Abstract
Mycoplasma hyopneumoniae mhp379 is a putative lipoprotein that shares significant amino acid sequence similarity with a family of bacterial thermostable nucleases. To examine the nuclease activity of mhp379, the gene was cloned and expressed in Escherichia coli following the deletion of the amino-terminal signal sequence and prokaryotic lipoprotein cleavage site and mutagenesis of the mycoplasma TGA tryptophan codons to TGG. The recombinant fusion protein yielded a 33-kDa thrombin cleavage product, corresponding in size to the mature mhp379 protein. Exonuclease activity was indicated by agarose gel electrophoresis analysis of the reaction products that were released when different nucleic acid substrates were used. Endonuclease activity was also indicated by the digestion of closed circular plasmid DNA. The recombinant mhp379 fusion protein completely digested single-stranded DNA, double-stranded DNA (dsDNA), and RNA. The optimal reaction conditions were determined with a novel nuclease assay based on the enhancement of fluorescence of SYBR green I bound to dsDNA. Optimal activity was observed in the presence of calcium ions at a concentration of 15 mM and a pH of 9.5. No nuclease activity was observed in the absence of calcium ions. Mycoplasmas do not have the ability to synthesize nucleic acid precursors, and thus, nucleases are likely to be important in the acquisition of precursors for the synthesis of nucleic acids. Homologs of an ATP-binding cassette (ABC) transport system were identified immediately downstream of the gene encoding mhp379, and two homologs of M. pneumoniae lipoprotein multigene family 2 were also identified immediately upstream. Homologs of mhp379 were identified in the sequenced genomes of a number of mycoplasma species, and in most cases the homologous ABC transport system was identified immediately downstream of the homologous gene; in several cases a homolog of M. pneumoniae lipoprotein multigene family 2 was also identified immediately upstream. These observations suggest that mhp379 comprises part of a conserved ABC transport operon in mycoplasmas and that the exonuclease activity of mhp379 may be associated with the conserved function of the ABC transport system in the import of nucleic acid precursors. This is the first study to identify the gene and characterize the activity of a mycoplasma exonuclease.
Neither the genes nor the enzymes for the biosynthesis of nucleic acid precursors in mycoplasmas have been identified (5, 21, 24, 34, 42, 46, 47, 51, 57, 59). However, genomic sequence analyses of a number of mycoplasma species have identified novel enzymes associated with salvage pathways for the biosynthesis of nucleic acids from nucleotides (5, 21, 24, 34, 42, 44, 51, 57-59). These salvage pathways have been functionally characterized in Mycoplasma mycoides subspecies mycoides (35-37, 58). Mycoplasma nucleases are thought to play a metabolic role in the production of nucleotide substrates from host or microbial nucleic acids released through natural and induced cell death. These nucleotide substrates may be produced following the import of small oligonucleotides into the cytoplasm or through the extracellular activity of nucleases attached to or secreted from the cell surface. One or more transport mechanisms for the import of intact exogenous nucleotides have been identified in M. mycoides subspecies mycoides (38, 39, 60). Membrane-associated nuclease activity has been identified in all mycoplasma species studied so far (33), and a comparatively large number of mycoplasmas have additionally been shown to produce extracellular nuclease activity (33).
The identification of intracellular, extracellular, and membrane-associated nuclease activities in a number of mycoplasma species suggests the involvement of nucleases in a variety of cellular processes (2, 25, 33, 45). This is supported by the identification of one or more nucleases in most mycoplasma species studied so far (33) and the concomitant identification of multiple genes encoding putative nucleases in genomic sequence analyses (5, 21, 24, 34, 42, 51, 57, 59). However, many of these nucleases do not appear to be conserved between mycoplasma species, based on differences in size and reaction conditions (2, 25, 33, 45). In particular, these nucleases differ in the requirement for specific divalent cations, which is an important criterion in their classification (6). Early studies of the biochemical properties of a number of mycoplasma nucleases used crude lysates of whole cells that contained multiple nuclease activities (43). For Mycoplasma pulmonis, the biochemical properties of membrane-associated nucleases have been partially characterized (25, 43) and the gene encoding the nuclease that is reportedly associated with the majority of this membrane-associated activity, designated mnuA, has been cloned and expressed (25). Only the major membrane-associated nuclease of Mycoplasma penetrans has been purified to homogeneity and the biochemical properties completely characterized (2). The nucleases of M. penetrans and M. pulmonis displayed endonuclease activity that was strictly dependent on the presence of calcium and/or magnesium ions (2, 43). Calcium and magnesium ion-dependent endonuclease activity has also been found in a number of other mycoplasma species (43). While the mnuA membrane nuclease gene of M. pulmonis encodes a putative prokaryotic lipoprotein cleavage site (25), preliminary studies have failed to identify a nuclease of M. pulmonis that is covalently modified by the fatty acid palmitate (33). Homologs of mnuA in a number of other mycoplasma species, including Mycoplasma hyopneumoniae, have been identified by nucleic acid hybridization and genomic sequence analyses (5, 25, 34, 42, 51).
