Abstract
The antibiotic kirromycin inhibits prokaryotic protein synthesis by immobilizing elongation factor Tu (EF-Tu) on the elongating ribosome. Streptomyces ramocissimus, the producer of kirromycin, contains three tuf genes. While tuf1 and tuf2 encode kirromycin-sensitive EF-Tu species, the function of tuf3 is unknown. Here we demonstrate that EF-Tu3, in contrast to EF-Tu1 and EF-Tu2, is resistant to three classes of EF-Tu-targeted antibiotics: kirromycin, pulvomycin, and GE2270A. A mixture of EF-Tu1 and EF-Tu3 was sensitive to kirromycin and resistant to GE2270A, in agreement with the described modes of action of these antibiotics. Transcription of tuf3 was observed during exponential growth and ceased upon entry into stationary phase and therefore did not correlate with the appearance of kirromycin in stationary phase; thus, it is unlikely that EF-Tu3 functions as a resistant alternative for EF-Tu1. EF-Tu3 from Streptomyces coelicolor A3(2) was also resistant to kirromycin and GE2270A, suggesting that multiple antibiotic resistance is an intrinsic feature of EF-Tu3 species. The GE2270A-resistant character of EF-Tu3 demonstrated that this divergent elongation factor is capable of substituting for EF-Tu1 in vivo.
Elongation factor Tu (EF-Tu) has a well-characterized role in bacterial protein synthesis by mediating the transport of aminoacyl-tRNA (aa-tRNA) to the ribosome. During this process, EF-Tu interacts sequentially with GTP, aa-tRNA, the ribosome, GDP, and EF-Ts (reviewed in references 18 and 29). Crystallographic structures exist for both nucleotide-bound states of EF-Tu (2, 4, 17, 31), for EF-Tu-GTP-aa-tRNA (24), and EF-Tu-EF-Ts (16). In addition, the binding site of immobilized EF-Tu-aa-tRNA on the ribosome has been visualized by cryoelectron microscopy (37). While most protein synthesis inhibitors are directed towards the ribosome, four types of antibiotics have EF-Tu as their target (reviewed in references 13 and 29). The structurally unrelated antibiotics pulvomycin and GE2270A both inhibit protein synthesis by impairing ternary complex formation between EF-Tu-GTP and aa-tRNA (3, 47) and thus interrupt the elongation cycle prior to interaction with the ribosome. A mixed population of resistant and sensitive EF-Tu species is therefore able to sustain polypeptide chain elongation in the presence of one of these antibiotics (22; unpublished results). Binding of either kirromycin or enacyloxin IIa to EF-Tu still allows the factor to interact sequentially with aa-tRNA and the ribosomal A site. However, after GTP hydrolysis, EF-Tu is no longer ejected from the ribosome, thus obstructing not only this ribosome but also all other ribosomes upstream on the same mRNA transcript. This phenomenon explains the recessive character of kirromycin- or enacyloxin IIa-resistant EF-Tu mutant proteins in a mixture with sensitive EF-Tu species (30, 51). Resistance to the four types of EF-Tu-targeted antibiotics is usually achieved by single amino acid substitutions at highly conserved positions of EF-Tu (Table 1).
TABLE 1.
Mutations in EF-Tu causing resistance to kirromycin, enacyloxin IIa, pulvomycin, or GE2270A (residue numbering according to E. coli EF-Tu)
| Antibiotic | Position | Amino acid change | Source of mutant EF-Tu (reference) |
|---|---|---|---|
| Kirromycin | 120 | Leu→Gln | Salmonella enterica serovar Typhimurium (1) |
| 124 | Gln→Arg, Glu | Salmonella enterica serovar Typhimurium (1) | |
| Gln→Lys | E. coli (21) | ||
| 160 | Tyr→Asn, Asp, Cys | Salmonella enterica serovar Typhimurium (1) | |
| 298 | Ile→ΔIle | Salmonella enterica serovar Typhimurium (1) | |
| 316 | Gly→Asp | Salmonella enterica serovar Typhimurium (1), E. coli (21) | |
| 329 | Gln→His | Salmonella enterica serovar Typhimurium (1) | |
| 375 | Ala→Ser, Thr, Val | Salmonella enterica serovar Typhimurium (1) | |
| Ala→Thr, Val | E. coli (21) | ||
| 378 | Glu→Lys | E. coli (21) | |
| 382 | Thr→Ser | E. coli (T. Plath, personal communication) | |
| Enacyloxin IIa | 124 | Gln→Lys | E. coli (51) |
| 316 | Gly→Asp | E. coli (51) | |
| 375 | Ala→Thr | E. coli (51) | |
| Pulvomycin | 230 | Arg→Cys | E. coli (50) |
| 233 | Arg→Ser | E. coli (5) | |
| 230/233 | Arg/Arg→Val/Phe | E. coli (5) | |
| 333 | Arg→Cys | E. coli (50) | |
| 334 | Thr→Ala | E. coli (50) | |
| GE2270A | 226 | Val→Ala | Bacillus subtilis (35) |
| 257 | Gly→Ser | E. coli (52) | |
| 275 | Gly→Ser | Bacillus subtilis (36) | |
| Gly→Ala | E. coli (52) |
The producers of the kirromycin class of antibiotics belong to the streptomycetes, a group of gram-positive, soil-dwelling, filamentous bacteria that exhibit complex multicellular development and morphological differentiation; branching, filamentous vegetative mycelia give rise to aerial hyphae bearing long chains of spores (6). The onset of morphological differentiation usually coincides with the production of antibiotics, mediated by complex “secondary metabolic” pathways. In liquid-grown cultures, antibiotic production is generally confined to stationary phase (15). One of the mechanisms used by streptomycetes to protect themselves against the toxic action of their own products is by duplication of the target protein or RNA so that both sensitive and resistant isoforms are produced, the latter usually at the onset of antibiotic production (9, 39).
The kirromycin producer Streptomyces ramocissimus is unique in containing three different tuf genes, designated tuf1, tuf2, and tuf3, which code for EF-Tus that are strikingly heterogeneous (44). The highly transcribed tuf1 gene encodes the constitutively expressed, kirromycin-sensitive EF-Tu1. Recently, we demonstrated that EF-Tu2 is indistinguishable from EF-Tu1 in its ability to promote protein biosynthesis in vitro and in its sensitivity to kirromycin (26). The physiological function of the divergent tuf3 gene product, which shares only about 65% amino acid identity with either EF-Tu1 or EF-Tu2, is unknown. In the genetically well-characterized Streptomyces coelicolor A3(2) two tuf genes have been identified (42), designated tuf1 and tuf3 by analogy to their homologues in S. ramocissimus. Studies revealed that tuf3 disruption did not noticeably affect growth or differentiation and that tuf3 transcription could be induced by amino acid starvation (43). Evidence that S. coelicolor EF-Tu3 is a functional, but less efficient, elongation factor was obtained in a Streptomyces in vitro translation system (25).
