Abstract
Cyanobacterial photosynthesis occurs in radically diverse habitats and utilizes various forms of a CO2-concentrating mechanism (CCM) featuring multiple inorganic carbon (Ci) transporters. Cyanobacteria from dynamic environments can transform CCM activity depending on Ci availability, and yet the molecular basis for this regulation is unclear, especially in coastal strains. LysR family transcription factors resembling the Calvin cycle regulator CbbR from proteobacteria have been implicated in the expression of Ci transporter genes in freshwater cyanobacteria. Our survey of related factors revealed a group of divergent CbbR-like sequences confined to freshwater and coastal or offshore cyanobacteria. Inactivation of the single gene (termed ccmR) from this variable cluster in the euryhaline (coastal) strain Synechococcus sp. strain PCC 7002 led to constitutive expression of a high-affinity CCM. Derepression of HCO3− transporter gene transcription, including that of BicA, a recently discovered HCO3− transporter (G. D. Price et al., Proc. Natl. Acad. Sci. USA 101:18228-18233, 2004), was observed. A unique CcmR-regulated operon containing bicA plus 9 open reading frames encoding likely Na+/H+ antiporters from the CPA1 and Mnh families was defined that is essential for maximal HCO3−-dependent oxygen evolution. The promoter region required for Ci-regulated transcription of this operon was defined. We propose that CcmR (and its associated regulon) represents a specialization for species inhabiting environments subject to fluctuating Ci concentrations.
Cyanobacteria inhabit a vast range of aquatic or damp environments including arctic, freshwater, hypersaline, deep-sea (euphotic), coastal, and soda lake environments plus lichen symbioses. Cyanobacteria are subject to a significant restriction on their rates of photosynthesis owing to the relatively low availability of CO2 in aquatic environments and have evolved several adaptations (collectively known as a “CO2-concentrating mechanism” [CCM]) to facilitate efficient capture and fixation of CO2 (reviewed in references 3, 19, and 23). Generically, the cyanobacterial CCM features multiple inorganic carbon transport systems for the active uptake of both CO2 and HCO3−. Other key features include the cytosolic accumulation of inorganic carbon (Ci) as HCO3− at significant disequilibrium from CO2 (cytosolic carbonic anhydrase [CA] is absent) and the localization of the CO2-fixing enzyme Rubisco inside microcompartments known as the carboxysomes. Inside the carboxysome, CO2 is generated from HCO3− in proximity to Rubisco by the activity of a specifically localized CA. However, there is considerable niche-related genetic diversity among the cyanobacteria with regard to the systems and components that comprise their CCM (4). An emerging area in this field is the study of the regulation of these systems. The extent to which the few known regulatory mechanisms are conserved or modified across ecotypes represents a new area of research in cyanobacterial comparative genomics.
The activity of the cyanobacterial CCM in the studied freshwater organisms Synechococcus sp. strain PCC 7942 and Synechocystis sp. strain PCC 6803 and the euryhaline (coastal/estuarine) strain Synechococcus sp. strain PCC 7002 is strongly Ci responsive and is fully engaged under Ci-limiting conditions. The apparent photosynthetic affinity for Ci, as measured by K0.5(Ci), ranges between a low-affinity, constitutive state (approximately 200 μM) under Ci-replete conditions and a high-affinity, induced state (10 to 15 μM; reviewed in references 3, 8, and 19) under Ci limitation. In freshwater strains, the transition between these states is characterized by dynamic and large changes in the expression and activity of inducible HCO3− and CO2 transport systems (11, 14-16, 20, 23, 28, 29). The signal response pathways that control CCM activity in response to Ci availability are incompletely understood, particularly in euryhaline and marine strains of cyanobacteria. In the freshwater species Synechococcus sp. strain PCC 7942 a strong correlation between CCM activity and transient fluctuations in the Ci pool has been established, as well as a dependence on oxygen for full expression of CCM activity (30). The downstream events in the Ci signaling pathway of freshwater species has been better characterized, and LysR-type transcription factors are known to be involved in the regulation of high-affinity Ci transporter gene expression as both activators and repressors (7, 15, 28).
In the euryhaline cyanobacterium Synechococcus sp. strain PCC 7002, two Na+-dependent inducible HCO3− transport activities have been characterized (20): a high-affinity, low-flux system, originally characterized in Synechocystis sp. strain PCC 6803 (23), which is encoded by the sbtA gene and a medium-affinity, high-flux transporter termed BicA, which is widely distributed across cyanobacterial species. A gene encoding a putative HCO3− porin (porB; the Synechocystis sp. strain PCC 6803 homologue [slr0042] is part of a known HCO3− transporter operon [15]) is strongly Ci responsive also in Synechococcus sp. strain PCC 7002 (20). The active transport of CO2 in cyanobacterial species including Synechococcus sp. strain PCC 7002 is associated with specialized NDH-1 dehydrogenase complexes that are proposed to convert CO2 to HCO3− within the cell. The ndhF4, NdhD4, and chpX (cupB) genes are required for a low-affinity constitutive CO2 transport activity termed NDH-14, whereas an inducible, high-affinity CO2 transporter, NDH-13, requires expression of the ndhD3, ndhF3, and chpY (cupA) genes (9, 10, 12, 13, 24). The role of LysR transcription factors in CCM regulation is currently unclear in this organism.
Euryhaline cyanobacterial strains such as Synechococcus sp. strain PCC 7002, which occur in coastal or estuarine environments, would certainly be subject to habitat fluctuations such as light, nutrients (especially Ci) and temperature owing to the injection of freshwater and nutrients from land, and tidal influences (4). Consequently, a regulatory system would be required to modulate CCM activity. In the present study, we have examined the genomic structure and transcriptional regulation of low-CO2 induced Ci uptake systems in Synechococcus sp. strain PCC 7002, revealing some unique and compact gene organization. Signifying the importance of transcriptional controls on CCM activity in this system, we generated a mutant of a LysR family transcriptional regulator that exhibited complete derepression of CCM activity under Ci-replete conditions. Based on this discovery, it would be reasonable for this LysR factor to be termed CcmR, for CCM regulator, as suggested by Wang et al. (26).
MATERIALS AND METHODS
Cyanobacterial strains and culture conditions.
Synechococcus sp. strain PCC 7002 cells (mutant and wild-type strains) were cultured as described by Price et al. (20) with a light intensity of approximately 85 μmol of photons m−2 s−1. For the ΔccmR and ccmR mutants, kanamycin was included at a final concentration of 150 μg ml−1. To induce a medium- to high-affinity CCM rapidly, exponentially growing cells (optical density at 730 nm of 0.3 to 0.4), which had been bubbled with an air-CO2 mixture containing 2% (vol/vol) CO2, were harvested by centrifugation at 4,800 × g for 6 min and resuspended in an equivalent volume of modified BG-11 medium containing a final concentration of approximately 0.1 mM Ci before transfer to bubbling with air or CO2-free air, respectively. The ΔccmR deletion mutant was constructed sequentially. First, a pUC18-based construct consisting of the 0.90- and 0.93-kb regions immediately upstream and downstream of ccmR was assembled. The primers for amplifying the upstream sequence were forward (5′-TTGAGCTCATCTAGGGCTTGGCGATC) and reverse (5′-TTGGATCCTCCTGAACTTGTGCTGTTATG) introducing the SacI and BamHI sites at the 5′ and 3′ ends, respectively. The primers for amplifying the downstream flanking sequence were forward (5′-TAGGATCCTTTCCCGTGCCTTTGGTAG) and reverse (5′-AAAAGCTTAATCAGCACCCAGGCTCCAG) introducing the BamHI and HindIII sites at the 5′ and 3′ ends, respectively, for assembly inside pUC18. A kanamycin resistance marker gene from the Tn903 transposon (GenBank X06404) was cloned as a BamHI fragment into the resulting BamHI site in both transcriptional orientations relative to the ccmR flanking sequences. The cartridge was used to transform wild-type Synechococcus strain PCC 7002 cells as described by Sültemeyer et al. (26), and segregation was confirmed by PCR analysis. The construct for generating a ccmR insertional mutant was assembled in pGEMT. The primers for amplifying the ccmR open reading frame (ORF) were forward (5′-AAGGATCCAAGTTCAGGAGTAAACCCATGATC) and reverse (5′-AGCGGCCGCCAACAATTTTGAGCTTTTAGGG). A kanamycin resistance marker from the Tn903 transposon was cloned into the XbaI site in the 5′ half of the ORF in the forward transcriptional orientation relative to the ccmR sequence. The cartridge was used for transformation of wild-type cells as described above.