In addition to their importance in the life cycle of mycoplasmas, nucleases attached to or secreted from the cell surface have been implicated as potential pathogenic determinants. Apoptotic changes characterized by the internucleosomal fragmentation of chromatin in epithelial cells cultured in the presence of Mycoplasma hyorhinis were due to the activity of a calcium and magnesium ion-dependent endonuclease (40, 41). A 40-kDa calcium and magnesium ion-dependent endonuclease of M. penetrans has also been shown to induce similar apoptotic changes in cultured lymphocytes (3). The activity of a family of phase-variable site-specific restriction endonucleases is reportedly associated with the colonization of the lower respiratory tract of infected rats by M. pulmonis (12, 17, 54). While site-specific restriction endonucleases are known to be important in protecting against the entry of foreign nucleic acids, the phase-variable restriction endonucleases of M. pulmonis appear to have an alternate, and as-yet-unknown, function (17). Putative restriction endonucleases in a number of mycoplasma genomes have been annotated (21, 24, 59), and a restriction endonuclease has been isolated from Mycoplasma fermentans (18). The indirect role of nucleases in both the life cycle and pathogenesis of mycoplasmas is a recurring theme in studies of mycoplasma-host interactions (49).
The aim of this study was to examine the function of M. hyopneumoniae mhp379. mhp379 is a putative lipoprotein with amino acid sequence similarity to a family of thermostable nucleases identified in other bacteria. The gene encoding mhp379 was cloned and expressed in Escherichia coli, and the nuclease activity, substrate specificity, and biochemical properties of the purified recombinant glutathione S-transferase (GST)-mhp379 protein were examined.
MATERIALS AND METHODS
Bacterial strains and media.
M. hyopneumoniae strains LKR (29) and Adelaide Beaufort (13) were grown at 37°C in Friis broth or on Friis agar (15). Cells were harvested at the late logarithmic phase of growth. Plasmid-transformed E. coli JM109 cells were grown at 37°C in Luria-Bertani broth or on Luria-Bertani agar containing 50 μg of ampicillin/ml (50).
Triton X-114 phase fractionation.
Triton X-114 (TX-114) detergent-soluble M. hyopneumoniae proteins were prepared as described previously (10) with some modifications. M. hyopneumoniae cells were resuspended in 0.5% (vol/vol) TX-114 (Sigma Aldrich) in phosphate-buffered saline (PBS) and incubated for 1 h at 4°C. Insoluble material was removed by centrifugation, and the supernatant was carefully added to a sucrose solution (0.6% [wt/vol] sucrose and 0.06% [vol/vol] TX-114 in PBS) and incubated for 10 min at 37°C. The solution was centrifuged at 37°C, and the supernatant containing the hydrophilic proteins was collected. Hydrophobic proteins in the TX-114 pellet were harvested by centrifugation following the addition of at least 3 volumes of a 1:4 mixture of chloroform and methanol. The pellet containing the hydrophobic proteins was dried under vacuum and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for analysis by SDS-PAGE and Western immunoblotting.
Trypsin treatment.
Intact M. hyopneumoniae cells were treated with trypsin to partially digest cell surface proteins, as described previously (10). A 20-ml culture of M. hyopneumoniae was resuspended in Tris-buffered saline (TBS) (50 mM Tris and 145 mM NaCl [pH 7.4]) and divided into five equal aliquots. Trypsin (Sigma Aldrich) was prepared fresh in TBS buffer at concentrations of 64, 16, 4, 1, and 0.25 μg/ml, and an equal volume of each dilution was added to each aliquot of cells. Samples were incubated for 30 min at 37°C, and trypsin digestion was stopped by the addition of soybean trypsin inhibitor (Sigma Aldrich) to a final concentration of 0.06% (wt/vol). Cells were harvested and resuspended in SDS-PAGE sample buffer for analysis by SDS-PAGE and Western immunoblotting.
SDS-PAGE and Western immunoblotting.
SDS-PAGE was performed as described previously (11, 28) with the Bio-Rad Mini Protean II gel system, according to the manufacturer's instructions. For Western immunoblotting, proteins were transferred from the electrophoresed gel to a nitrocellulose membrane (Hybond-C; GE Healthcare) with the Bio-Rad Western transfer apparatus, according to the manufacturer's instructions. The immunological detection of the transferred proteins was performed as described previously (11) with some modifications. Nitrocellulose membranes were incubated for at least 2 h in 10% (wt/vol) skim milk (Difco) and washed in 0.5% (vol/vol) Tween 20 (Sigma Aldrich) in PBS (PBS-T) between each step. The primary antibody and horseradish peroxidase (HRP)-conjugated secondary antibody were diluted 1:500 and 1:2,000, respectively, in PBS-T and incubated with the nitrocellulose membranes for 1 h. The HRP-conjugated secondary antibody was detected with FAST 3,3′-diaminobenzidine tablet sets (Sigma Aldrich) or the ECL detection kit (GE Healthcare), according to the manufacturers' instructions. The optimal dilution of primary antibody was determined by titration. HRP-conjugated secondary antibodies used in this study were rabbit anti-rat (Dako) and goat anti-pig (Kirkegaard & Perry Laboratories) immunoglobulins.