In this report we tackle the long-standing and obvious question of whether S. ramocissimus EF-Tu3 might be a kirromycin-resistant EF-Tu isoform and thus be involved in the self-protection mechanism of S. ramocissimus. Furthermore, we present evidence that this divergent EF-Tu3, despite being an outsider in the family of EF-Tu proteins, can substitute for EF-Tu1 in vivo.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and vectors.
Escherichia coli JM101 (32) and ET12567 (20), grown and transformed by standard procedures (32), were used for routine subcloning. All DNA manipulations were performed by following standard protocols (27).
S. ramocissimus B7 was obtained from Gist-brocades NV (Delft, The Netherlands) and was grown as liquid culture in S medium for the isolation of EF-Tu1. SFM medium (containing per liter 20 g mannitol, 20 g soy flour, and 20 g agar, dissolved in tap water and autoclaved twice) is a modified version of that reported by Hobbs et al. (12) and was used to make high-titer spore suspensions of S. ramocissimus B7. Conditions for reproducible dispersed growth of S. ramocissimus B7 in NMMP medium (14) containing 1% (wt/vol) glucose were as described elsewhere (40). S. ramocissimus B7 spores were plated on cellophane disks on AMMAT medium (40) to facilitate harvesting of the mycelium for RNA isolation. Morphology of the surface-grown cultures was determined by phase-contrast microscopy, while kirromycin secretion into the agar was detected by using E. coli JM101 as the indicator strain.
S. coelicolor M145 (14) was obtained from the John Innes Centre, Norwich, United Kingdom; the construction of the S. coelicolor J1501 derivatives J1501ΔglkAΔtuf3 and LT2 has been described by Van Wezel et al. (43) and Olsthoorn-Tieleman et al. (25), respectively. S. coelicolor strains were grown in YEME medium and on R5 plates, when necessary supplemented with 1% (wt/vol) mannitol, 7.5 μg/ml uracil, and 50 μg/ml histidine, as described previously (14). MSP (2% [wt/vol] mannitol, 2% [wt/vol] soy peptone) was used to grow S. coelicolor LT2 for in vitro translation experiments. Protoplast preparation and transformation were performed as described by Hopwood et al. (14).
Vectors are described in Table 2.
TABLE 2.
Plasmids and constructs
| Plasmid | Description | Reference |
|---|---|---|
| pUC18 | E. coli multicopy plasmid | 49 |
| pUSRT3-1 | pUC18 with a 3.9-kb SacI/PstI fragment harboring tuf3 | 44 |
| pUSRT3-3 | pUC18 with 2.8-kb fragment harboring tuf3, preceded by an introduced EcoRI site and an 0.2-kb tuf1 upstream coding sequence | 44 |
| pUSRT3-6 | pUC18 with 2.8-kb EcoRI/PstI fragment harboring tuf3, lacking any upstream coding sequence | This work |
| pIJ4070 | pUC18 with 0.3-kb fragment harboring the ermE promoter region | M. J. Bibb, unpublished data |
| pUSRT3-7 | pUC18 with 2.8-kb EcoRI/PstI fragment harboring tuf3, preceded by an 0.3-kb fragment harboring the ermE promoter region | This work |
| pIJ487 | Streptomyces multicopy plasmid | 45 |
| pIJ4083 | pIJ487 with DNA fragment harboring Pseudomonas putida xylE, lacking any upstream coding sequence | 7 |
| pISRT3xylE-1 | pIJ4083 with DNA fragment harboring the tuf3 upstream region | This work |
| pISRT3-1 | pIJ487 with 3.1-kb KpnI/PstI fragment from pUSRT3-7, harboring tuf3, preceded by an 0.3-kb fragment harboring the ermE promoter region | This work |
Promoter probing experiments.
pISRT3xylE-1 was constructed by cloning the 0.8-kb SacI/FspI fragment of tuf3-containing plasmid pUSRT3-1 (44) via pUC18 into xylE-based promoter probe vector pIJ4083 (7). Transformants containing pISRT3xylE-1 were grown on R5 in the presence of 5 μg/ml thiostrepton (a gift from Squibb, Princeton, NJ). Plates were sprayed with 0.5 M catechol after 2 to 5 days of growth, and the amount of catechol converted into yellow 2-hydroxymuconic semialdehyde by catechol 2,3-dioxygenase was assessed visually.
Nuclease S1 protection assays.
RNA was isolated from liquid- and surface-grown S. ramocissimus B7 cultures as described by Hopwood et al. (14), except that DNase I treatment was used in addition to salt precipitation to eliminate DNA from the nucleic acid preparations. RNA concentrations were determined spectrophotometrically, and the quality of the preparations was checked by gel electrophoresis. Hybridization of 30 μg RNA with the tuf3 probe was performed in sodium trichloroacetate-based buffer (23) at 45°C overnight after denaturation at 70°C for 15 min. All subsequent steps were carried out as described previously (26) with an excess of probe. The 950-bp PvuII/StyI fragment from pUSRT3-1, 32P end labeled at the 5′ end of the StyI site, was used for mapping tuf3 transcripts. The tuf1 and tuf2 genes have no homology with this probe, thus excluding the possibility that these mRNAs would contribute to the protection pattern. Products were analyzed on denaturing 6% polyacrylamide gels, using 32P-end-labeled HpaII fragments of pBR322 as size markers.
Construction of tuf3 overexpression vector pISRT3-1.
The S. ramocissimus tuf3 gene was isolated from plasmid pUSRT3-3 (44) as a 2.8-kb EcoRI/PstI fragment and cloned into the HincII/PstI sites of pUC18, yielding pUSRT3-6. The 0.3-kb KpnI/BamHI fragment from pIJ4070 was then cloned into the corresponding restriction sites of pUSRT3-6, resulting in pUSRT3-7. In this way tuf3 was placed under the control of the strong and constitutive Streptomyces ermE promoter. Finally pISRT3-1 was constructed by inserting the tuf3 gene and upstream ermE promoter as a 3.1-kb KpnI/PstI fragment into the KpnI/PstI sites of Streptomyces high-copy-number vector pIJ487 (46).
Production and purification of S. ramocissimus EF-Tu1 and EF-Tu3.
Exponentially growing S. ramocissimus B7, cultured in S medium, was used as a source for EF-Tu1. EF-Tu3 overproduction was achieved by inoculation of fresh spores from S. coelicolor LT2 harboring pISRT3-1 into YEME containing 5 μg/ml thiostrepton and growth for 40 h at 30°C. The mycelium was washed twice with ice-cold TuGly buffer (50 mM Tris-HCl, pH 7.6, 7 mM MgCl2, 60 mM NH4Cl, 1 mM dithiothreitol, 10 μM GDP, 10 μM phenylmethylsulfonyl fluoride, and 10% [vol/vol] glycerol) and kept frozen at −80°C. For the purification of S. ramocissimus EF-Tu1 and EF-Tu3, the method described by Olsthoorn-Tieleman et al. (26) for S. ramocissimus EF-Tu2 was used. The protein solutions were concentrated over Amicon Centriflo ultrafiltration cones and stored at −80°C. Protein concentrations were determined with Coomassie protein assay reagent (Pierce) by using bovine serum albumin as a standard.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis.