The Δnha mutant was made by assembling a construct in vector pGEMT (Invitrogen, Carlsbad, CA). A region from 860 bp upstream to 810 bp downstream of the SmaI and XbaI sites in nhaS3 and mnhD1, respectively (see Fig. 5A), was amplified by PCR, and the section internal to the SmaI and XbaI sites (Fig. 5A) was replaced by cloning of a chloramphenicol acetyltransferase gene (conferring chloramphenicol resistance). The primers used to amplify the flanking sequences were as follows: forward, 5′-AATTCTATGCTAAGTCCCACAATCTTTTC, and reverse, 5′-AATTGCGGCTGGATAACTGTCC. Segregation of the Δnha allele from wild-type alleles after three rounds of chloramphenicol selection was confirmed by PCR (results not shown).
FIG. 5.
Structure, regulation, and function of the bicA genomic region in Synechococcus strain PCC 7002. (A) Map of the bicA genomic region. Cotranscribed regions are indicated with arrows (dashed lines represent the range of 5′ and 3′ delimits for each mRNA as determined by RT-PCR; see Table 6). nhaS3, Na+/H+ antiporter (HMM PF00999); mnhC (HMM PF00420), mnhD1/2 (HMM PF00361), and mnhB (HMM PF04039), NADH:ubiquinone oxidoreductase-like subunits from a putative multisubunit Na+/H+ antiporter; T'glycosylase, transglycosylase (PFAM03562); recB, nuclease (COG2251.1); HC, hypothetical conserved protein. (B) Relative mRNA abundance of the ORFs downstream of bicA in wild-type (WT) cells transferred from bubbling with 2% CO2 in air to bubbling with air. Transcript changes were determined by quantitative RT-PCR (n = 4), and symbols represent transcript expression ± the SE relative to the housekeeping gene, rnpA (set at 1.0). (C) Relative mRNA abundance of ORFs in the bicA region in wild-type (WT) or ΔccmR mutant cells bubbled continuously with 2% (vol/vol) CO2 in air. Transcript changes were determined by quantitative RT-PCR (n = 4), and symbols represent transcript expression relative to the wild-type 0-min amount (set at 100%) ± the SE. (D) HCO3−-dependent oxygen evolution in air-grown wild-type cells and cells containing a deletion between the indicated SmaI and XbaI sites (see panel A) of the nhaS3 and mnhD1 genes (Δnha). Cells were analyzed by membrane inlet mass spectrometry at the indicated Ci concentrations in the presence of added CA or the CO2 uptake inhibitor, EZ. (E) Relative photosynthetic affinity for Ci for cells (part D) as measured by the K0.5(Ci) (Ci concentration for half maximal rate). Note the break in the y axis in panel B.
RIF treatment of cyanobacterial cultures.
High-Ci cells in mid-exponential growth phase were transferred to CO2-free air for 30 min to induce high-affinity Ci transporter gene expression as previously described (20). At this point (designated time zero), two cultures were supplemented with 5 mM NaHCO3 and swapped to bubbling with 1.7% CO2 in the presence or absence of 200 μg of the transcriptional inhibitor rifampin (RIF) ml−1 for a further 2 h. The remaining two cultures were supplemented with 5 mM NaCl to balance sodium and bubbled with CO2-free air with or without RIF as described above. All four cultures were sampled throughout this period.
Use of genome and protein databases.
Synechococcus sp. strain PCC 7002 sequences were accessed from the complete genome database constructed by Donald Bryant and Tao Li (Pennsylvania State University) and Jindong Zhao (College of Life Sciences, Peking University) and are available at GenBank (for accession numbers, see Table 3). For other cyanobacterial sequences, gene object identifiers (GOIs) from the Integrated Microbial Genomes database (DOE, Joint Genome Institute; http://img.jgi.doe.gov/cgi-bin/pub/main.cgi) were used in preference to GenBank accession numbers if locus tags were unavailable. Information about the databases used to detect conserved protein domains is available at http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml.
TABLE 3.
Sequences of the gene-specific primers used in quantitative RT-PCR assays for Synechococcus strain PCC 7002a
| Clone | GenBank accession no. | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|
| ndhF3 | U97516 | ACCGCGATGTATCTTTATCTC | ACAAAATAGCGGTCTAACCAG |
| ndhD3 | U97516 | CGTGTTCCAAGGTAGCTTTAG | AAAAGGATAATGACCGTCAAG |
| chpY | U97516 | AAATTATTCCCAAATCCAAAG | GCATACAAAATACCGTAGTGG |
| ORF133 | U97516 | TGCCAAGTATTGTGGATATTG | TAAGTGAGAATACGCGCTAAC |
| ORF133+ | U97516 | CGCGTATTCTCACTTATCACG | TGAAACCGGAAAATTTGAAAC |
| sbtA | DQ632590 | CAGCGATTGTTGTAGCTAGTC | AATTTCAACCCGATTAGTAGC |
| porB | DQ632588 | TTACAAGCCTACAATTTTTCG | CCGTTAATTTATCTGTGATCG |
| bicA | AF381039 | GTTCTTTTGTTTCAACTCAGTG | GCTTAAAGAGCTTGAGTTTTTC |
| nhaS3 | AF381039 | GATTTCGTTCCTTCCCTGTC | GAGACCCCGACTAGAACACC |
| mnhC | AF381039 | ATCGCGTATTACATTCTCGTG | ATTATCCCGTGACAACTTCATC |
| mnhD1 | AF381039 | GGGATAATCCGACCTTAGAAAC | TCCAAGAGTTCTAGGGTAAAAGG |
| mnhD2 | AF381039 | AGAGCGGCTATTTTATTTTGAC | AACGTAGAGGCTAATCAGATCG |
| HCP 1 | AF381039 | GATTAAAGATTGGCTCCATGTG | ACAAAATTAGGACACCGACAAC |
| HCP 2 | AF381039 | GCGTTACTACTCCCGATTTATG | ACAAAATTAGGACACCGACAAC |
| HCP 3 | AF381039 | CGTTCTGTGTTGTTTAAATTGC | CACATACCCCAGTACCGTATTC |
| HCP 4 | AF381039 | CCTTGTTATTGCCCGTGAC | AACATCGTCCCCACTAACG |
| mnhB | AF381039 | CTTTATTTGTGAAAATGCTCGTC | GGGAATCGGTAAATTGCTTAAC |
| mnhB+ | AF381039 | TGCCATTTATAAAAAGTGGAATG | CAAACCGTATGCTGGTATGAC |
| rbcS | AF015889 | TGATGGATCAAGGCTATATTCC | AACAACACGGATGTAGCAGTC |
| rbcX | AF015889 | AACAACACGGATGTAGCAGTC | ACTCACCAGGGTTAGGTTCAG |
| rbcL | AF015889 | TACTTGGACCACTGTATGGAC | TCAAAACGTTGGTTACAGAAC |
| ccmK1 | AF015889 | CTATTGCAGTCGGAATGGTAG | GGAAGCTTGGACTTCAGAAAC |
| ccmK2 | AF015889 | CGTGGTGATGTTTCTGAAGTC | ACTTCTTCGGTGTAGCGAATC |
| ccmL | AF015889 | CAGCACCTACAAAGCAGAAAG | GTATCAATGATGCCAATGACC |
| ccmM | AF015889 | TCGAATTTAATTGGTGATGTC | ATCCAGACAGAATAGTCATGG |
| ccmN | AF015889 | AAATGCCTGCTTAGGTTATGG | TTGTGGCAGTCTGGTTAGTTG |
| ccmO | To be assigned | ACAACTGGCCTCCTATGAAAC | TTGGTTCTGCCTTATCTAGCC |
| ccaA | To be assigned | TATGCCTTTGGTTTACGATTG | ATCCAGCCATAGATTTTCAGC |
| p450 | U97516 | CTGACCCGATGGACATTTAC | AGCAGATAAATTGACCCCATC |
HCP, hypothetical conserved protein from the bicA operon (nomenclature as per Fig. 5).
Reverse transcription-PCR (RT-PCR) assays.
TAQ-Ti DNA polymerase kit components (Fisher-Biotech, West Perth, Australia) were used in 20-μl reactions containing 1.5 mM MgCl2, 200 μM concentrations each of dATP, dTTP, dCTP and dGTP, 0.5 μM concentrations each of forward and reverse primer, 0.5 U of Taq, and cDNA template equivalent to 25 ng of total RNA. First-strand cDNA synthesis from normalized total RNA (derived from wild-type cells bubbled with air for 60 min and isolated according to the method of McGinn et al. [11]), was synthesized as described in Woodger et al. (29) using the set of reverse primers listed in Tables 1, 2, and 3. Thermocycling was performed in a TGradient thermal cycler (BIOMETRA, Goettingen, Germany) for 28 to 30 cycles consisting of denaturation for 15 s at 94°C, annealing at 54°C for 30 s, and extension at 72°C for 60 s. Cycling was preceded by a 5-min 94°C activation step. Primer pair sequences are listed in Tables 1, 2, and 3. For every reaction an RNA sample without reverse transcriptase was included to control for genomic DNA contamination, along with a positive control containing 5 to 50 ng of genomic DNA as a template. Products were separated by electrophoresis of 10 μl of each reaction on a 1.2% agarose gel containing 0.09 M Tris-borate-0.002 M EDTA (pH 8.0) with 0.5 mg of ethidium bromide ml−1 and visualized on a UV transilluminator.