DNA cloning, sequencing, and expression of the mhp379 gene.
Genomic DNA was purified from M. hyopneumoniae as described previously (30). The DNA sequence of the mhp379 gene from M. hyopneumoniae strain 232 (GenBank accession no. NC006360) was used to design oligonucleotide primers for the PCR amplification of mhp379 from strain LKR. The mhp379 gene was amplified downstream of the predicted prokaryotic lipoprotein cleavage site, and the mycoplasma TGA tryptophan codons were mutagenized to TGG by overlap extension PCR to enable the expression of the truncated mhp379 sequence in E. coli. Briefly, separate PCRs were performed with 1 μl of genomic DNA template for each of the oligonucleotide primer pairs BAMH1F-1WR, 1WF-2WR, and 2WF-SAL1R (Table 1) in a reaction volume of 50 μl containing 2 mM MgSO4, 100 μM concentrations of each deoxynucleoside triphosphate, 0.4 μM concentrations of each primer, and 1.5 U of Platinum Taq thermopolymerase (Life Technologies Inc.). Touchdown PCRs were performed in a thermocycler (Hybaid) under the following conditions: for primer pair BAMH1F-1WR, 95°C for 5 min, then 18 cycles of 95°C for 1 min, 62.5°C lowered to 52.5°C (approximately 1.25°C every two cycles) for 1 min, and 68°C for 2 min, followed by 25 cycles of 95°C for 1 min, 52.5°C for 1 min, and 68°C for 2 min, with a final extension at 68°C for 10 min; and for primer pairs 1WF-2WR and 2WF-SAL1R, 95°C for 5 min, then 18 cycles of 95°C for 1 min, 65°C lowered to 55°C (approximately 1.25°C every two cycles) for 1 min, and 68°C for 2 min followed by 25 cycles of 95°C for 1 min, 55°C for 1 min, and 68°C for 2 min, with a final extension at 68°C for 10 min. The PCR products were resolved by agarose gel electrophoresis and purified with the QIAquick gel extraction kit (QIAGEN) according to the manufacturer's instructions. Approximately equimolar amounts of all three PCR products were used as templates in an overlap extension touchdown PCR with the oligonucleotide primer pair BAMH1F-SAL1R (Table 1) under the following conditions: 95°C for 5 min, then 18 cycles of 95°C for 1 min, 65°C lowered to 55°C (approximately 1.25°C every two cycles) for 1 min, and 68°C for 2 min, followed by 25 cycles of 95°C for 1 min, 55°C for 1 min, and 68°C for 2 min, with a final extension at 68°C for 10 min. The PCR product was purified with the QIAquick PCR purification kit (QIAGEN), digested with BamHI and SalI, and ligated into the expression vector pGEX-4T-1 (GE Healthcare) according to the manufacturer's instructions. E. coli JM109 cells were transformed with the ligation mixture, and clones containing the mutagenized mhp379 gene were selected. The pGEX-4T-1 plasmid construct containing the mutagenized mhp379 gene was purified with the Plasmid Midi kit (QIAGEN) according to the manufacturer's instructions and was used as a template in DNA sequencing reactions using the BigDye Terminator cycle sequencing kit (Applied Biosystems) and the oligonucleotides pGEXfwd and pGEXrev (Table 1). The expression of the recombinant mhp379 fusion protein in E. coli was induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 2 mM. The recombinant GST-mhp379 protein was purified by affinity chromatography with a glutathione-Sepharose column (GE Healthcare) and dialyzed against PBS overnight at 4°C. The GST fusion partner was removed by enzymatic cleavage with thrombin (GE Healthcare) according to the manufacturer's instructions. The thrombin cleavage products were analyzed by SDS-PAGE.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′)a |
|---|---|
| BAMH1F | TATTggatccTATCAAAATGAAGCAACTCG |
| 1WR | GTTTATCTTGTTGAGTcCATTGATAATTTTG |
| 1WF | GATCAAAATTATCAATGgACTCAACAAGATAAAC |
| 2WR | GTATCTCCATCTTTcCATTCAACAATTTCGGC |
| 2WF | GCCGAAATTGTTGAATGgAAAGATGGAGATAC |
| SAL1R | TAGgtcgacTATTTTTTTTCCTTTAATTCTCGACC |
| pGEXfwd | GGGCTGGCAAGCCACGTTTGGTG |
| pGEXrev | CCGGGAGCTGCATGTGTCAGAGG |
Lowercase letters indicate nucleotide differences from the published sequences. These changes were introduced to create a restriction endonuclease cleavage site or to mutate a TGA tryptophan codon to TGG. Boldface letters indicate an introduced in-frame stop codon.
Preparation of antiserum.