Protein expression and purification were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using the Mini Protean II system (Bio-Rad), and Western blotting was conducted as described in reference 44, using a 1:5,000 dilution of antibodies. Nonradioactive detection was performed using Western blot chemiluminescence reagent (NEN Life Science Products). The rabbit polyclonal antibodies used were raised against the S. ramocissimus tuf3 gene product expressed in E. coli (44).
In vitro translation assays in the absence and presence of antibiotics.
S30 cell extracts of S. coelicolor LT2 (if necessary harboring pISRT3-1 or pIJ487), from which His6-tagged EF-Tu1 was removed by treatment with Ni2+-nitrilotriacetic acid (NTA)-agarose beads, were obtained as described previously (25). The extracts, supplemented with purified EF-Tu species and antibiotics at various concentrations, were incubated in translation buffer {final concentrations, 50 mM Tris-HCl [pH 7.6], 9 mM MgCl2, 60 mM NH4Cl, 1 mM dithiothreitol, 1 mM ATP, 1 mM GTP, 6 mM phosphoenolpyruvate, 50 μg/ml pyruvate kinase, 0.1 mg/ml poly(U), 0.2 mg/ml E. coli tRNA, and 13.2 μM [14C]Phe (specific activity, 531 mCi/mmol) } at 30°C for 10 min. The total volume was 50 μl. The reaction was stopped by the addition of 15 μl of 1 M NaOH and further incubation at 30°C for 10 min. After precipitation with 5% (wt/vol) trichloroacetic acid and filtration, the incorporation of [14C]Phe was determined by liquid scintillation counting.
The antibiotics kirromycin and pulvomycin were generous gifts from Gist-brocades NV (Delft, The Netherlands) and A. Parmeggiani (Palaiseau, France), respectively. GE2270A was isolated by V. G. Möhrle (Leiden University, Leiden, The Netherlands) as described previously (33).
RESULTS
Growth phase-dependent transcription of tuf3.
The apparent absence of detectable tuf3 expression in total protein extracts of S. ramocissimus (44) suggests that tuf3 is expressed at very low levels under normal growth conditions or that tuf3 might be a silent gene. To distinguish between these two possibilities, we determined whether the tuf3 upstream region possessed promoter activity and if tuf3 transcripts could be observed using the sensitive S1 nuclease protection assay. To determine the presence and approximate location of the tuf3 promoter(s), S. coelicolor M145 was transformed with pISRT3xylE-1, which contains the 0.8-kb SacI/FspI fragment (−757/+21 relative to the start site of tuf3; Fig. 1) of pUSRT3-1 cloned into the promoter-probe vector pIJ4083 (7). Transformants containing pISRT3xylE-1 yielded colonies with bright-yellow aerial hyphae when sprayed with catechol after 3 days of growth on R5 agar plates, indicative of at least one promoter in the 0.8-kb tuf3 upstream region.
FIG. 1.
Restriction map of the S. ramocissimus tuf3 region. Only the relevant SacII restriction site is shown (#). The probe used for S1 nuclease mapping of tuf3 transcripts (the asterisk indicates the 32P-labeled 5′ end) is shown above the restriction map. Dashed line, nonhomologous pUC18-derived extension.
To establish the level and timing of tuf3 transcription in vivo, RNA was isolated from mycelium harvested from S. ramocissimus B7 cultures grown in NMMP with 1% (wt/vol) glucose. RNA samples were analyzed by high-resolution nuclease S1 mapping. The 950-bp PvuII/StyI fragment from pUSRT3-1, uniquely 32P labeled at the 5′ end of the StyI site, was used as a probe (Fig. 1); the nonhomologous pUC18-derived extension allowed discrimination between full-length RNA-protected fragments and reannealed probe. A band corresponding to tuf3 transcripts was detected in RNA isolated during early exponential growth but was absent during kirromycin production (Fig. 2a and b). The tuf3 transcript corresponds to a transcription start site located approximately 190 nucleotides (nt) upstream of the tuf3 translation start site, similar to the situation for S. coelicolor tuf3 (43). The putative transcription start site is preceded by the sequence 5′ TCGACG-17 nt-AATGAT 3′, which shows similarity to the consensus sequence for the major class of Streptomyces promoters (38) and is almost identical to the proposed promoter region for S. coelicolor tuf3 (43). To confirm that the RNA-protected fragment assigned to tuf3p represented a transcription start site, the 340-bp SacII/StyI fragment of pUSRT3-1 was used as a template for in vitro transcription assays using partially purified RNA polymerase isolated from S. coelicolor M145. The runoff transcript of approximately 295 nt corresponded to the expected size for tuf3p predicted from the S1 nuclease protection assays (data not shown).
FIG. 2.
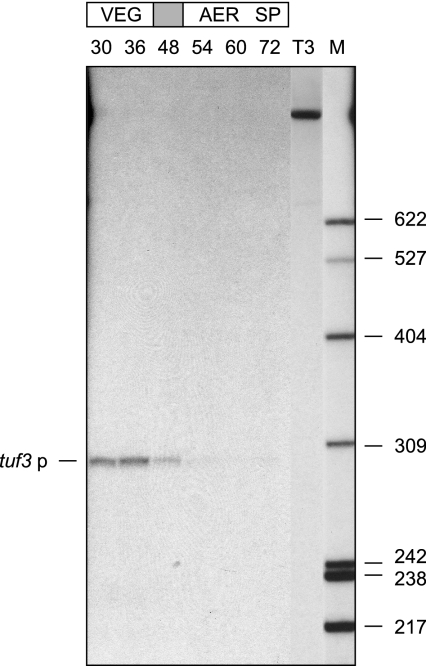
(a) Growth curve of S. ramocissimus B7. Arrowheads indicate the time points at which samples were taken for RNA isolation and kirromycin quantitation. The shaded box labeled kirromycin denotes the presence of the antibiotic in the filtrate. The doubling time during exponential growth was 2.5 h. (b) S1 nuclease protection analysis of the tuf3 transcripts in RNA isolated at the time points indicated in panel a (30 μg of RNA per sample). EXP and STAT indicate exponential and stationary growth phases, respectively, and the shaded box indicates the transition phase. tuf3p, a transcript initiated at tuf3p. Lane T3 shows the location of the 950-nt full-length tuf3 probe. Lane M contains end-labeled HpaII-digested pBR322 size markers (in nucleotides).
Transcription of tuf3 was also monitored during growth of S. ramocissimus on solid medium (Fig. 3). As expected, a signal was detected in RNA isolated during the formation of vegetative hyphae, with transcription initiating from the same start site as that found for the liquid culture. A drop in the amount of tuf3 transcripts occurred in the transition phase from vegetative growth to aerial mycelium development, coinciding with the production of kirromycin, followed by complete absence of tuf3 transcripts during the stage of aerial mycelium formation. At the later stages of spore development, transcripts corresponding to tuf3p reappeared again, as they do for tuf1 (40) and tuf2 (26). Since no temporal correlation was observed between tuf3 transcription and kirromycin production in either liquid- or surface-grown cultures, it seemed unlikely that S. ramocissimus EF-Tu3 was involved in conferring resistance to kirromycin.