TABLE 1.
Sequences of the gene-specific primers used in RT-PCR assays of the ccmR genomic region of Synechococcus strain PCC 7002a
| Primer | Sequence (5′-3′) |
|---|---|
| 1350F | CTTTTGTCCAGAGAACCATACTC |
| 2002R | GCAAATTACTCAGGATGTAGAGATC |
| 1521F | GCACAAGTTCAGGAGTAAACCC |
| 2876R | AATGCTGGAAATATCGAGGTC |
| 3215R | CAAAATTCCAAGTACCAGCAAG |
| 4670R | ATACCAGCATCTTGGAGGTTG |
| 1950F | GATCGGATGGCGGATAATC |
| 4228F | ACACAGTTATTTTTGCCGTTG |
| 5806F | GACAGTCACACCGCCTATTAC |
| 6253R | ACTTGTTCCGCAATGTACAAC |
| 7172F | GTCGATGTACAACGTCACCTC |
| 7602R | AGTGAGAATACGCGCTAACTG |
| 7589F | CGCGTATTCTCACTTATCACG |
| 8288R | CGTTTATGGCTCAAAACAGTG |
| 8273F | GCCAAAGGAAAGTGCTTATAG |
| 8726R | GAAGCGCAATTTTTATTGAAG |
TABLE 2.
Sequences of the gene-specific primers used in RT-PCR assays of the bicA genomic region of Synechococcus strain PCC 7002a
| Primer | Sequence (5′-3′) |
|---|---|
| 1279F | AGTTTTGCCATTGTAACTCG |
| 1980R | TGACATGAAGCCAGAAATCAC |
| 1462F | GATATTAAACTATGCTCCTGTTGC |
| 1561F | TGATCTGAAAAATGCGTAACC |
| 3198F | GGAAGAAGCCCTCAAGAATG |
| 3700R | GAGACCCCGACTAGAACACC |
| 4697F | TGATCTTGACCACTTTCTTGG |
| 5247R | ATTATCCCGTGACAACTTCATC |
| 5061F | GGTGATCGCGTATTACATTCTC |
| 5462R | TCCAAGAGTTCTAGGGTAAAAGG |
| 6718F | TGGCTTAGGACAACTCTCAGTG |
| 7128R | AACGTAGAGGCTAATCAGATCG |
| 8014F | TATCTCGAAGCCTATCAACTGG |
| 8486R | GAGAAAAATTAAGCCTGGAGTC |
| 8333F | GATTAAAGATTGGCTCCATGTG |
| 8774R | ACAAAATTAGGACACCGACAAC |
| 8613F | GCGTTACTACTCCCGATTTATG |
| 9081R | CACATACCCCAGTACCGTATTC |
| 8923F | CGTTCTGTGTTGTTTAAATTGC |
| 9325R | GAGGGTAATGGATAACATCGTC |
| 9510F | AAATGTGCACTGTATTTTGCAG |
| 9976R | GGGAATCGGTAAATTGCTTAAC |
| 10140F | TGCCATTTATAAAAAGTGGAATG |
| 10539R | CAAACCGTATGCTGGTATGAC |
| 10181F | TTTCGGTACTGGTCTTTTTCC |
| 10692R | AGCACCGATTTTTCATTTTTC |
Real-time quantitative RT-PCR assays.
First-strand cDNA synthesis from normalized total RNA, isolated according to the method of McGinn et al. (11), was conducted as described in Woodger et al. (29). Quantitative real-time RT-PCR assays, using primer pairs specific to the bicA operon, sbtA, ndhR, and the ndhF3 ndhD3 chpY orf133 operon and incorporating SYBR green I to monitor product formation, were performed as described by Woodger et al. (29). Primer sequences are listed in Table 3 or as described by Price et al. (20). The rnpA housekeeping gene was used in normalization as described by Price et al. (20). All reactions were carried out in quadruplicate, and errors were determined by standard methods.
Determination of TSPs.
Transcription start points (TSPs) for bicA and ndhF3 were determined by RNA ligase-mediated amplification of full-length 5′ cDNA ends (RLM-RACE) using the Generacer kit (L1500-1502; Invitrogen, Carlsbad, CA) according to the instructions of the manufacturer. In brief, total RNA was isolated from wild-type cells bubbled with air for 60 min (as described above). First-strand cDNA was generated from 3 μg of dephosphorylated RNA with full-length mRNA subsequently decapped and ligated to a specific RNA oligonucleotide. Nested PCR was performed, and specific products cloned and sequenced in the vector pCR4Blunt-TOPO. The following primers were used: ndhF3 reverse, 5′-GCCCATTCAGCCAGACCATCAAA; ndF3 reverse nested, 5′-CCACAGCATAGAGTTGGGCGAGTAA; bicA reverse, 5′ CAGAAATCACGGTGTAGGGCATCAT; and bicA reverse nested, ATGGGCAATCACAGCGGTCATAA.
Luciferase assays and luxAB fusions.
Putative promoter elements were amplified by PCR with the primers indicated in Table 4, cloned to pENTR (Invitrogen, Carlsbad, CA), and fused to Vibrio luciferase genes (luxAB) by Gateway LR recombination in a version of the Synechococcus sp. strain PCC 7002 neutral-site-integration reporter vector, pMBB, modified for lambda site-specific recombination. The pMBB vector (kindly provided by G. Bullerjahn) is a modified version of the Synechococcus sp. strain PCC 7942 luciferase reporter construct, pAM1414 (1), and contains a neutral integration site encoding omega-3 desaturase (desB) (21). Luminescence in liquid cyanobacterial cultures was measured in triplicate in freshly harvested 2-ml aliquots after the rapid addition of 4 μl of 0.025 to 0.05% (vol/vol) Decanal (D7384; Sigma, St. Louis, MO) in a TD-20/20 luminometer. Substrate was diluted in 100% dimethyl sulfoxide. The optimal decanal concentration was determined for each experiment. The average raw luminescence was normalized on a chlorophyll a basis and is expressed as a percentage of the empty vector control (high-Ci cells). The chlorophyll a concentration was determined as previously described (17).
TABLE 4.
Sequences of the primers used to amplify Synechococcus sp. strain PCC 7002 DNA fragments from the upstream regions of the ndhF3 [ndhF3(P550)] and bicA genes (722F/R and clones 2 to 12) for analysis as luxAB fusions in the integration vector pMBB
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| ndhF3(P550) | GAAAAATCTCCCTCTGTCTGTGTTGGATG | CACCATGACGATGACGCTGAAATAATTTG |
| bicA(P722F) | GCTATGGTCGATGGCGTTGAGTAAC | CACCTGTGGGTTCGGAAATGAG |
| bicA(P722R) | CACCGTCGATGGCGTTGAGTAAC | CTGTGGGTTCGGAAATGAGGG |
| Clone 2 | GCTATGGTCGATGGCGTTGAGTAAC | CACCTCTGCATGGTTACGCATTTTTC |
| Clone 4 | GCCCAGGGTGGCAATGGTTAGAC | CACCTCTGCATGGTTACGCATTTTTC |
| Clone 5 | GCCCAGGGTGGCAATGGTTAGAC | CACCAGTGCAACAGGAGCATAGTTTAATATC |
| Clone 7 | CATAGGCGATCGCCACCTATGTTTTAG | CACCTCTGCATGGTTACGCATTTTTC |
| Clone 8 | AGGGGGCTATGGTTAGATGTTAATCAATAC | CACCTCTGCATGGTTACGCATTTTTC |
| Clone 9 | TATGCTCCTGTTGCACTGACAACTTAAC | CACCTCTGCATGGTTACGCATTTTTC |
| Clone 10 | AATTCATCAATAAAGATAGTTTTCCCAATG | CACCTATCTATGAAAATTTTATTTTGTATTG |
| Clone 12 | CATAGGCGATCGCCACCTATGTTTTAG | CACCAGTGCAACAGGAGCATAGTTTAATATC |
Mass spectrometric measurements.
Cells were prepared and analyzed by mass spectrometry as previously described (2, 25). Assays were performed in 4-ml volumes in a thermostat-controlled (30°C) mass spectrometer cuvette allowing membrane inlet analysis of O2 (mass 32) and CO2 (mass 44). For measurements of the photosynthetic affinity for Ci, cells were assayed at a chlorophyll density of 2 μg ml−1 in BG11 medium buffered with 50 mM BisTrisPropane-HCl (pH 7.9) in which NaNO3 had been replaced with 20 mM NaCl. A light intensity of 700 μmol of photons m−2 s−1 was used. The maximum rate of net O2 evolution (Vmax) was measured in the presence of at least 1 mM NaHCO3, and the photosynthetic affinity for Ci was determined as K0.5(Ci), that is, the Ci concentration required to reach half the maximum rate of net O2 evolution. Measurements at low levels of Ci were initiated at around 25 μM O2 and allowed to progressively increase throughout the Ci range.