The thrombin-cleaved recombinant mhp379 protein was used to immunize rats, as described previously (20). Briefly, thrombin-cleaved recombinant mhp379 was separated by SDS-PAGE and stained with 0.05% (wt/vol) Coomassie brilliant blue until the corresponding band was visible. The stained thrombin-cleaved recombinant mhp379 protein band was excised, and the polyacrylamide gel slice was emulsified in an equal volume of Freund's complete adjuvant and injected subcutaneously into PVG/c rats. Three further booster immunizations were given at 1-month intervals with the thrombin-cleaved recombinant mhp379 polyacrylamide gel slice emulsified in Freund's incomplete adjuvant. Serum was collected 10 days after each booster, and the titer was assessed by Western immunoblotting. Antibodies were purified by affinity chromatography with a protein L agarose column (Sigma Aldrich), as described previously (20), and sterilized by passage through a 0.22-μm filter. Immune serum was heat inactivated by incubation at 56°C for 20 min.
Polyclonal rabbit antiserum raised against M. hyopneumoniae strain LKR whole cells was obtained from A. Kanci (School of Veterinary Science, The University of Melbourne, Melbourne, Australia). Polyclonal antiserum from a pig infected with M. hyopneumoniae strain Adelaide Beaufort was obtained from T. Czaja (School of Veterinary Science, The University of Melbourne, Melbourne, Australia) and has been described previously (8). Polyclonal rabbit antiserum raised against purified recombinant GST was obtained from C.-J. Chiu (School of Veterinary Science, The University of Melbourne, Melbourne, Australia).
Assays for nuclease activity.
The nuclease activity of recombinant GST-mhp379 was analyzed by agarose gel electrophoresis. Approximately 4 μg of recombinant GST-mhp379 was incubated at 37°C in 100-μl nuclease reaction buffer (25 mM Tris-HCl [pH 8.8] and 10 mM CaCl2) containing 1 to 5 μg of nucleic acid substrate. A 10-μl aliquot was removed at different time intervals, and EDTA was added to a final concentration of 20 mM. The reaction products were analyzed by 1% agarose gel electrophoresis, and the DNA was visualized by staining with ethidium bromide. Exonuclease and endonuclease activities were analyzed with double-stranded (ds) λ phage DNA (New England BioLabs) and closed circular plasmid DNA (pGEX-4T-1 containing the mutagenized mhp379 gene purified as described above [pGEX-mhp379]) as substrates, respectively. Substrate specificity was analyzed with single-stranded (ss) M13 phage DNA purified according to the method of Sambrook et al. (50), and total RNA was purified from E. coli with the RNeasy RNA purification kit (QIAGEN). The mode of nuclease action was further analyzed by using a DNA substrate that was PCR amplified in the presence of a 5′ digoxigenin (DIG)-labeled primer that was obtained from C.-W. Tseng (School of Veterinary Science, The University of Melbourne, Melbourne, Australia). The reaction products were subjected to 1% agarose gel electrophoresis and Southern transferred to a nylon membrane (Hybond-N+) (GE Healthcare), as described previously (50). The DIG-labeled reaction products were detected with the DIG chemiluminescence detection kit (Roche) and autoradiographed with Biomax film (Kodak) according to the manufacturers' instructions. GST was expressed and affinity purified from E. coli transformed with pGEX-4T-1, as described above, and used as a negative control in all assays.
The optimal nuclease reaction conditions were determined with a real-time nuclease assay based on the enhancement of fluorescence of SYBR green I bound to dsDNA. Approximately 4 μg of recombinant GST-mhp379 was added to 50 μl of nuclease reaction buffer containing 1× SYBR green I (Molecular Probes) and 50 ng of herring sperm DNA (Roche), and the mixture was incubated at 37°C in an Mx3000P fluorescence detection PCR instrument (Stratagene). Fluorescence data were collected every 1 min. The optimal nuclease reaction conditions were determined by comparing the initial reaction velocity, as measured by the relative change in fluorescence under different experimental conditions. This nuclease assay was used to examine the effects of pH, ionic strength (sodium and potassium ions), concentration of divalent cations (calcium and magnesium), and temperature. EDTA was added to a final concentration of 5 mM to examine the nuclease activity of recombinant GST-mhp379 in the absence of exogenously supplied divalent cations. The effect of temperature was examined in terms of both the nuclease activity and the stability of recombinant GST-mhp379 at different temperatures. The temperature stability was examined by first incubating the reaction mixture for 10 min at the designated temperature in the absence of the DNA substrate. All reactions were repeated in triplicate. GST was prepared as described above and used as a negative control in all assays.
Sequence analysis.
Amino acid sequence similarity searches were performed with BLASTP (1). ClustalX was used to align similar amino acid sequences. Putative amino-terminal signal sequences and prokaryotic lipoprotein cleavage sites were identified with SignalP (4) and LipoP (26), respectively. The TNASE_3 thermonuclease domain profile was identified by searching the PROSITE database (14).
RESULTS
Cloning and expression of the gene encoding mhp379.