FIG. 3.
S1 nuclease protection analysis of the tuf3 transcripts in RNA isolated from surface-grown cultures at the time points (h) indicated, using 40 μg of RNA per sample. VEG, AER, and SP indicate the appearance of vegetative mycelium, aerial mycelium, and spores, respectively, and the shaded area corresponds to the transition phase. tuf3p indicates a transcript initiated at tuf3p; lanes T3 and M are as in Fig. 2b.
S. ramocissimus tuf3 encodes a functional, but less efficient, EF-Tu.
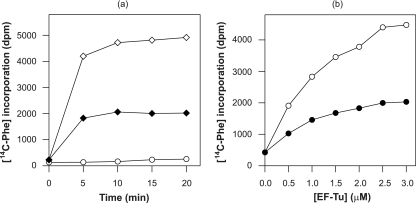
Previously we demonstrated that S. coelicolor EF-Tu3 is able to stimulate protein synthesis in vitro, although not as efficiently as S. coelicolor EF-Tu1 (25). On the basis of the homology between the two EF-Tu3 proteins (91% amino acid identity), we expected that this would also be the case for the S. ramocissimus tuf3 gene product. To verify this assumption, in vitro translation experiments were performed with complete cell S30 extracts from S. coelicolor LT2 harboring the S. ramocissimus tuf3 expression vector pISRT3-1. S. coelicolor LT2 is a J1501 derivative modified in both tuf genes; tuf3 is disrupted and tuf1 is replaced by tuf1His, encoding His6-tagged EF-Tu1 (25). After removal of EF-Tu1His by selective adsorption to Ni2+-NTA-agarose beads, residual translational activity was observed in the resulting extract, containing plasmid-borne S. ramocissimus EF-Tu3 as the only possible EF-Tu (Fig. 4a). A control experiment was performed with a Ni2+-NTA-treated extract of LT2 harboring pIJ487 as the parental vector without tuf3, and virtually no activity was detected.
FIG. 4.
Translational activity of S. ramocissimus EF-Tu3. (a) Translational activity of cell extracts of S. coelicolor LT2 harboring expression vector pISRT3-1 with (⋄) and after removal of (♦) endogenous EF-Tu1His. A Ni2+-NTA-treated cell extract of S. coelicolor LT2 harboring pIJ487, the parental vector without tuf3, was used as a control (○). (b) In vitro translation of an EF-Tu-depleted S. coelicolor LT2 cell extract supplemented with S. ramocissimus EF-Tu3 (•) and S. ramocissimus EF-Tu1 (○). The translation of the poly(U) messenger was studied by measuring the incorporation of [14C]Phe at 30°C as a function of time (a) and of the concentrations of the EF-Tu species during a 10-min incubation (b).
To investigate its translational capacity in more detail, S. ramocissimus EF-Tu3 was purified from S. coelicolor LT2 harboring pISRT3-1. EF-Tu1 and EF-Tu3 were used to complement an EF-Tu-dependent Streptomyces in vitro translation system (25), and their translational capacities were studied as a function of the EF-Tu concentration. The purified EF-Tu3 clearly participated in poly(Phe) synthesis (Fig. 4b), but the extent of [14C]Phe incorporation was significantly lower than that of EF-Tu1 at all concentrations, including near-saturation levels, thus paralleling the observations made previously in S. coelicolor.
S. ramocissimus EF-Tu3 is resistant to three EF-Tu-specific antibiotics.
The presence of three tuf genes in the kirromycin producer S. ramocissimus, each encoding a functional EF-Tu, suggested that one of these EF-Tu species might be involved in conferring kirromycin resistance. Previous studies with S. ramocissimus EF-Tu1 and EF-Tu2 revealed that the two proteins are equally sensitive to kirromycin (26). Thus, we investigated whether S. ramocissimus EF-Tu3 was able to stimulate protein synthesis in the presence of kirromycin. Given that EF-Tu3 contains a large number of amino acid substitutions at highly conserved positions (25), we also assessed the resistance of S. ramocissimus EF-Tu3 to the EF-Tu inhibitors pulvomycin and GE2270A.
As monitored using the Streptomyces in vitro translation system, EF-Tu3 was clearly resistant to kirromycin, retaining 25% of activity at concentrations as high as 100 μM (Fig. 5a). In agreement with previous results (26), S. ramocissimus EF-Tu1 was fully inhibited by a kirromycin concentration more than 100-fold lower, with a concentration at which 50% inhibition is observed (IC50) of about 0.04 μM. In similar experiments with pulvomycin EF-Tu3 was about five times more resistant to this antibiotic than EF-Tu1, with IC50 values of about 3.5 μM and 0.7 μM for EF-Tu3 and EF-Tu1, respectively (Fig. 5b). In addition, we found that EF-Tu3 was insensitive to the thiazolyl peptide antibiotic GE2270A at concentrations as high as 41 μM, while EF-Tu1 has an IC50 value of about 0.07 μM (Fig. 5c). Summarizing, in contrast to the antibiotic-sensitive EF-Tu1, EF-Tu3 is highly resistant to kirromycin, completely resistant to GE2270A, and somewhat resistant to pulvomycin.
FIG. 5.
Translational activities of S. ramocissimus EF-Tu1 and EF-Tu3 in the presence of antibiotics. In vitro translation of an EF-Tu-depleted S. coelicolor cell S30 extract supplemented with 0.5 μM S. ramocissimus EF-Tu1 (○) or S. ramocissimus EF-Tu3 (•). The translation of the poly(U) messenger was studied by measuring the incorporation of [14C]Phe at 30°C as a function of the concentrations of kirromycin (a), pulvomycin (b), and GE2270A (c). For both EF-Tu1 and EF-Tu3, activity in the absence of the antibiotics was normalized to 100% (2,700 dpm for EF-Tu1 and 1,800 dpm for EF-Tu3).
The kirromycin resistance of S. ramocissimus EF-Tu3 is recessive to sensitive EF-Tu1.
The observation that EF-Tu3 is a kirromycin-resistant elongation factor raised the possibility that this protein was responsible for catalyzing protein biosynthesis in S. ramocissimus during kirromycin production. Since sensitive EF-Tu1 is present throughout the entire growth cycle (40, 44), we examined the effect of kirromycin on poly(Phe) synthesis mediated by a mixed population of purified EF-Tu1 and EF-Tu3. The combination of the two EF-Tu species was as sensitive to 10 μM kirromycin as EF-Tu1 was (Fig. 6), demonstrating that the sensitive fraction of the mixed population is dominant over the kirromycin-resistant EF-Tu3. As a control, a similar experiment was performed using GE2270A. In the presence of 4.1 μM GE2270A, the mixed population displayed the same activity in poly(Phe) synthesis as EF-Tu3 alone did, whereas the sensitive EF-Tu1 was totally inhibited. Thus, these experiments predict that during kirromycin production by S. ramocissimus the kirromycin-resistant character of EF-Tu3 is likely be suppressed by the continuing presence of the sensitive EF-Tu1. However, it should be noted that these experiments were carried out using S. coelicolor EF-Tu-depleted extracts, and it is conceivable that S. ramocissimus ribosomes possess a marked preference for EF-Tu3 over EF-Tu1, thus allowing phenotypic expression of the apparently recessive kirromycin resistance.
