3% agar, our data suggest that this phenomenon might also be extended to swarming conditions. Also consistent with our data is the finding that E. coli strains locked in the “tumbling” chemotaxis mode by mutation had a reduced ability to attach to an abiotic surface compared to the WT or mutants locked in the “running” mode (28). In addition to controlling flagellar function, by coordinating the production of the Pel polysaccharide, SadB can modulate another facet of biofilm initiation and swarming. Work presented here and recent published studies (24, 48) show that a functional pel locus contributes to early biofilm formation, and we show here that mutating the pel locus promotes swarming motility (Fig. 4D). This inverse relationship between polysaccharide production and motility has been noted in several other studies of P. aeruginosa (5, 9, 18, 43).
Our studies may provide a mechanistic basis for a recent exciting report by Shrout and colleagues (43). They proposed that early in biofilm formation, the extent of swarming motility helps dictate the final structure of the biofilm. That is, under conditions that promote swarming early in biofilm formation, the resulting mature biofilm is flat, while under conditions inhibitory to swarming motility, a biofilm with aggregates (distinct macrocolonies) will result. Based on their experimental work and accompanying mathematical simulations, they also postulated a role for a polysaccharide-containing matrix in the formation of the aggregates during biofilm development (43). One interpretation of the study of Shrout et al. is that P. aeruginosa must be able to integrate several important cell functions early in biofilm formation, namely, swarming motility and matrix production. The data presented in our report suggest that SadB and the CheIV cluster provide a molecular means for coregulating these functions.
We propose that P. aeruginosa inversely regulates the surface-associated behaviors of biofilm formation and swarming by controlling both flagellar reversals and the production of the Pel polysaccharide. Flagellar reversal rates in E. coli are largely regulated nontranscriptionally via the Che signal transduction pathway (25), and given the high sequence similarity of the cheIV cluster components to their E. coli counterparts, this is likely also the case in P. aeruginosa. CR binding and SEM data, together with the pelA and pelG transcriptional analysis presented here, indicate that production of the pel-encoded EPS may also be controlled by SadB and the CheIV cluster via a mechanism other than transcriptional control. Given that PilJ is also involved in twitching motility, the CheIV cluster may coordinate three different surface behaviors: swarming motility, twitching motility and biofilm formation. An appealing aspect of this regulatory strategy for coregulating surface behaviors is that in adapting to a surface-associated lifestyle, P. aeruginosa may be able to seamlessly and rapidly transition among its surface behaviors as it encounters ever-changing substratum properties and environmental conditions.
Acknowledgments
We thank C. Daghlian and the Ripple Electron Microscopy facility at Dartmouth College for assistance with the SEM studies.
This work was supported by NIH training grant T32 AI007519 (predoctoral fellowship) to N.C.C., predoctoral fellowship T32 GM08704 to J.H.M., and grant AI51360-01 to G.A.O.
Footnotes
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caiazza, N. C., and G. A. O'Toole. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 186:4476-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caiazza, N. C., R. M. Shanks, and G. A. O'Toole. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187:7351-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrandez, A., A. C. Hawkins, D. T. Summerfield, and C. S. Harwood. 2002. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J. Bacteriol. 184:4374-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 8.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett, E. S., D. Perlegas, and D. J. Wozniak. 1999. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J. Bacteriol. 181:7401-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gjermansen, M., P. Ragas, C. Sternberg, S. Molin, and T. Tolker-Nielsen. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7:894-906. [DOI] [PubMed] [Google Scholar]
- 11.Harshey, R. M. 1994. Bees aren't the only ones: swarming in gram-negative bacteria. Mol. Microbiol. 13:389-394. [DOI] [PubMed] [Google Scholar]
- 12.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 102:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 14.Ishimoto, K. S., and S. Lory. 1989. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc. Natl. Acad. Sci. USA 86:1954-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson, K. D., M. Starkey, S. Kremer, M. R. Parsek, and D. J. Wozniak. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadouri, D., and G. A. O'Toole. 2005. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol. 71:4044-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazmierczak, B. I., M. B. Lebron, and T. S. Murray. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 60:1026-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirisits, M. J., L. Prost, M. Starkey, and M. R. Parsek. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:4809-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klausen, M., M. Gjermansen, J. U. Kreft, and T. Tolker-Nielsen. 2006. Dynamics of development and dispersal in sessile microbial communities: examples from Pseudomonas aeruginosa and Pseudomonas putida model biofilms. FEMS Microbiol. Lett. 261:1-11. [DOI] [PubMed] [Google Scholar]