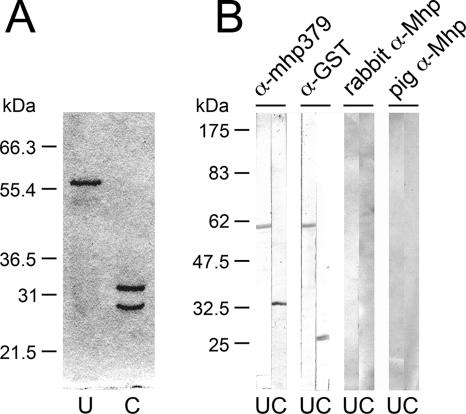
Two TGA tryptophan codons identified by sequence analysis of the mhp379 gene in M. hyopneumoniae strain 232A were mutagenized to TGG by overlap extension PCR. The mutagenized mhp379 gene encoding the mature protein immediately downstream of the cysteine residue of the prokaryotic lipoprotein cleavage site was cloned into the expression vector pGEX-4T-1. SDS-PAGE analysis of the expressed recombinant GST-mhp379 protein identified a band of approximately 59 kDa (Fig. 1A, lane U). Thrombin cleavage of recombinant GST-mhp379 produced two bands of approximately 26 and 33 kDa (Fig. 1A, lane C), corresponding in size to the GST fusion partner and the mhp379 protein fragment, respectively. Western immunoblotting using rat antiserum against thrombin-cleaved recombinant mhp379 identified proteins corresponding in size to the expressed recombinant GST-mhp379 (59 kDa) and thrombin-cleaved mhp379 (33 kDa) proteins (Fig. 1B, α-mhp379 lanes U and C). Conversely, while rabbit antiserum against GST also detected the expressed recombinant GST-mhp379 protein (59 kDa), it detected only the GST fusion partner (26 kDa) in the thrombin cleavage products (Fig. 1B, α-GST lanes U and C). Western immunoblotting using rabbit antiserum against M. hyopneumoniae whole cells and pig antiserum against live M. hyopneumoniae did not bind the expressed recombinant GST-mhp379 protein or thrombin cleavage products (Fig. 1B, rabbit α-Mhp and pig α-Mhp lanes U and C).
FIG. 1.
SDS-PAGE analysis and Western immunoblotting of recombinant GST-mhp379 expressed in E. coli and purified by affinity chromatography. (A) Coomassie brilliant blue-stained gel of recombinant GST-mhp379 (lane U) and thrombin-cleaved recombinant GST-mhp379 (lane C) fusion products separated by SDS-10% PAGE with molecular mass markers (Novex). (B) Western blots of uncleaved (lanes U) and thrombin-cleaved (lanes C) recombinant GST-mhp379 probed with rat antiserum raised against gel-purified thrombin-cleaved recombinant mhp379 (α-mhp379), rabbit antiserum raised against GST (α-GST), rabbit antiserum raised against M. hyopneumoniae strain LKR whole-cell proteins (rabbit α-Mhp), and antiserum from a pig infected with M. hyopneumoniae strain Adelaide Beaufort (pig α-Mhp). Proteins were separated by SDS-10% PAGE with prestained molecular mass markers (New England BioLabs) and Western transferred.
Characterization of the expression of mhp379 in M. hyopneumoniae.
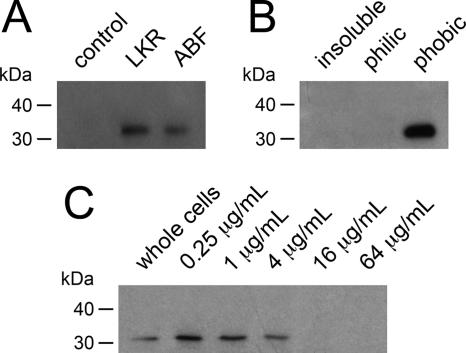
While rabbit and pig antiserum against M. hyopneumoniae did not bind the recombinant mhp379 protein, rat antiserum against recombinant mhp379 identified a 33-kDa protein in Western immunoblots of M. hyopneumoniae strains LKR and Adelaide Beaufort (Fig. 2A). The 33-kDa protein partitioned exclusively into the hydrophobic phase following TX-114 fractionation (Fig. 2B) and was also shown to be susceptible to proteolytic cleavage after the treatment of intact M. hyopneumoniae cells with increasing concentrations of trypsin (Fig. 2C).
FIG. 2.
Analysis of the expression of mhp379 in M. hyopneumoniae by Western immunoblotting using rat antiserum raised against gel-purified thrombin-cleaved recombinant mhp379. (A) Whole-cell proteins of M. hyopneumoniae strains LKR and Adelaide Beaufort (ABF). Whole-cell proteins of M. hyopneumoniae strains LKR were used in a control reaction performed in the absence of rat antiserum raised against thrombin-cleaved recombinant mhp379 (control). (B) TX-114-fractionated proteins of M. hyopneumoniae strain LKR. TX-114-insoluble proteins (insoluble), hydrophilic-phase proteins (philic), and hydrophobic-phase proteins (phobic) are shown. (C) Trypsin treatment of M. hyopneumoniae strain LKR whole cells. From left to right are untreated whole cells and cells treated with 0.25, 1, 4, 16, or 64 μg of trypsin/ml. All proteins were separated by SDS-10% PAGE with molecular mass markers (Cell Signaling Technology).
Sequence analysis.