FIG. 6.
Translational activity of a mixture of S. ramocissimus EF-Tu1 and EF-Tu3 in the presence of the antibiotics kirromycin and GE2270A. In vitro translation of an EF-Tu-depleted S. coelicolor LT2 cell S30 extract supplemented with 0.5 μM EF-Tu3 (black bars), with 0.5 μM EF-Tu1 (white bars), or with a mixture of 0.5 μM EF-Tu3 and 0.5 μM EF-Tu1 (hatched bars). Poly(U) translation was studied by measuring the incorporation of [14C]Phe in the presence of 10 μM kirromycin (K) or 2 μM GE2270A (G). Activity of the EF-Tu mixture in the absence of antibiotics was taken as 100%.
EF-Tu3 from S. coelicolor is also resistant to kirromycin and GE2270A.
Since the EF-Tu3 proteins from S. ramocissimus and S. coelicolor contain similar deviations from the common eubacterial EF-Tu sequence (25), we studied the antibiotic resistance profile of S. coelicolor EF-Tu3 as well. In vitro translation experiments were performed with complete cell S30 extracts from S. coelicolor LT2 harboring the S. coelicolor tuf3 expression vector pISCT3-1. As illustrated in Fig. 7, 30% of the original translational activity was observed in the presence of 4.1 μM GE2270A, whereas in the presence of 10 μM kirromycin the activity dropped to background levels. After removal of EF-Tu1His from the extract, again about 30% residual activity was observed, attributable to the presence of overexpressed EF-Tu3, and no drop in activity was measured after the addition of either kirromycin or GE2270A. A control experiment was performed with an extract of LT2 containing pIJ487 as the parental vector without tuf3, and virtually no activity was measured in the presence of either kirromycin or GE2270A, demonstrating that S. coelicolor EF-Tu1His is fully inhibited at the antibiotic concentrations used. These experiments reveal that S. coelicolor EF-Tu3, like its counterpart in S. ramocissimus, possesses high-level resistance to kirromycin and GE2270A. Similar tests with pulvomycin (data not shown) were ambiguous, probably reflecting interference with EF-Tu3 function of the relatively high concentrations of pulvomycin required to inhibit EF-Tu1: S. coelicolor EF-Tu3 is either sensitive to pulvomycin or, like S. ramocissimus EF-Tu3, moderately resistant.
FIG. 7.
Translational activity of cell extracts of S. coelicolor LT2 harboring the S. coelicolor tuf3 expression vector pISCT3-1 with (hatched bars) and after removal of (black bars) endogenous EF-Tu1His by Ni2+-NTA treatment (NTA). A cell extract of S. coelicolor LT2 containing pIJ487, the parental vector without tuf3, was used as a control (white bars). Poly(U) translation was studied by measuring the incorporation of [14C]Phe in the presence of 10 μM kirromycin (K) or 2 μM GE2270A (G). Activity of the cell extract in the absence of antibiotics was taken as 100%.
We tentatively conclude that the multiple antibiotic resistance profile of S. ramocissimus EF-Tu3 is a general feature of this class of variant EF-Tu proteins and that it is not functionally related to kirromycin production by this strain.
The variant EF-Tu3 can substitute for the regular EF-Tu1 in vivo.
The above results indicate that the divergent EF-Tu3 is able to sustain the whole elongation process in vitro in a poly(U)-dependent poly(Phe) system. To demonstrate that the protein is also active in vivo, we used our newfound knowledge that EF-Tu3 is insensitive to the antibiotic GE2270A and that this resistance is dominant in the presence of the sensitive EF-Tu1. Protoplasts of the GE2270A-sensitive strain S. coelicolor J1501ΔglkAΔtuf3 harboring pISRT3-1, pISCT3-1 (43), or pIJ487 were tested for their GE2270A resistance by plating them on R5 supplemented with uracil, histidine, 1% (wt/vol) mannitol, 25 μg/ml thiostrepton, and 200 μg/ml crude GE2270A (32 μM pure GE2270A). Successful transformation of the three different vectors was confirmed by plating the protoplasts on the same medium containing thiostrepton but lacking GE2270A. Both EF-Tu3-overproducing strains were able to grow in the presence of GE2270A, while transformants harboring the parental vector pIJ487 were not. Thus, overproduction of the plasmid-borne EF-Tu3 in S. coelicolor J1501ΔglkAΔtuf3 results in a GE2270A-resistant phenotype. Since the antibiotic is known to inhibit only the sensitive EF-Tu (in this case chromosomally encoded EF-Tu1) in a mixed population (22), the change in phenotype of the transformants from GE2270A sensitive to resistant indicates that EF-Tu3 also functions as an elongation factor in vivo.
DISCUSSION
The presence of three tuf genes in S. ramocissimus, producer of the EF-Tu-specific inhibitor kirromycin, has been intriguing ever since their discovery in the early 1990s. In this paper we focused on the role of tuf3 in kirromycin resistance. Self-protection of antibiotic producers by duplication of genes encoding target proteins, one producing a sensitive and the other producing a resistant isoform, the latter usually at the onset of antibiotic production, is a mechanism used by other streptomycetes, including Streptomyces sphaeroides and Streptomyces arenae, the producers of novobiocin and pentalenolactone, respectively (9, 39). In the case of kirromycin, such a mechanism would be complicated by the recessive character of kirromycin resistance in a mixed population of resistant and sensitive EF-Tu species, although Tubulekas et al. (41) reported that certain error-restrictive streptomycin-resistant ribosomes (mutated in ribosomal protein S12) could overcome their expected recessive phenotype. Remarkably, the same type of S12 mutations activated actinorhodin production in Streptomyces lividans, demonstrating that the onset and extent of secondary metabolism in Streptomyces spp. are significantly influenced by the translational machinery (34).
In this work we demonstrated that transcription of S. ramocissimus tuf3 shows a growth phase dependence similar to that of S. ramocissimus tuf1 (40), although tuf1 and tuf3 transcript levels differ by at least several orders of magnitude during growth. tuf3 is transcribed neither shortly before nor during kirromycin production and does not respond to kirromycin induction. These results argue against a role for the EF-Tu3 isoform in the kirromycin resistance in S. ramocissimus.