RESULTS
Turnover of mRNA abundance for inducible Ci transporter genes.
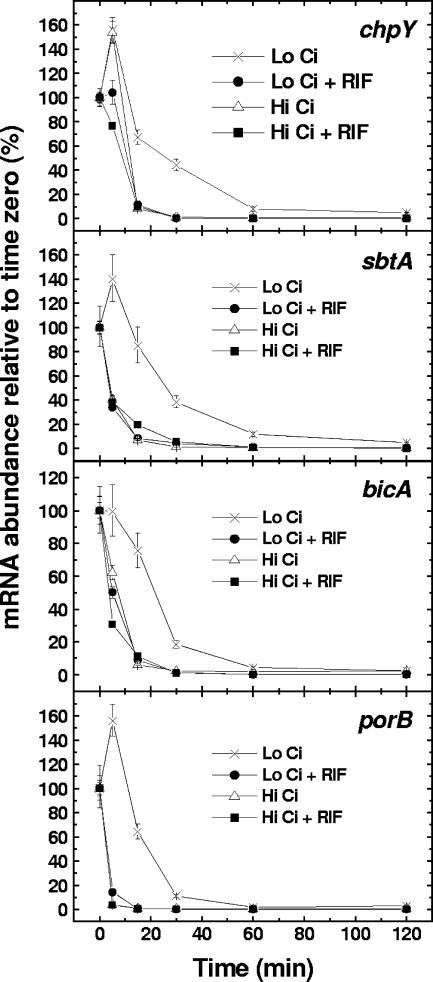
We have previously shown that transcripts encoding high-affinity Ci transport systems in Synechococcus sp. strain PCC 7002 are rapidly and strongly induced upon the transition to Ci limitation (20). To assess the importance of any transcriptional control of mRNA pool size, quantitative RT-PCR was used to monitor mRNA turnover in wild-type cells at high Ci or low Ci with or without the transcriptional inhibitor, RIF. The mRNA half-lives of bicA and sbtA (encoding inducible, high-affinity HCO3− transporters), chpY (encoding part of an inducible, high-affinity CO2 uptake system), and porB (encoding a putative HCO3− porin) were measured (Fig. 1).
FIG. 1.
Turnover of mRNA encoding Ci transporters in Synechococcus strain PCC 7002 cells. Exponentially growing high-Ci cells were swapped to buffer containing approximately 100 μM Ci and bubbled with CO2-free air for 30 min. At this time (time zero) cells were transferred to either CO2-free air or 2.0% CO2 in the presence or absence of 200 μg of RIF ml−1. The relative abundance of chpY, sbtA, bicA, and porB mRNA was determined by quantitative RT-PCR (n = 4). Symbols represent mRNA abundance relative to the 0-min amount (set at 100%) ± the standard error (SE).
All transcript half-lives, estimated from the rate of mRNA degradation in the presence of RIF, were relatively short (approximately 5 to 10 min), irrespective of the Ci concentration. This suggests that any posttranscriptional control is not exerted differentially according to Ci concentration. Furthermore, cells transferred from low Ci to high Ci in the absence of RIF had similarly rapid transcript turnover rates. That is, when transcriptional control in cells was maintained, transcripts were still equally rapidly turned over. Similar to previous findings in Synechococcus sp. strain PCC 7942 (29), these results suggest that control of the abundance of these transcript pools in response to Ci concentration is primarily transcriptional rather than posttranscriptional.
Deletion of a CbbR-like transcription factor (CcmR) derepresses CCM activity in Synechococcus sp. strain PCC 7002.
We found that CbbR-like proteins from diverse cyanobacterial species cluster into two main groups (Fig. 2A). The less-divergent sequences (group 1) may represent proteins with an essential function, such as the Calvin cycle gene regulator CbbR, since this cluster contains the Synechocystis sp. strain PCC 6803 sll0998 locus which cannot be stably inactivated (7, 15, G. D. Price, unpublished data). A more divergent grouping (group 2) containing CmpR and NdhR/CcmR from Synechocystis sp. strain PCC 6803 (abbreviated as 6803) is evident. NdhR/CcmR_6803 would appear to repress the transcription of both ndhF3 ndhD3 chpY (cupA) and sbtA (7, 28), whereas CmpR_6803 appears to activate transcription of the cmpABCD operon (15). The freshwater strain Synechococcus sp. strain PCC 7942 has a single group 2 representative, CmpR_7942 (15). A single group 2 representative is also apparent in Synechococcus sp. strain PCC 7002.
FIG. 2.
CbbR-like genes in Synechococcus sp. strain PCC 7002. (A) Phylogenetic tree of proteins with >27% amino acid identity to Rhodobacter sphaeroides CbbR from cyanobacteria representing diverse habitats. Proteins were aligned by using CLUSTAL W and the dendrogram generated in the TREEVIEW program. Species name abbreviations: alr/all, Nostoc sp. strain PCC 7120; Avar, Anabaena variabilis ATCC 29413; Cwat, Crocosphaera watsonii WH8501; gll/glr, Gloeobacter violaceus (strain PCC 7421); Npun, Nostoc punctiforme ATCC 29133; PMM, Prochlorococcus marinus (subsp. pastoris, strain CCMP 1378/MED4); Selo, Synechococcus sp. strain PCC 7942 (elongatus); slr/sll, Synechocystis sp. strain PCC 6803; SynW8102, Synechococcus sp. strain WH8102; 7002, Synechococcus sp. strain PCC 7002; tlr/tll, Thermosynechococcus elongatus strain BP-1; Tery, Trichodesmium erythraeum IMS101; YSA′ and YSB′, Synechococcus sp. strain Yellowstone A-Prime and B-Prime. Locus tags are given for the Anabaena, Synechocystis strain PCC 6803, and Gloeobacter violaceus genes. GOIs from the Integrated Microbial Genomes database (DOE, Joint Genome Institute; http://img.jgi.doe.gov/cgi-bin/pub/main.cgi) are used elsewhere except for the Yellowstone species, for which the GenBank accession numbers are given. (B) Physiological phenotype of the group 2 “ccmR” deletion mutant. Rates of Ci-dependent O2 evolution for wild-type (WT) and ΔccmR cells grown continuously with 2% (vol/vol) CO2 in air (two replicates are shown). (C) Structure of the ccmR genomic region in Synechococcus sp. strain PCC 7002. Possible cotranscribed regions are indicated with arrows (dashed lines represent the range of 5′ and 3′ delimits for each mRNA as indicated by RT-PCR; see Table 5). Predicted stem and loop structures are shown. ndhF3/D3, NAD(P)H-dehydrogenase (NDH-13) subunit genes; chpY/cupA, gene for putative CO2 hydration protein associated with NDH-1; orf133, conserved gene of unknown function associated with NDH-1 genes; cytP450, cytochrome p450 (PFAM00067); DSBA, DSBA thioredoxin (PFAM 03123).
We hypothesized that the single group 2 protein from Synechococcus sp. strain PCC 7002 might exert predominant control over the activity of the CCM. Although originally designated RbcR by Klughammer et al. (9), we tentatively renamed this factor CcmR (CcmR_7002), for the high-affinity CCM regulator, following the suggestion of Wang et al. (28). To test this idea, the CcmR_7002 ORF was deleted by transforming Synechococcus sp. strain PCC 7002 cells with a construct consisting of the flanking regions of ccmR fused to a kanamycin resistance marker. The segregation of the ΔccmR allele from wild-type alleles after three rounds of kanamycin selection was confirmed by PCR (results not shown). As shown in Fig. 2B, mutant cells grown continuously at high Ci had a striking physiological phenotype. The cells have a very high photosynthetic affinity for Ci, with a K0.5(Ci) of approximately 10 μM, compared to the wild type, which has a K0.5(Ci) of approximately 250 μM. This result suggests that CcmR ordinarily functions to repress high-affinity CCM activity under nonlimiting Ci conditions.
CcmR represses expression of high-affinity Ci transporters in Synechococcus sp. strain PCC 7002 under nonlimiting Ci conditions.