The cloned mhp379 gene fragment was 855 bp in length and coded for a mature peptide of 285 amino acids with a predicted molecular mass of 33.2 kDa. The predicted molecular mass was consistent with that determined by SDS-PAGE (Fig. 1A, lane C). Two substitutions were identified in the nucleotide sequence of the cloned mhp379 gene fragment from M. hyopneumoniae strain LKR compared to the published nucleotide sequence of mhp379 from strain 232. One of these substitutions was in the third position of the codon and was not associated with a change in the translated amino acid residue. The other nucleotide substitution changed the translated amino acid residue from aspartate to asparagine at position 28 of the mature peptide sequence.
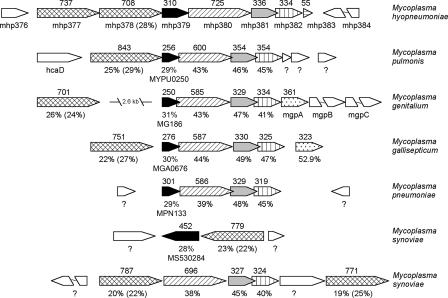
In M. hyopneumoniae, the gene encoding mhp379 is located immediately upstream of three genes that comprise the four domains of an ATP-binding cassette (ABC) transport system (Fig. 3). The two ATP-binding domains that couple ATP hydrolysis to transport are predicted to be encoded by one gene, mhp380, and the two transmembrane domains that are associated with the passage of the substrate into the cell are encoded by two separate genes, mhp381 and mhp382. The amino acid sequence of mhp380 reportedly shows similarity to the PotA spermidine/putrescine ATP-binding protein, and mhp381 and mhp382 show similarity to the UgpA and UgpE glycerol-3-phosphate transport system permease proteins, respectively. It is important to note that this similarity is based on the presence of conserved ABC transporter family signature sequences and conserved domains and that significant sequence similarity could also be identified with almost every other class of ABC transporter. Two paralogous genes encoding mhp378 and mhp377 are located immediately upstream of mhp379 and are homologs of M. pneumoniae lipoprotein multigene family 2 (19). mhp378 is a membrane-associated protein that is immunogenic in pigs infected with M. hyopneumoniae (31). The hypothetical protein mhp383 has been annotated only in the complete genome of M. hyopneumoniae strain 232, not in strain J or strain 7448 (57). However, the nucleotide sequence of the region between mhp377 and mhp383 in M. hyopneumoniae strain 232 is nearly identical to that of the homologous regions in strains J and 7448 (data not shown). The differences in the reported sizes of the homologous proteins in M. hyopneumoniae strains J and 7448 are due to differences in the predicted translational initiation sites.
FIG. 3.
Physical map of mycoplasma homologs of the putative mhp379-mhp380-mhp381-mhp382 ABC transport system. Homologous proteins are shaded in the same pattern, and their amino acid identities to the respective M. hyopneumoniae strain LKR proteins are indicated below each row. Amino acid identities indicated in parentheses refer to mhp377. Amino acid identity was determined by BLASTP search of the GenBank nonredundant bacterial database. Numbers above orthologous genes indicate the size of the product expressed as the number of amino acid residues. Hypothetical or unknown proteins are indicated by a question mark (?).
A BLASTP search of the GenBank database identified homologs of mhp379 in M. pulmonis (MYPU0250), Mycoplasma genitalium (MG186), Mycoplasma gallisepticum (MGA0676), Mycoplasma pneumoniae (MPN133), and Mycoplasma synoviae (MS530284). A homolog of mhp379 could not be identified in the sequenced genomes of Mycoplasma capricolum, M. mobile, M. mycoides subspecies mycoides SC, or M. penetrans. However, nuclease activity in M. capricolum and M. penetrans has been identified previously (2, 33). No sequence similarity could be identified between mhp379 and the MnuA major membrane nuclease of M. pulmonis (MYPU6930). All mycoplasma homologs of mhp379 contained a hydrophobic amino-terminal signal sequence and prokaryotic lipoprotein cleavage site. More importantly, all mycoplasma homologs of mhp379, with the exception of M. synoviae MS530284, were located immediately upstream of homologs of the mhp380-mhp381-mhp382 ABC transport system, and most of the genes encoding the conserved putative mhp379-mhp380-mhp381-mhp382 operon were partially overlapping (Fig. 3). While M. synoviae MS530284 is also located immediately downstream of a homolog of mhp377-mhp378, the mhp380-mhp381-mhp382 ABC transport system homologs are located approximately 59 kb from these genes (Fig. 3). Homologs of mhp377-mhp378 have also been identified upstream and downstream of the mhp380-mhp381-mhp382 ABC transport system homologs in M. synoviae (Fig. 3).