Purified EF-Tu3 was able to sustain poly(Phe) synthesis in a Streptomyces in vitro translation system, albeit with lower efficiency than EF-Tu1, possibly reflecting suboptimal interactions with aa-tRNA, EF-Ts, or the ribosome. Intriguingly, we observed that EF-Tu3 displays remarkably high resistance to kirromycin. In agreement with the described mode of action of this antibiotic, the resistance of EF-Tu3 is suppressed by the simultaneous presence of the kirromycin-sensitive EF-Tu1. Since high amounts of EF-Tu1 are present in S. ramocissimus throughout the entire growth cycle (40, 44), this observed dominance of sensitivity again argues against a role for EF-Tu3 as the kirromycin resistance determinant in vivo. However, it should be noted that the in vitro translation experiments were carried out using S. coelicolor EF-Tu-depleted extracts. Therefore, we cannot rule out the possibility that (transiently altered) S. ramocissimus ribosomes might favor the resistant EF-Tu3 for translation, thus allowing phenotypic expression of the apparently recessive kirromycin resistance, analogous to the situation reported for certain error-restrictive streptomycin-resistant ribosomes and a mixture of kirromycin-sensitive and -resistant EF-Tu species in E. coli (41).
The comparable genetic organization of S. coelicolor and S. ramocissimus tuf3 also indicates that the latter is not linked to the kirromycin biosynthesis cluster and that it very likely fulfills the same function as its homologue in S. coelicolor. The observation that S. coelicolor EF-Tu3 is highly resistant to kirromycin also suggests that this might be a general feature of EF-Tu3 proteins, previously shown to be the most divergent subset of eubacterial EF-Tu species (25).
Organisms that synthesize antibiotics to which they are potentially sensitive must have a resistance mechanism to protect themselves, at least during the phase of antibiotic biosynthesis. Since self-protection by expression of the resistant isoform EF-Tu3 is hampered by the presence of the sensitive EF-Tu1, it seems very likely that S. ramocissimus possesses other resistance mechanisms such as intracellular inactivation of the antibiotic or limitation of intracellular drug concentration to subinhibitory levels via efflux and/or exclusion. The kirromycin biosynthetic cluster from Streptomyces collinus has been cloned and sequenced (T. Weber and W. Wohlleben, unpublished results). None of the tuf genes is located near the cluster. Interestingly, while the exact resistance mechanism is yet unknown, heterologous expression of (part of) the cluster in Streptomyces results in enhanced kirromycin resistance, suggesting that resistance genes are located within the kirromycin biosynthetic cluster (W. Wohlleben, personal communication).
In addition to their kirromycin-resistant character, S. coelicolor and S. ramocissimus EF-Tu3 displayed complete resistance to GE2270A, and EF-Tu3 from S. ramocissimus also exhibited moderate resistance to pulvomycin. Dual resistance to the EF-Tu inhibitors kirromycin and pulvomycin was known for the wild-type EF-Tu proteins of Staphylococcus aureus (10), Bacillus stearothermophilus (19), and some lactobacilli (48), while simultaneous resistance to GE2270A and kirromycin was observed for Planobispora rosea EF-Tu1 (22). However, S. ramocissimus EF-Tu3 is the first example of a eubacterial EF-Tu with resistance to all three antibiotics, each with different binding sites on EF-Tu, reflecting the structural (and possibly functional) divergence of EF-Tu3 from the major class of EF-Tu proteins. This multiple antibiotic resistance phenotype must reflect some of the many deviations of EF-Tu3 from the common eubacterial EF-Tu amino sequence (25). Mutations leading to kirromycin resistance cluster at the interface of domains 1 and 3 of EF-Tu-GTP (1, 21), and crystallographic data for the EF-Tu-kirromycin complex reveal that the antibiotic indeed binds to this region (45). The kirromycin resistance of EF-Tu3 could be explained by the replacement of a conserved Tyr residue at position 160 with His. This Y160H substitution would abolish the strong hydrogen bond between the pyridone ring of kirromycin and the tyrosine side chain of EF-Tu, as well as disrupting hydrophobic interactions with the aromatic ring of Tyr and the linker between the tetrahydrofuran and pyridone rings. However, P. rosea EF-Tu1, which contains the Y160Q substitution, is only about 1 order of magnitude more resistant to kirromycin than S. coelicolor EF-Tu1, whereas we noted a difference of 2 orders of magnitude (Fig. 2b). The substitutions I92V and R373G are part of the kirromycin binding pocket (Fig. 8a) and might also contribute to the kirromycin-resistant character of EF-Tu3. The binding site of enacyloxin IIa partially overlaps with the kirromycin binding site (28). The strong salt bridge between Lys313 and the antibiotic is no longer possible due to the K313A substitution in EF-Tu3. In addition, the Y160H and R373G substitutions lead to the loss of several hydrogen bonds between EF-Tu3 and enacyloxin IIa. Therefore, it seems very likely that EF-Tu3 is also resistant to enacyloxin IIa. The binding site of pulvomycin overlaps the binding site of the 3′ end of aa-tRNA and the three-domain junction interface of EF-Tu-GTP (Fig. 8c). Ala230 of EF-Tu3 is the most likely candidate for conferring moderate pulvomycin resistance, analogous to the R230C mutation in E. coli EF-Tu. Other remarkable substitutions are I92V, F218L, and R373G. GE2270A resistance mutations cluster in or near the binding site for the 3′ end of aa-tRNA on EF-Tu-GTP, and crystallographic data for the EF-Tu-GE2270A complex suggested that the antibiotic binds to this region (11) (Fig. 8d). The observed unusual GE2270A resistance of both EF-Tu3 proteins can be attributed to the presence of Ala at the position of the conserved Gly275. Introduction of this G275A mutation in E. coli EF-Tu yielded a completely resistant protein, which explains the resistant character of wild-type P. rosea EF-Tu1 (52). Other notable amino acid substitutions are D216N and R262G, where Asp and Arg form hydrogen bonds with a hydroxyl group and sulfur, respectively, of GE2270A.
FIG. 8.
Location of conspicuous residues on the crystal structures of Thermus thermophilus EF-Tu-GDP complexed with aurodox (N-methyl derivative of kirromycin) (a), E. coli EF-Tu-GDPNP complexed with enacyloxin IIa (b), T. thermophilus EF-Tu-GDPNP complexed with pulvomycin (c), and T. thermophilus EF-Tu-GDPNP complexed with GE2270A (d). The antibiotics are shown as stick figures. Hydrogen bonds are shown as black dotted lines, and the salt bridge in panel b is shown as a cyan dotted line. Selected side chains and stretches of the EF-Tu backbone that are likely involved in conferring antibiotic resistance are shown in green on the schematic representation. Carbon atoms are either in beige (antibiotics) or in green (side chains changed in EF-Tu3 that are very likely involved in conferring antibiotic resistance). Oxygen atoms are red, nitrogens are blue, the chlorines in enacyloxin IIa are purple, and the sulfurs in GE2270A are yellow. Numbering of the residues is according to E. coli EF-Tu. The figures were prepared using Pymol (8).