The expression of CcmR_7002 gene-flanking genes (Fig. 2C), high-affinity Ci transporter genes, and carboxysome-associated genes was assessed in ΔccmR and wild-type cells bubbled with high Ci or air (Fig. 3). The mRNA abundance for the divergently oriented cytochrome p450 gene was largely unaffected by the replacement of the ccmR gene with a kanamycin resistance marker, irrespective of orientation (Fig. 3A). Thus, the mutant that contained the resistance marker in the same transcriptional orientation as ccmR was used in all subsequent experiments. The close juxtaposition in Synechococcus sp. strain PCC 7002 of ccmR and the genes for high-affinity CO2 uptake—ndhF3, ndhD3, chpY (cupA), and orf133—is unique among the sequenced cyanobacterial genomes (Fig. 2C). Unlike the freshwater strain Synechocystis sp. strain PCC 6803 (7, 28), transcriptional derepression of this region was not observed in ΔccmR. Instead, a low-level constitutive expression of the region was found, irrespective of the Ci concentration or transgene orientation. It should be noted that the Tn903 kanamycin cassette lacks a typical terminator and is likely to provide basal expression of the ndhF3, ndhD3, chpY, and orf133 genes downstream of ccmR and be nonresponsive to CO2. In contrast, and consistent with the increased photosynthetic affinity for Ci in the mutant (Fig. 2B), ΔccmR cells grown at high Ci had relatively abundant mRNA pools for the inducible HCO3− transporter genes bicA and sbtA and the putative HCO3− porin gene, porB, compared to wild-type cells (Fig. 3B). These levels approached the maximal levels in air-induced wild-type cells (Fig. 3B). We also investigated whether carboxysome-associated genes are targets for CcmR action but found little difference in the mRNA abundance for these genes between mutant and wild-type cells (Fig. 3D). Given that these genes are not strongly Ci responsive in the wild type, this result is not surprising. An exception was the mRNA pool encoding the carboxysomal CA, ccaA, which was more strongly induced under air limitation in ΔccmR cells. This mild induction of ccaA mRNA has been noted in other CCM-deficient mutants (6).
FIG. 3.
Gene expression in ΔccmR. The relative abundance of mRNA for ccmR-neighboring genes (A), bicarbonate transporter genes (B), and carboxysomal genes (C and D) in wild-type and ΔccmR cells after transfer from bubbling with 2% (vol/vol) CO2 in air to bubbling with air alone for 60 min was determined. Transcript changes were determined by quantitative RT-PCR (n = 4), and symbols represent transcript expression relative to the wild-type (WT) 0-min amount (set at 100%) ± the SE. Note the break in the y axis in panels A and B. The experiment was independently replicated, and representative data are shown.
Transcriptional regulation of the CcmR region in Synechococcus sp. strain PCC 7002.
The proximity of the CcmR_7002 gene to the ndhF3 ndhD3 chpY (cupA) orf133 gene cluster (Fig. 2C), plus the lack of transcriptional Ci responsiveness of this region in the ΔccmR mutant (Fig. 3B), led us to investigate the transcriptional regulation of this region. We generated an insertional ccmR mutant at the XbaI site (Fig. 2C) located 420 bp from the start of the ccmR ORF; however, the phenotype was the same as the deletion mutant (results not shown). This result indicates that it is difficult to engineer mutations at this locus that do not interfere with transcription of the downstream gene cluster. The expression of the 5′ half of the ccmR transcript, upstream of the insertion site for the antibiotic resistance marker, was monitored in the insertional mutant by quantitative RT-PCR and found to be elevated at high Ci (Fig. 4B). This result signifies that CcmR ordinarily autorepresses ccmR transcription, as previously found for NdhR/CcmR in the freshwater strain Synechocystis sp. strain PCC 6803 (7).
FIG. 4.
Transcriptional regulation of the ccmR region in Synechococcus strain PCC 7002. (A) Relative mRNA abundance of the ORFs downstream from ccmR in wild-type cells transferred from bubbling with 2% CO2 in air to bubbling with normal air. Transcript changes were determined by quantitative RT-PCR (n = 4), and symbols represent transcript expression ± the SE relative to the housekeeping gene, rnpA (set at 1.0). (B) Relative mRNA abundance of ccmR in wild-type (WT) or ccmR insertional mutant cells after transfer from bubbling with 2% (vol/vol) CO2 in air to bubbling with normal air. Transcript changes were determined by quantitative RT-PCR (n = 4), and symbols represent transcript expression relative to the wild-type high-Ci amount (set at 100%) ± the SE. Note the break in the y axis in panel A. (C) Predicted promoter structure of the ndhF3 upstream region. Consensus LysR binding sites are in boldface, and putative −35 and −10 elements (as predicted by BPROM) are also shown. The most 5′ TSP determined experimentally is marked with an arrow. (D) Luciferase activity directed by the 550-bp ndhF3 upstream region fused to luxAB genes (pF3:P550) and stably integrated into the genome of Synechococcus sp. strain PCC 7002 cells. Cells were grown at high Ci or with air bubbling for 5 h. A positive Ci-responsive control (pbicA:P722F) consisting of a 722-bp fragment from the upstream region of the bicA gene fused to luxAB, is shown (see Fig. 6A). Relative luminescence units are expressed on a chlorophyll a basis as a percentage of the value obtained for high-Ci cells containing the empty vector control ± the standard deviation.
An RT-PCR and quantitative RT-PCR approach was taken to define the limits of the mRNA(s) which encompasses ccmR and the downstream ORFs [ndhF3, ndhD3, chpY (cupA), and orf133]. The transcription of the entire region was coinduced in response to Ci limitation in wild-type Synechococcus sp. strain PCC 7002 cells (Fig. 4A); however, only very weak transcription of the sequence downstream from orf133 was detected. The results of RT-PCR, using first-strand cDNA generated from the set of reverse primers described in Tables 1, 2, and 3 and primer pairs that span the junctions of the ORFs in this region of the genome (Table 1), are summarized in Table 5. As previously speculated (9), the ndhF3, ndhD3, chpY (cupA), and orf133 ORFs probably form part of an operon, since it was possible to amplify at least 250 bp beyond the junctions of all of these ORFs. It was also possible to amplify significantly overlapping fragments of the ccmR and ndhF3 ORFs from first-stand cDNA, although based solely on this analysis the two ORFs are not necessarily part of the same mRNA (see Table 5, the 1521F-2876R and 1521F-3215R primer pairs).
TABLE 5.
Amplification of mRNA sequences from the ccmR region of Synechococcus strain PCC 7002 by RT-PCRa
| Primer pair coordinates | Junction | Product | Primer pair coordinates | Junction | Productb | |
|---|---|---|---|---|---|---|
| 1350F-2002R | Upstream/ccmR | N | 4228F-4670R | ndhF3/ndhD3 | Y | |
| 1521F-2002R | ccmR/ndhF3 | Y | 5806F-6253R | ndhD3/chpY | Y | |
| 1521F-2876R | ccmR/ndhF3 | Y | 7172F-7602R | chpY/ORF133 | Y | |
| 1521F-3215R | ccmR/ndhF3 | N | 7589F-7963R | ORF133/downstream | Y* | |
| 1521F-4670R | Upstream/F3 | N | 7589F-8288R | ORF133/downstream | Y* | |
| 1950F-4670R | Upstream/F3 | Y | 82737F-8726R | ORF133/downstream | Weak* |
The sequence map coordinates of primers (according to Fig. 4; GenBank accession no. U97516) and the presence (Y) or absence (N) of an amplified product are indicated. First-strand cDNA template was generated from RNA derived from Synechococcus strain PCC 7002 cells transferred from 2% CO2 to air bubbling for 30 min.
*, Not Ci responsive.
An analysis of the region upstream of the start codon of ndhF3 revealed putative −35 and −10 elements and LysR binding sites (TnA{n7/8}TnA; Fig. 4C). Using RLM-RACE, a TSP was mapped to a point that supports the predicted promoter structure (Fig. 4C). To determine whether the region 5′ to ndhF3 can function as a promoter, the 550 bp upstream of the ndhF3 start codon was fused to genes encoding Vibrio luciferase (luxAB) in a neutral-site integration reporter vector, pMBB. However, no significant luciferase activity was detected in wild-type cells transformed with the resulting construct at high Ci or after 5 h of air bubbling (Fig. 4D), suggesting that this region does not contain a functional promoter.
The bicA gene is part of a large CcmR-regulated operon.
Our finding that Synechococcus sp. strain PCC 7002 CcmR represses full expression of the bicA gene at nonlimiting Ci (Fig. 3B) led us to examine the regulation and function of neighboring genes. Figure 5A shows the genomic organization of the region surrounding the bicA locus (GenBank accession no. AF381039). Suggestive of the existence of an operon, nine ORFs occur immediately downstream of bicA without significant intergenic sequences. Immediately downstream of bicA is an ORF that encodes a protein with domains of a Na+/H+ antiporter from the CPA1 family, HMM PF00999, which we have termed nhaS3 after the Synechocystis sp. strain PCC 6803 homologue (sll0689). The subsequent eight ORFs encode hydrophobic sequences resembling a class of cation/proton antiporter proposed to function as a hetero-oligomer (27). The nomenclature of these transporters is unresolved. We have chosen Mnh for multisubunit Na+/H+ antiporter (alternatively Mrp, Mnh, Pha, Sha 27), since Nostoc strain PCC 7120 mutants at a similar locus display Na+ sensitivity (5). Several of these Mnh genes in Synechococcus sp. strain PCC 7002 encode proteins with similarity to the hydrophobic subunits of proton-pumping NADH:ubiquinone oxidoreductases, including MnhC (HMM PF00420), MnhD1/2 (HMM PF00361), and MnhB (HMM PF04039); however, their general role, if any, in transporter energization is obscure (27). This tripartite clustering of genes encoding transporters—bicA, nhaS3, and mnh—is unique within the sequenced cyanobacterial genomes.