A search for protein family signature sequences and conserved domains in the PROSITE database showed that the region between amino acid positions 94 and 255 in mhp379 had significant identity and similarity with the thermonuclease domain profile (TNASE_3). The TNASE_3 thermonuclease domain profile was first described for the Nuc thermonuclease of Staphylococcus aureus (55) and was found to be conserved in all of the mycoplasma homologs of mhp379. All mycoplasma homologs of mhp379 belong to the micrococcal nuclease (thermonuclease) cluster of orthologous proteins (COG1525). Multiple sequence alignments indicated that much of the similarity between mycoplasma homologs of mhp379 was associated with the identification of the TNASE_3 thermonuclease domain profile (Fig. 4). More importantly, the amino acid residues aspartate, aspartate, and tyrosine, located in mhp379 at positions 108, 132, and 133, respectively, were strictly conserved. These residues are reported to be involved in the binding of calcium ions (23). The amino acid residues arginine, glutamate, and arginine, located in mhp379 at positions 127, 135, and 188, respectively, are also strictly conserved and comprise the active catalytic site (23). In comparison to other mycoplasma homologs of mhp379, M. synoviae MS530284 had an amino-terminal extension of approximately 149 amino acids.
FIG. 4.
Multiple sequence alignment of M. hyopneumoniae strain LKR mhp379 with homologous proteins identified in other mycoplasma species and the Nuc thermonuclease of Staphylococcus aureus. The complete amino acid sequence of each homologous protein downstream of the putative prokaryotic lipoprotein cleavage site is shown. The sequence of M. synoviae MS530284 from 149 amino acid residues downstream of the putative prokaryotic lipoprotein cleavage site is shown. Numbers on the right indicate the position of the adjacent amino acid residue. Identical amino acids are shaded in gray. An asterisk (*) indicates an amino acid that is conserved in all aligned sequences, a colon (:) indicates that there is at least one substitution with a very similar amino acid, and a full stop (.) indicates that there is at least one substitution with a similar amino acid. Dashed lines (—) indicate gaps in the amino acid sequence alignment. The TNASE_3 thermonuclease domain profile identified by searching the PROSITE database is underlined. The conserved amino acid residues aspartate, aspartate, and tyrosine, located in mhp379 at positions 108, 132, and 133, respectively, are believed to be involved in the binding of calcium ions and are shown in boldface. The conserved active-site residues arginine, glutamate, and arginine, located in mhp379 at positions 127, 135, and 188, respectively, are also shown in boldface.
Nuclease activity of recombinant GST-mhp379.
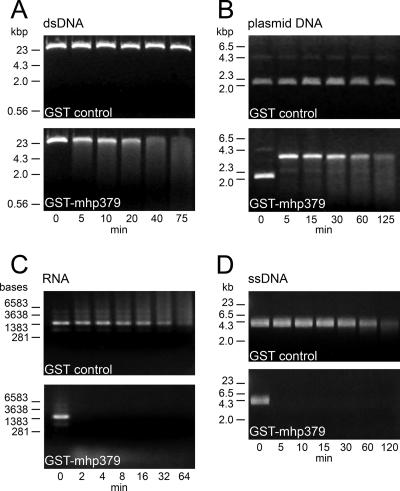
Recombinant GST-mhp379 was incubated in solutions containing different nucleic acid substrates, and the reaction products were analyzed by agarose gel electrophoresis. In the presence of recombinant GST-mhp379, the amount of linear dsDNA substrate reduced significantly over time; this was associated with the appearance of a DNA smear (Fig. 5A). The size of the DNA smear decreased as the reaction time increased, and this occurred more rapidly when the reaction was repeated with a higher concentration of recombinant GST-mhp379 (data not shown). This indicated that recombinant GST-mhp379 had exonuclease activity, and this was confirmed by the concomitant increase in the relative absorbance at 260 nm (27) (data not shown). The endonuclease activity of recombinant GST-mhp379 was demonstrated by using closed circular plasmid DNA as the substrate. In the presence of recombinant GST-mhp379, the plasmid DNA was cleaved into an open circular form and was subsequently digested over time in association with the appearance of a DNA smear (Fig. 5B). Recombinant GST-mhp379 thus displays an initial endonuclease nicking activity against closed circular plasmid DNA. The substrate specificity of recombinant GST-mhp379 was further analyzed by using RNA and ssDNA as substrates. Both the RNA and ssDNA substrates were completely digested after only 2 and 5 min of incubation in the presence of 4 μg of recombinant GST-mhp379, respectively (Fig. 5C and D). In comparison, under the same reaction conditions, it took more than 20 min before the dsDNA substrate was completely digested (Fig. 5A). Recombinant GST-mhp379 thus appears to be far more active against RNA and ssDNA than against dsDNA. No nuclease activity was observed in parallel experiments wherein GST was substituted for recombinant GST-mhp379 (Fig. 5). Rat antiserum against recombinant mhp379 did not appear to inhibit the nuclease activity of recombinant GST-mhp379, as observed by agarose gel electrophoresis and confirmed with the SYBR green I real-time nuclease assay (data not shown).
FIG. 5.
Nuclease activity and substrate specificity of recombinant GST-mhp379. Approximately 4 μg of recombinant GST-mhp379 (GST-mhp379) was incubated with phage λ dsDNA (A), closed circular plasmid DNA (plasmid DNA [pGEX-mhp379]) (B), total RNA isolated from E. coli (RNA) (C), or phage M13 ssDNA (D). The reaction products were analyzed by 1% agarose gel electrophoresis with molecular mass markers (HindIII-digested phage λ DNA or RNA markers [Promega]), and the nucleic acid was visualized by staining with ethidium bromide. An aliquot of each reaction mixture was analyzed at different times, as indicated for each lane. The nuclease activity of an equimolar concentration of GST was analyzed in parallel (GST control).