Overexpression of EF-Tu3 in S. coelicolor J1501ΔglkAΔtuf3 alters the phenotype of this strain from GE2270A sensitive to resistant, implying that EF-Tu3 can substitute for the sensitive EF-Tu1 in vivo. It would be interesting to determine whether down-regulation of tuf1 expression (e.g., by placing the chromosomal tuf1 under control of the thiostrepton-inducible tipA promoter) would lead to a compensatory increase in tuf3 expression. Answers about the role of tuf3 in vivo might also be found by succeeding in tuf gene disruption experiments in S. ramocissimus, which are currently hampered by the presence of an efficient restriction modification system (L. N. Olsthoorn-Tieleman, unpublished results).
In summary, we conclude that S. ramocissimus EF-Tu3, although highly resistant to kirromycin, is not responsible for conferring kirromycin resistance in the producing strain. Other physiological roles for this unusual elongation factor should therefore be considered.
Acknowledgments
We are grateful to V. G. Möhrle for isolating GE2270A and to W. Wohlleben for sharing unpublished data. We thank B. Kraal for the supervision prior to his retirement.
This work was in part supported by grants from the Council for Chemical Sciences of The Netherlands Organization for Scientific Research (CW-NWO) and from the Commission of the European Community in the framework of the Human Capital and Mobility Programme (contract ERBCHRXCT 940510).
Footnotes
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Abdulkarim, F., L. Liljas, and D. Hughes. 1994. Mutations to kirromycin resistance occur in the interface of domains I and III of EF-Tu.GTP. FEBS Lett. 352:118-122. [DOI] [PubMed] [Google Scholar]
- 2.Abel, K., M. D. Yoder, R. Hilgenfeld, and F. Jurnak. 1996. An α to β conformational switch in EF-Tu. Structure 4:1153-1159. [DOI] [PubMed] [Google Scholar]
- 3.Anborgh, P. H., and A. Parmeggiani. 1991. New antibiotic that acts specifically on the GTP-bound form of elongation factor Tu. EMBO J. 10:779-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berchtold, H., L. Reshetnikova, C. O. A. Reiser, N. K. Schirmer, M. Sprinzl, and R. Hilgenfeld. 1993. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365:126-132. [DOI] [PubMed] [Google Scholar]
- 5.Boon, K., I. Krab, A. Parmeggiani, L. Bosch, and B. Kraal. 1995. Substitution of Arg230 and Arg233 in Escherichia coli elongation factor Tu strongly enhances its pulvomycin resistance. Eur. J. Biochem. 227:816-822. [DOI] [PubMed] [Google Scholar]
- 6.Chater, K. F. 1998. Taking a genetic scalpel to the Streptomyces colony. Microbiology 144:1465-1478. [DOI] [PubMed] [Google Scholar]
- 7.Clayton, T. M., and M. J. Bibb. 1990. Streptomyces promoter-probe plasmids that utilise the xylE gene of Pseudomonas putida. Nucleic Acids Res. 18:1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific LLC, San Carlos, CA.
- 9.Frohlich, K. U., M. Wiedmann, F. Lottspeich, and D. Mecke. 1989. Substitution of a pentalenolactone-sensitive glyceraldehyde-3-phosphate dehydrogenase by a genetically distinct resistant isoform accompanies pentalenolactone production in Streptomyces arenae. J. Bacteriol. 171:6696-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, C. C., J. D. Watkins, and N. H. Georgopapadakou. 1991. Comparison of the Tu elongation factors from Staphylococcus aureus and Escherichia coli: possible basis for elfamycin insensitivity. Antimicrob. Agents Chemother. 35:2366-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heffron, S. E., and F. Jurnak. 2000. Structure of an EF-Tu complex with a thiazolyl peptide antibiotic determined at 2.35 Å resolution: atomic basis for GE2270A inhibition of EF-Tu. Biochemistry 39:37-45. [DOI] [PubMed] [Google Scholar]
- 12.Hobbs, G., C. M. Frazer, D. C. J. Gardner, F. Flett, and S. G. Oliver. 1989. Dispersed growth of Streptomyces in liquid culture. Appl. Microbiol. Biotechnol. 31:272-277. [Google Scholar]
- 13.Hogg, T., J. R. Mesters, and R. Hilgenfeld. 2002. Inhibitory mechanisms of antibiotics targeting elongation factor Tu. Curr. Protein Pept. Sci. 3:121-131. [DOI] [PubMed] [Google Scholar]
- 14.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. John Innes Foundation, Norwich, United Kingdom.
- 15.Hopwood, D. A., K. F. Chater, and M. J. Bibb. 1995. Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Biotechnology 28:65-102. [DOI] [PubMed] [Google Scholar]
- 16.Kawashima, T., C. Berthet-Colominas, M. Wulff, S. Cusack, and R. Leberman. 1996. The structure of the Escherichia coli EF-Tu.EF-Ts complex at 2.5 Å resolution. Nature 379:511-518. [DOI] [PubMed] [Google Scholar]
- 17.Kjeldgaard, M., P. Nissen, S. Thirup, and J. Nyborg. 1993. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure 1:35-50. [DOI] [PubMed] [Google Scholar]
- 18.Krab, I. M., and A. Parmeggiani. 1998. EF-Tu, a GTPase odyssey. Biochim. Biophys. Acta 1443:1-22. [DOI] [PubMed] [Google Scholar]
- 19.Krásny, L., J. R. Mesters, L. N. Tieleman, B. Kraal, V. Fucik, R. Hilgenfeld, and J. Jonák. 1998. Structure and expression of elongation factor Tu from Bacillus stearothermophilus. J. Mol. Biol. 283:371-381. [DOI] [PubMed] [Google Scholar]
- 20.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 21.Mesters, J. R., L. A. H. Zeef, R. Hilgenfeld, J. M. de Graaf, B. Kraal, and L. Bosch. 1994. The structural and functional basis for the kirromycin resistance of mutant EF-Tu species in Escherichia coli. EMBO J. 13:4877-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Möhrle, V. G., L. N. Tieleman, and B. Kraal. 1997. Elongation factor Tu1 of the antibiotic GE2270A producer Planobispora rosea has an unexpected resistance profile against EF-Tu targeted antibiotics. Biochem. Biophys. Res. Commun. 230:320-326. [DOI] [PubMed] [Google Scholar]
- 23.Murray, M. G. 1986. Use of sodium trichloroacetate and mung bean nuclease to increase sensitivity and precision during transcript mapping. Anal. Biochem. 158:165-170. [DOI] [PubMed] [Google Scholar]
- 24.Nissen, P., M. Kjeldgaard, M. Thirup, G. Polekhina, L. Reshetnikova, B. F. Clark, and J. Nyborg. 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270:1464-1472. [DOI] [PubMed] [Google Scholar]