Quantitative RT-PCR analysis (Fig. 5B) showed that nhaS3 plus the mnh-like genes in Synechococcus sp. strain PCC 7002 are upregulated in response to Ci limitation and, as with bicA, nhaS3 and mnh gene expression is also derepressed at high Ci in ΔccmR (Fig. 5C). Previously, similar regulation of mnh genes in the freshwater strains Synechocystis sp. strain PCC 6803 (28) and Nostoc sp. strain PCC 7120 (5) has been demonstrated. RT-PCR was carried out with primers that span the junctions of the ORFs in the bicA region in Synechococcus sp. strain PCC 7002 (Table 2). Consistent with the cotranscription of these genes as an operon (Table 6), products that span the intergenic regions were successfully amplified from first-strand cDNA. The failure to detect products at 300 bp upstream of the presumptive bicA start codon suggests the 5′ untranslated region is contained in the intervening region (Table 6, primer pair 1279F-1980R). Relatively weak, Ci-responsive transcription was detected up to 172 bp beyond the presumptive stop codon for MnhB (Fig. 5A); however, quantitative RT-PCR showed that, beyond this point, transcripts were not detectable above background levels (results not shown), even though very weak bands were detected using a nonquantitative RT-PCR endpoint approach (Table 6, primer pair 10181F-10692R).
TABLE 6.
Amplification of mRNA sequences from the bicA region of Synechococcus strain PCC 7002 by RT-PCRa
| Primer pair coordinates | Junction | Product | Primer pair coordinates | Region or junction | Product | |
|---|---|---|---|---|---|---|
| 1279F-1980R | Upstream/bicA | N | 8014F-8486R | ORF5/6 | Y | |
| 1462F-1980R | Upstream/bicA | Y | 8333F-8774R | ORF6/7 | Y | |
| 1561F-1980R | Upstream/bicA | Y | 8613F-9081R | ORF7/8 | Y | |
| 3198F-3700R | bicA/napA | Y | 8923F-9325R | ORF8/9 | Y | |
| 4697F-5247R | napA/ORF3 | Y | 9510F-9976R | ORF9/10 | Y* | |
| 5061F-5462R | ORF3/4 | Y | 10140F-10539R | ORF10/downstream | Y* | |
| 6718F-7128R | ORF4/5 | Y | 10181F-10692R | ORF10/downstream | Med* |
The sequence map coordinates of primers (according to Fig. 5A; GenBank accession no. AF381039) and the presence (Y) of an amplified product are indicated. First-strand cDNA template was generated from RNA derived from Synechococcus strain PCC 7002 cells transferred from 2% CO2 to air bubbling for 30 min. *, Not Ci responsive.
The bicA operon is essential for full bicarbonate transport activity.
To assess the potential role of the nhaS3 mnh gene cluster in HCO3− uptake, the first three ORFs in this region were inactivated by engineering a deletion between the SmaI and XbaI sites noted in Fig. 5A. This strategy was adopted given the possibility that this gene cluster encodes redundant functions. Segregation of the resulting Δnha allele from wild-type alleles was confirmed by PCR after three rounds of chloramphenicol selection (results not shown). The HCO3−-dependent oxygen evolution (a surrogate measure for net HCO3− uptake) was determined in air-grown wild-type and Δnha cells incubated with the known CO2 uptake inhibitor, ethoxyzolamide (EZ) (18, 20). Control cells were incubated with 10 μg of bovine CA/ml to ensure rapid equilibrium of Ci species. As shown in Fig. 5D, at Ci concentrations between approximately 20 and 130 μM, oxygen evolution in mutant cells incubated with 300 μM EZ was markedly reduced compared to the wild type. That is, when active HCO3− uptake was the major route for Ci uptake, oxygen evolution was diminished in the mutant but not in the wild type. The relative photosynthetic affinity for Ci in the presence of EZ, as measured by the K0.5(Ci), was 15.5 ± 2.6 μM in the wild type but only 74.9 ± 1.5 μM in the mutant (Fig. 5E). This suggests that the nhaS3-mnh region ordinarily has a role in supporting maximal HCO3− uptake in this cyanobacterium. The growth of the mutant was investigated at high pH, under air limitation, to maximize the contribution of HCO3− transport to growth. However, no difference in growth rate compared to the wild type was detected (data not shown).
Analysis of the structure of the bicA promoter.
To identify potential Ci-responsive elements within the region immediately upstream and downstream of the bicA start codon, a series of DNA fragments from this region (Fig. 6A) were fused to genes encoding Vibrio luciferase (luxAB) in a version of the Synechococcus sp. strain PCC 7002 neutral-site integration reporter vector, pMBB, modified for lambda site-specific recombination. Elements were selected according to proximity to consensus LysR binding sites and predicted −35 and −10 elements (Fig. 6B). Using RLM-RACE, several TSPs were mapped to a region that supports this predicted promoter structure (Fig. 6B).
FIG. 6.
Analysis of the bicA promoter region in Synechococcus sp. strain PCC 7002. (A) Luciferase activity directed by bicA upstream elements fused to luxAB genes and stably integrated into the genome of Synechococcus sp. strain PCC 7002 cells. Cells were grown at high Ci or with air bubbling for 5 h. Relative luminescence units are expressed on a chlorophyll a basis as a percentage of the value obtained for high-Ci cells containing the empty vector control ± the standard deviation. Asterisks indicate the location of putative LysR-binding motifs. (B) Predicted promoter structure of the bicA upstream region in Synechococcus sp. strain PCC 7002. Putative LysR binding sites are underlined, and putative −35 and −10 elements (as predicted by BPROM at www.softberry.com) are shown in boldface. The TSPs detected by using RLM-RACE are marked with arrowheads; although it is usual to place functional bias on the longest start points relative to the start codon, the shorter products may be genuine alternative TSPs. Note that the first base of the putative −10 box is a G and not the more usual T.
The Ci-responsive luciferase activity of bicA promoter-luxAB transformants was measured (Fig. 6A). The region approximately 500 nucleotides upstream and 200 nucleotides downstream of the bicA start codon directed low-Ci responsive luciferase activity. The reverse orientation of this same element produced a low, constitutive luciferase activity, perhaps from elements contained in the divergently oriented promoter region from the neighboring transglycosylase gene (Fig. 5A). A series of 5′ and 3′ truncations (clones 2, 4, 5, 7, 8, and 12) suggest that the basal promoter sequences and Ci response elements are contained between positions −95 and −239 (clone 12) relative to the bicA start codon (note the difference in activity between clone 12 and clone 8, Fig. 6A). This small region directs inducible luciferase activity at low Ci but not to the same degree as the longest region investigated (−515 to +208), which would appear to contain enhancer elements (compare clone 722F with clone 12). Interestingly, the deletion of two putative LysR binding sites from the 5′ and 3′ regions of clone 12 (yielding clone 10) resulted in an apparent inversion in the Ci responsiveness of this region. That is, clone 10 produced strongly derepressed luciferase activity at high Ci (to levels exceeding those of air-induced clone 12), but this activity was apparently suppressed under Ci limitation (Fig. 6A). However, the luciferase activity in the negative controls (empty vector and 722R) was also repressed to a similar degree at low Ci, suggesting the existence of a generalized repression of luciferase activity under this stress condition. We ascribe this to a general downturn in protein synthesis under Ci limitation.
DISCUSSION
We investigated the organization and regulation of CO2-concentrating mechanism genes in a euryhaline, coastal/estuarine cyanobacterium, Synechococcus sp. strain PCC 7002. It was found that mRNA abundance for high-affinity Ci transporter genes (20) is controlled primarily at the transcriptional level (Fig. 1). We inactivated a gene encoding a LysR transcription factor that resembles the proteobacterial Calvin cycle regulator CbbR and which in phylogenetic analyses is found in a divergent grouping of cyanobacterial CbbR-like factors containing the NdhR/CcmR and CmpR factors from freshwater cyanobacteria (group 2; Fig. 2A). Consistent with an elevated mRNA abundance for medium and high-affinity HCO3− transporter systems (Fig. 3), the mutant strains grown at high Ci had fully induced CCMs at a physiological level (Fig. 2B). Accordingly, since the gene product regulates the transition from a basal CCM to a CCM with high affinity for Ci, we renamed this gene ccmR, for CCM regulator as suggested by Wang et al. (28). We also found that CcmR_7002 regulates a unique 10-gene operon containing the low-affinity HCO3− transporter gene, bicA, and genes for two likely Na+/H+ antiporters that are essential for full HCO3− uptake activity (Fig. 5). The minimal bicA promoter was defined, along with Ci response elements contiguous with consensus LysR binding sites (Fig. 6).