- 20.Kohler, T., L. K. Curty, F. Barja, C. van Delden, and J. C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuchma, S. L., J. P. Connolly, and G. A. O'Toole. 2005. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187:1441-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulasakara, H., V. Lee, A. Brencic, N. Liberati, J. Urbach, S. Miyata, D. G. Lee, A. N. Neely, M. Hyodo, Y. Hayakawa, F. M. Ausubel, and S. Lory. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. USA 103:2839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence, J. R., P. J. Delaquis, D. R. Korber, and D. E. Caldwell. 1987. Behavior of Pseudomonas fluorescens within the hydrodynamic boundary layers of surface microenvironments. Microb. Ecol. 14:1-14. [DOI] [PubMed] [Google Scholar]
- 24.Ma, L., K. D. Jackson, R. M. Landry, M. R. Parsek, and D. J. Wozniak. 2006. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188:8213-8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 26.Marshall, K. C., R. Stout, and R. Mitchell. 1971. Mechanism of the initial events in the sorption of marine bacteria to surfaces. J. Gen. Microbiol. 68:337-348. [Google Scholar]
- 27.Matsukawa, M., and E. P. Greenberg. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClaine, J. W., and R. M. Ford. 2002. Reversal of flagellar rotation is important in initial attachment of Escherichia coli to glass in a dynamic system with high- and low-ionic-strength buffers. Appl. Environ. Microbiol. 68:1280-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merritt, J. H., D. E. Kadouri, and G. A. O'Toole. 2005. Growing and analyzing static biofilms, p. 1B.1.1-1B.1.17. In R. Coico, T. Kowalik, J. Quarles, B. Stevenson, and R. Taylor (ed.), Current protocols in microbiology. J. Wiley & Sons, Hoboken, NJ. [DOI] [PMC free article] [PubMed]
- 30.Monds, R. D., P. D. Newell, J. A. Schwartzman, and G. A. O'Toole. 2006. Conservation of the Pho regulon in Pseudomonas fluorescens Pf0-1. Appl. Environ. Microbiol. 72:1910-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montrone, M., M. Eisenbach, D. Oesterhelt, and W. Marwan. 1998. Regulation of switching frequency and bias of the bacterial flagellar motor by CheY and fumarate. J. Bacteriol. 180:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 33.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 34.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to the study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 35.Pardee, A. B., F. Jacob, and J. Monod. 1959. The genetic control and cytoplasmic expression of “inducibility” in the synthesis of β-galactosidase in E. coli. J. Mol. Biol. 1:165-178. [Google Scholar]
- 36.Prasad, K., S. R. Caplan, and M. Eisenbach. 1998. Fumarate modulates bacterial flagellar rotation by lowering the free energy difference between the clockwise and counterclockwise states of the motor. J. Mol. Biol. 280:821-828. [DOI] [PubMed] [Google Scholar]
- 37.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 38.Ritchings, B. W., E. C. Almira, S. Lory, and R. Ramphal. 1995. Cloning and characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 63:4868-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 103:6712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 103:109-112. [DOI] [PubMed] [Google Scholar]
- 42.Shanks, R. M., N. C. Caiazza, S. M. Hinsa, C. M. Toutain, and G. A. O'Toole. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shrout, J. D., D. L. Chopp, C. L. Just, M. Hentzer, M. Givskov, and M. R. Parsek. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264-1277. [DOI] [PubMed] [Google Scholar]
- 44.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 45.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 46.Totten, P. A., J. C. Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toutain, C. M., M. E. Zegans, and G. A. O'Toole. 2005. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 187:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasseur, P., I. Vallet-Gely, C. Soscia, S. Genin, and A. Filloux. 2005. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151:985-997. [DOI] [PubMed] [Google Scholar]
- 49.Whitchurch, C. B., A. J. Leech, M. D. Young, D. Kennedy, J. L. Sargent, J. J. Bertrand, A. B. Semmler, A. S. Mellick, P. R. Martin, R. A. Alm, M. Hobbs, S. A. Beatson, B. Huang, L. Nguyen, J. C. Commolli, J. N. Engel, A. Darzins, and J. S. Mattick. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52:873-893. [DOI] [PubMed] [Google Scholar]
- 50.Wolfe, A. J., and H. C. Berg. 1989. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. USA 86:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zobell, C. E. 1943. The effects of solid surfaces upon bacterial activity. J. Bacteriol. 46:39-56. [DOI] [PMC free article] [PubMed] [Google Scholar]