- 25.Olsthoorn-Tieleman, L. N., I. J. Plooster, and B. Kraal. 2001. The variant tuf3 gene of Streptomyces coelicolor A3(2) encodes a real elongation factor Tu, as shown in a novel Streptomyces in vitro translation system. Eur. J. Biochem. 268:3807-3815. [DOI] [PubMed] [Google Scholar]
- 26.Olsthoorn-Tieleman, L. N., S. E. J. Fischer, and B. Kraal. 2002. The unique tuf2 gene from the kirromycin producer Streptomyces ramocissimus encodes a minor and kirromycin-sensitive elongation factor Tu. J. Bacteriol. 184:4211-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parmeggiani, A., I. M. Krab, S. Okamura, R. C. Nielsen, J. Nyborg, and P. Nissen. 2006. Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu. Biochemistry 45:6846-6857. [DOI] [PubMed] [Google Scholar]
- 28.Parmeggiani, A., I. M. Krab, T. Watanabe, R. C. Nielsen, C. Dahlberg, J. Nyborg, and P. Nissen. 2006. Enacyloxin IIa pinpoints a binding pocket of elongation factor Tu for development of novel antibiotics. J. Biol. Chem. 281:2893-2900. [DOI] [PubMed] [Google Scholar]
- 29.Parmeggiani, A., and P. Nissen. 2006. Elongation factor Tu-targeted antibiotics: four different structures, two mechanisms of action. FEBS Lett. 580:4576-4581. [DOI] [PubMed] [Google Scholar]
- 30.Parmeggiani, A., and G. W. M. Swart. 1985. Mechanism of action of kirromycin-like antibiotics. Annu. Rev. Microbiol. 39:557-577. [DOI] [PubMed] [Google Scholar]
- 31.Polekhina, G., S. Thirup, M. Kjeldgaard, P. Nissen, C. Lippmann, and J. Nyborg. 1996. Helix unwinding in the effector region of elongation factor EF-Tu.GDP. Structure 4:1141-1151. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Selva, E., G. Beretta, N. Montanini, G. S. Saddler, L. Gastaldo, P. Ferrari, R. Lorenzetti, P. Landini, F. Ripamonti, B. P. Goldstein, L. Montanaro, and M. Denaro. 1991. Antibiotic GE2270A: a novel inhibitor of bacterial protein synthesis. I. Isolation and characterization. J. Antibiot. 44:693-701. [DOI] [PubMed] [Google Scholar]
- 34.Shima, J., A. Hesketh, S. Okamoto, S. Kawamoto, and K. Ochi. 1996. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 178:7276-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimanaka, K., H. Iinuma, and M. Hamada. 1995. Novel antibiotics, amythiamicins. IV. A mutation in the elongation factor Tu gene in a resistant mutant of B. subtilis. J. Antibiot. 48:182-184. [DOI] [PubMed] [Google Scholar]
- 36.Sosio, M., G. Amati, C. Cappellano, E. Sarubbi, F. Monti, and S. Donadio. 1996. An elongation factor Tu (EF-Tu) resistant to the EF-Tu inhibitor GE2270 A in the producing organism Planobispora rosea. Mol. Microbiol. 22:43-51. [DOI] [PubMed] [Google Scholar]
- 37.Stark, H., M. V. Rodnina, J. Rinke-Appel, R. Brimacombe, W. Wintermeyer, and M. van Heel. 1997. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389:403-406. [DOI] [PubMed] [Google Scholar]
- 38.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiara, A. S., and E. Cundliffe. 1989. Interplay of novobiocin-resistant and -sensitive DNA gyrase activities in self-protection of the novobiocin producer, Streptomyces sphaeroides. Gene 81:65-72. [DOI] [PubMed] [Google Scholar]
- 40.Tieleman, L. N., G. P. van Wezel, M. J. Bibb, and B. Kraal. 1997. Growth phase-dependent transcription of the Streptomyces ramocissimus tuf1 gene occurs from two promoters. J. Bacteriol. 179:3619-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tubulekas, I., R. H. Buckingham, and D. Hughes. 1991. Mutant ribosomes can generate dominant kirromycin resistance. J. Bacteriol. 173:3635-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Wezel, G. P., L. P. Woudt, R. Vervenne, M. L. A. Verdurmen, E. Vijgenboom, and L. Bosch. 1994. Cloning and sequencing of the tuf genes of Streptomyces coelicolor A3(2). Biochim. Biophys. Acta 1219:543-547. [DOI] [PubMed] [Google Scholar]
- 43.Van Wezel, G. P., E. Takano, E. Vijgenboom, L. Bosch, and M. J. Bibb. 1995. The tuf3 gene of Streptomyces coelicolor A3(2) encodes an inessential elongation factor Tu that is apparently subject to positive stringent control. Microbiology 141:2519-2528. [DOI] [PubMed] [Google Scholar]
- 44.Vijgenboom, E., L. P. Woudt, P. W. H. Heinstra, K. Rietveld, J. van Haarlem, G. P. van Wezel, S. Shochat, and L. Bosch. 1994. Three tuf-like genes in the kirromycin producer Streptomyces ramocissimus. Microbiology 140:983-998. [DOI] [PubMed] [Google Scholar]
- 45.Vogeley, L., G. J. Palm, J. R. Mesters, and R. Hilgenfeld. 2001. Conformational change of elongation factor Tu (EF-Tu) induced by antibiotic binding. J. Biol. Chem. 276:17149-17155. [DOI] [PubMed] [Google Scholar]
- 46.Ward, J. M., G. R. Janssen, T. Kieser, M. J. Bibb, M. J. Buttner, and M. J. Bibb. 1986. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol. Gen. Genet. 203:468-475. [DOI] [PubMed] [Google Scholar]
- 47.Wolf, H., D. Assmann, and E. Fischer. 1978. Pulvomycin, an inhibitor of protein biosynthesis preventing ternary complex formation between elongation factor Tu, GTP, and aminoacyl-tRNA. Proc. Natl. Acad. Sci. USA 75:5324-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wörner, W., C. Glöckner, M. Mierzowski, and H. Wolf. 1983. On heterogeneity of elongation factor Tu among eubacteria. FEMS Microbiol. Lett. 18:69-73. [Google Scholar]
- 49.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13 mp18 and pUC19 vectors. Gene 3:103-119. [DOI] [PubMed] [Google Scholar]
- 50.Zeef, L. A. H., L. Bosch, P. H. Anborgh, R. Cetin, A. Parmeggiani, and R. Hilgenfeld. 1994. Pulvomycin-resistant mutants of E. coli elongation factor Tu. EMBO J. 13:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuurmond, A.-M., L. N. Olsthoorn-Tieleman, J. M. de Graaf, A. Parmeggiani, and B. Kraal. 1999. Mutant EF-Tu species reveal novel features of the enacyloxin IIa inhibition mechanism on the ribosome. J. Mol. Biol. 294:627-637. [DOI] [PubMed] [Google Scholar]
- 52.Zuurmond, A.-M., J. M. de Graaf, L. N. Olsthoorn-Tieleman, B. Y. van Duyl, V. G. Möhrle, F. Jurnak, J. R. Mesters, R. Hilgenfeld, and B. Kraal. 2000. GE2270A-resistant mutations in elongation factor Tu allow productive aminoacyl-tRNA binding to EF-Tu.GTP.GE2270A complexes. J. Mol. Biol. 304:995-1005. [DOI] [PubMed] [Google Scholar]