Prior to the present study, little physiological data has been reported for LysR regulators of CCM genes (NdhR/CcmR and CmpR), and there is an apparent complexity of function within the group 2 factors (i.e., both activators and repressors; Fig. 2A). The striking physiological phenotype of the Synechococcus sp. strain PCC 7002 group 2 ΔccmR mutant, in which CCM activity was fully induced at high Ci (reported here for the first time in cyanobacteria), was consistent with the types of genes that were shown to be constitutively upregulated (Fig. 3). Identified gene targets of CcmR repressor function include the known loci for high-affinity HCO3− transporters, sbtA and bicA, as well as that for the presumptive HCO3− porin, porB. Putative LysR binding motifs (TnA{n7/8}TnA, 22) exist within 500 bp of the sbtA, porB, and bicA start codons in the Synechococcus sp. strain PCC 7002 genome (Fig. 6 and 7). Interestingly, bicA was not previously identified as a target for NdhR/CcmR in Synechocystis sp. strain PCC 6803 on microarrays (28), since the gene appears to be constitutively expressed in this strain. Indeed, there is no recognizable bicA-like gene in the genome of the much-studied Synechococcus sp. strain PCC 7942.
FIG. 7.
Putative promoter structure of the sbtA (A), porB (B), and ccmR (C) upstream regions in Synechococcus sp. strain PCC 7002. Putative LysR binding sites are underlined, −35 and −10 elements are in boldface, and predicted TSPs are marked with an arrowhead (as predicted by BPROM).
Owing to the parallel orientation and proximate location of ccmR, we were unable to confirm whether the ndhF3 ndhD3 chpY (cupA) orf133 operon is also a target for CcmR repression, as was found for CcmR_6803 (7, 28). However, LysR factors are often located in proximity to target genes (22), and a tight relationship exists in cyanobacterial genomes between the presence of a ccmR-like gene and specific genes of the NDH-13 complex (see below). These observations are highly suggestive of a functional relationship. From the present study it is unclear whether ccmR is expressed as a monocistronic mRNA or whether this gene is cotranscribed from a single promoter upstream of ccmR together with the ndhF3, ndhD3, chpY (cupA), and orf133 genes under some or all growth conditions. The latter option (cotranscription) is supported by our finding that the region 550 bp upstream of the start codon of ndhF3 does not encode a functional promoter under standard growth conditions (Fig. 4D) and also that there were similar levels of mRNA detected for each ORF in this region at both high Ci and low Ci (Fig. 4A). That mRNA for CcmR (a repressor) may be cotranscribed with part of the CcmR regulon is counterintuitive. LysR repressors are commonly transcribed divergently from neighboring target genes, as is the case for CcmR (NdhR) and Slr1727 in Synechocystis sp. strain PCC 6803 (7). Significantly, a predicted stem-loop structure is found within the ccmR-ndhF3 intergenic region (Fig. 2C). It is possible that this structure is a target for a regulatory mechanism to produce nonstoichiometric levels of mRNA or protein from this region, for example, via antitermination factors for RNA polymerase, or masking of ribosome entry sites. Alternatively, posttranslational regulation of CcmR by a Ci-related signal such as cytosolic HCO3−, production of which is directly linked to the physiological activity of the associated ndhF3 regulon, might constitute a sufficient mechanism to ensure adequate modulation of mRNA for the high-affinity CO2 uptake system. Certainly, mRNA for ccmR and high-affinity Ci uptake systems is initially strongly induced but returns rapidly (60 to 90 min) to a low, constitutive level after Ci depletion (20). In addition, studies with the freshwater strain Synechococcus sp. strain PCC 7942 suggest that proteins encoding the high-affinity Ci uptake systems are only very slowly turned over (29). Hence, functional transporters could persist after an initial burst of transcription. More work is required to resolve this question.
Our finding that bicA is a target for CcmR function in Synechococcus sp. strain PCC 7002 prompted a closer examination of the regulation and function of the bicA genomic region (Fig. 5). An operon of 10 ORFs was defined: it commences with bicA and encodes two likely Na+/H+ antiporters, a CPA1 family member similar to NhaS3 from Synechocystis sp. strain PCC 6803 and a multigene Na+/H+ transporter that we have termed Mnh with components resembling NADH:ubiquinone oxidoreductases. This tripartite clustering of transporter activities is unique among known cyanobacterial genomes; however, some clustering of bicA-like genes with likely Na+/H+ antiporter genes can be discerned in other organisms, including Crocosphaera watsonii WH8501 (GOI 400892570) and Nostoc sp. strain PCC 7120 (GOI 4222050). Our deletion of a region downstream of bicA showed that it encodes factors essential to full HCO3− utilization (Fig. 5D). Although a Ci-responsive mnh-like operon is known in both Synechocystis sp. strain PCC 6803 and Nostoc sp. strain PCC 7120 (5, 28) and implicated in salt stress in Nostoc sp. strain PCC 7120, the potential role of the complex, if any, in HCO3− uptake was uncertain: the Mnh complex may be a primary extrusion mechanism to maintain the Na+ gradient required for HCO3− uptake, or it may harness an existing proton gradient to mitigate the cytosolic alkalinization resulting from HCO3− uptake (the subsequent conversion to and fixing of CO2 would necessitate effective proton exchange systems). This might be particularly significant in the case of BicA, which sustains a relatively high flux rate compared to other inducible systems (20). Our new data (Fig. 5D and E) showing that the genes from the bicA operon are required for maximal HCO3−-uptake activity are consistent with both ideas, and further work is needed to more fully understand the physiological functions encoded by this operon.
We examined the structure of the bicA promoter by fusing a series of upstream elements to genes encoding vibrio luciferase and by measuring the resultant Ci-responsive luciferase activity of transformants. We identified a minimal 144-bp region required for Ci-responsive expression (Fig. 6A), as well as two LysR consensus-binding sites flanking the presumptive −35 and −10 elements that, when deleted, result in derepression of the promoter at high Ci. We speculate that, at high Ci, one or more of these sites ordinarily binds CcmR, suppressing futile expression of the operon, not unlike the situation for control of the ndhF3 promoter in Synechocystis sp. strain PCC 6803 (7). We are currently exploring the nature of any physical interaction between these elements and CcmR_7002 given the strongly positive evidence obtained in the present study that there is a functional link between CcmR and bicA regulation.
Our survey of CbbR-like factors in cyanobacteria revealed several interesting relationships (Fig. 2). All genomes investigated encoded a single CbbR-like protein belonging to a highly conserved family (group 1), which may indicate an essential function such as Calvin cycle gene regulation. Consistent with this idea, group 1 contains the Synechocystis sp. strain PCC 6803 sll0998 locus, which cannot be stably inactivated (7, 15; Price, unpublished). Based on Rubisco subtype (4), and exclusively open-ocean dwelling, the “alpha” strains are not represented in the more divergent group 2. With the exception of Trichodesmium erythraeum, the “beta” strains (freshwater species and some coastal or offshore marine strains) contain one or two group 2 proteins. The group 2 genomes also encode recognizable NDH-13 complexes for high-affinity CO2 transport (4) and, with the exception of Gloeobacter violaceus, all have two or three likely high-affinity HCO3− transporters (4). Conversely, strains that do not contain genes for group 2 proteins lack the genes for a recognizable NDH-13 complex, including Trichodesmium erythraeum. Owing to a habitat typified by fluctuating Ci concentrations, we speculate that group 2 CbbR-like proteins are a habitat-specific adaptation that is related to a requirement for high-affinity CO2 transport. Intriguingly, Synechococcus sp. strain PCC 7942 has no recognizable CcmR-like protein and yet it possess functional transporters encoded by cmpABCD, sbtA, and ndhF3 ndhD3 chpY (cupA) (19). Instead, it contains a single group 2 protein, CmpR, which is believed to function as an activator of the cmp operon, which encodes a high-affinity HCO3− transporter (15). This raises questions about how the CCM is regulated in Synechococcus sp. strain PCC 7942.
In conclusion, we have shown the importance of a “derepression” mechanism for CCM regulation in a coastal/estuarine cyanobacterium. Under growth conditions in which Ci is not limiting, a single LysR transcription factor, CcmR, is essential for strong and coordinate transcriptional repression of multiple genes encoding high-affinity Ci transporters. We hypothesize that under limiting Ci, CcmR is transiently inactivated, even though the mRNA for this factor is still abundant. Further work in this species could profitably focus on the role of metabolite binding (possibly HCO3−) in the regulation of CcmR activity. Our survey of CbbR-like genes in cyanobacterial genomes shows that related genes are present only in the beta cyanobacteria that likely use high-affinity CO2 transport systems. Consequently, we propose that CcmR is a habitat-specific specialization for species that exist in environments subject to widely fluctuating Ci concentrations. Given the divergence of the NdhR/CmpR subgroup of LysR factors in the beta cyanobacteria and the fact that Synechococcus sp. strain PCC 7942 encodes only a more distant form of CcmR, a species-specific approach to characterizing CCM regulation is warranted. Considering the long evolutionary history of the cyanobacteria, it should not be surprising that organisms from differing ecological niches have evolved independent solutions to the problem of optimizing their Ci acquisition systems. The differences we have uncovered in the present study between Synechococcus sp. strain PCC 7002 and the freshwater models are a case in point.
ADDENDUM IN PROOF
Details of the construction of the pMBB reporter plasmid can be found in an online Ph.D. thesis by R. Boyanapalli at www.ohiolink.edu/etd/.
Acknowledgments
L. Tucker, J. Janek, and L. Sheridan provided excellent technical assistance. We also thank Tao Li (Pennsylvania State University) and Jindong Zhao (College of Life Sciences, Peking University) for providing access to the Synechococcus sp. strain PCC 7002 draft genome database.
This study was supported by a Discovery Grant from the Australian Research Council to F.J.W. and G.D.P. (DP0556115) and by grants from the National Institutes of Health (GM31625) and National Science Foundation (MCB-0519743) to D.A.B.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Andersson, C. R., N. F. Tsinoremas, J. Shelton, N. V. Lebedeva, J. Yarrow, H. T. Min, and S. S. Golden. 2000. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 305:527-542. [DOI] [PubMed] [Google Scholar]
- 2.Badger, M. R., K. Palmqvist, and J. W. Yu. 1994. Measurement of CO2 and HCO3− fluxes in cyanobacteria and microalgae during steady-state photosynthesis. Physiol. Plant 90:529-536. [Google Scholar]
- 3.Badger, M. R., and G. D. Price. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54:609-622. [DOI] [PubMed] [Google Scholar]
- 4.Badger, M. R., G. D. Price, B. M. Long, and F. J. Woodger. 2006. The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J. Exp. Bot. 57:249-265. [DOI] [PubMed] [Google Scholar]
- 5.Blanco-Rivero, A., F. Leganés, E. Fernández-Valiente, P. Calle, and F. Fernádez-Piñas. 2005. mrpA, a gene with roles in resistance to Na+ and adaptation to alkaline pH in the cyanobacterium Anabaena sp. PCC 7120. Microbiol. 151:1671-1682. [DOI] [PubMed] [Google Scholar]
- 6.Emlyn-Jones, D., F. J. Woodger, T. J. Andrews, G. D. Price, and S. M. Whitney. 2006. A Synechococcus PCC 7942 ΔccmM (Cyanophyceae) mutant pseudoreverts to air growth without regaining carboxysomes. J. Phycol. 42:769-777. [Google Scholar]
- 7.Figge, R. M., C. Cassier-Chauvat, F. Chauvat, and R. Cerff. 2001. Characterization and analysis of an NAD(P)H dehydrogenase transcriptional regulator critical for the survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol. Microbiol. 39:455-468. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan, A., and L. Reinhold. 1999. CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:539-570. [DOI] [PubMed] [Google Scholar]
- 9.Klughammer, B., D. Sultemeyer, M. R. Badger, and G. D. Price. 1999. The involvement of NAD(P)H dehydrogenase subunits, NdhD3 and NdhF3, in high-affinity CO2 uptake in Synechococcus sp. PCC 7002 gives evidence for multiple NDH-1 complexes with specific roles in cyanobacteria. Mol. Microbiol. 32:1305-1315. [DOI] [PubMed] [Google Scholar]
- 10.Maeda, S., M. R. Badger, and G. D. Price. 2002. Novel gene products associated with NdhD3/D4-containing NDH-1 complexes are involved in photosynthetic CO2 hydration in the cyanobacterium, Synechococcus sp. PCC 7942. Mol. Microbiol. 43:425-435. [DOI] [PubMed] [Google Scholar]
- 11.McGinn, P. J., G. D. Price, R. Maleszka, and M. R. Badger. 2003. Inorganic carbon limitation and light control the expression of transcripts related to the CO2-concentrating mechanism in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 132:218-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohkawa, H., H. B. Pakrasi, and T. Ogawa. 2000. Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC 6803. J. Biol. Chem. 275:31630-31634. [DOI] [PubMed] [Google Scholar]
- 13.Ohkawa, H., G. D. Price, M. R. Badger, and T. Ogawa. 2000. Mutation of ndh genes leads to inhibition of CO2 uptake rather than HCO3− uptake in Synechocystis sp. strain PCC 6803. J. Bacteriol. 182:2591-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohkawa, H., M. Sonoda, H. Katoh, and T. Ogawa. 1998. The use of mutants in the analysis of the CO2-concentrating mechanism in cyanobacteria. Can. J. Bot. 76:1035-1042. [Google Scholar]
- 15.Omata, T., S. Gohta, Y. Takahashi, Y. Harano, and S. Maeda. 2001. Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J. Bacteriol. 183:1891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omata, T., G. D. Price, M. R. Badger, M. Okamura, S. Gohta, and T. Ogawa. 1999. Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechococcus sp. strain PCC 7942. Proc. Natl. Acad. Sci. USA 96:13571-13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porra, P. J. 1990. A simple method for extracting chlorophylls from the recalcitrant alga, Nannochloris atomus, without formation of spectroscopically different magnesium-rhodochlorin derivatives. Biochim. Biophys. Acta 1019:137-141. [Google Scholar]
- 18.Price, G. D., and M. R. Badger. 1989. Ethoxyzolamide inhibition of CO2 uptake in the cyanobacterium Synechococcus PCC 7942 without apparent inhibition of internal carbonic anhydrase activity. Plant Physiol. 89:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price, G. D., S. Maeda, T. Omata, and M. R. Badger. 2002. Modes of active inorganic carbon uptake in the cyanobacterium Synechococcus sp. PCC 7942. Funct. Plant Biol. 29:131-149. [DOI] [PubMed] [Google Scholar]
- 20.Price, G. D., F. J. Woodger, M. R. Badger, S. M. Howitt, and L. Tucker. 2004. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc. Natl. Acad. Sci. USA 101:18228-18233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakamoto, T., G. Z. Shen, S. Higashi, N. Murata, and D. A. Bryant. 1998. Alteration of low-temperature susceptibility of the cyanobacterium Synechococcus sp. PCC 7002 by genetic manipulation of membrane lipid unsaturation. Arch. Microbiol. 169:20-28. [DOI] [PubMed] [Google Scholar]
- 22.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 23.Shibata, M., H. Katoh, M. Sonoda, H. Ohkawa, M. Shimoyama, H. Fukuzawa, A. Kaplan, and T. Ogawa. 2002. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: function and phylogenetic analysis. J. Biol. Chem. 277:18658-18664. [DOI] [PubMed] [Google Scholar]
- 24.Shibata, M., H. Ohkawa, T. Kaneko, H. Fukuzawa, S. Tabata, A. Kaplan, and T. Ogawa. 2001. Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc. Natl. Acad. Sci. USA 98:11789-11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sültemeyer, D., G. Amoroso, and H. Fock. 1995. Induction of intracellular carbonic anhydrases during the adaptation to low inorganic carbon concentrations in wild-type and ca-1 mutant cells of Chlamydomonas reinhardtii. Planta 196:217-224. [Google Scholar]
- 26.Sültemeyer, D., B. Klughammer, M. Ludwig, M. R. Badger, and G. D. Price. 1997. Random insertional mutagenesis used in the generation of mutants of the marine cyanobacterium Synechococcus sp. strain PCC 7002 with an impaired CO2 concentrating mechanism. Aust. J. Plant Physiol. 24:317-327. [Google Scholar]
- 27.Swartz, T. H., S. Ikewada, O. Ishikawa, M. Ito, and T. A. Krulwich. 2005. The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles 9:345-354. [DOI] [PubMed] [Google Scholar]
- 28.Wang, H. L., B. L. Postier, and R. L. Burnap. 2004. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 279:5739-5751. [DOI] [PubMed] [Google Scholar]
- 29.Woodger, F. J., M. R. Badger, and G. D. Price. 2003. Inorganic carbon limitation induces transcripts encoding components of the CO2-concentrating mechanism in Synechococcus sp. PCC 7942 through a redox-independent pathway. Plant Physiol. 133:2069-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodger, F. J., M. R. Badger, and G. D. Price. 2005. Sensing of inorganic carbon limitation in Synechococcus PCC 7942 is correlated with the size of the internal inorganic carbon pool and involves oxygen. Plant Physiol. 139:1959-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]