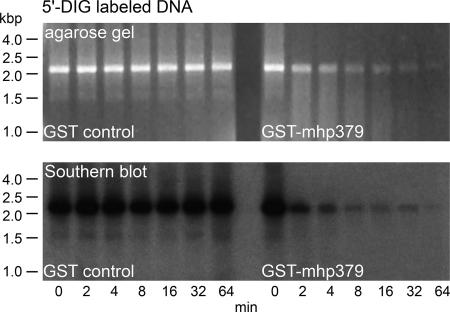
To determine whether the exonuclease activity of recombinant GST-mhp379 was specifically associated with the cleavage of nucleotides from the 5′ or 3′ end, a 2.2-kb 5′ DIG-labeled PCR product was used as the substrate and the reaction products were analyzed by agarose gel electrophoresis and Southern immunoblotting. In the presence of recombinant GST-mhp379, the amount of 5′ DIG-labeled DNA substrate was reduced significantly over time, and this was associated with the appearance of a DNA smear, as previously observed (Fig. 6). While a DNA smear was also observed at 4 and 8 min in a Southern immunoblot of the agarose gel using anti-DIG antibodies, this observation appears to be an artifact, as the amount and size of the DNA smear were no greater than those observed at 0 min before recombinant GST-mhp379 was added to the reaction mixture (Fig. 6). A DNA smear could not be identified in the Southern immunoblot of the agarose gel that was specifically associated with the cleavage of the 5′ DIG-labeled substrate by recombinant GST-mhp379. In particular, the Southern immunoblot of the agarose gel showed that both the amount of 5′ DIG-labeled substrate and the amount and size of the DNA smear were significantly reduced over time. These observations indicate that DIG-labeled nucleotides were being cleaved from the 5′ end of the DNA substrate. No nuclease activity was observed in a parallel experiment when GST was substituted for recombinant GST-mhp379 (Fig. 6).
FIG. 6.
Mode of nuclease activity of recombinant GST-mhp379. Approximately 4 μg of recombinant GST-mhp379 was incubated with PCR-amplified 5′ DIG-labeled DNA substrate. The reaction products were analyzed by 1% agarose gel electrophoresis with molecular mass markers (HindIII-digested phage λ DNA), and the DNA was visualized by staining with ethidium bromide. The DIG-labeled reaction products were detected by Southern immunoblotting of the agarose gel. An aliquot of each reaction mixture was analyzed at different times, as indicated for each lane. The nuclease activity of an equimolar concentration of GST was analyzed in parallel (GST control).
Biochemical characterization of nuclease activity.
The effects of pH, ionic strength (sodium and potassium ions), concentration of divalent cations (calcium and magnesium), and temperature on the nuclease activity of recombinant GST-mhp379 were determined with a real-time nuclease assay based on the enhancement of fluorescence of SYBR green I bound to dsDNA. A similar assay using PicoGreen DNA dye was described previously (56). SYBR green I did not appear to inhibit the nuclease activity of recombinant GST-mhp379 under the reactions conditions used (data not shown). The relative nuclease activities of recombinant GST-mhp379 were similar in all replicate assays, which also indicates that the real-time nuclease assay was reproducible under the reaction conditions used. The effect of pH on the relative activity of recombinant GST-mhp379 was shown to be dependent on the concentration of calcium ions (Fig. 7A). However, the optimal pH did not change in the presence of 5 or 50 mM Ca2+ and was estimated to be approximately 9.5. An increase in the concentration of calcium ions from 5 to 50 mM increased the relative nuclease activity of recombinant GST-mhp379 at pH values above and below the optimum. At pH 9.5, the optimum concentration of calcium ions was estimated to be 15 mM, and the nuclease activity of recombinant GST-mhp379 decreased significantly as the concentration of calcium ions increased above 15 mM (Fig. 7B). No nuclease activity was observed in the absence of calcium ions or in the presence of only magnesium ions (data not shown). At optimal pH and concentration of calcium ions, the nuclease activity of recombinant GST-mhp379 did not significantly increase or decrease following the addition of magnesium ions up to a concentration of 15 mM (data not shown). The nuclease activity of recombinant GST-mhp379 was optimal at low ionic strengths and decreased significantly as the concentration of sodium or potassium ions increased above 10 or 15 mM, respectively (Fig. 7C). When the nuclease assay was performed at different temperatures, the greatest activity was observed between 35 and 45°C (Fig. 7D). While the nuclease activity of recombinant GST-mhp379 increased slightly as the temperature increased to 45°C, only 30% activity was observed at 50°C. Recombinant GST-mhp379 was most stable between 35 and 40°C (Fig. 7E). Thus, while the nuclease activity of recombinant GST-mhp379 was greater at approximately 45°C, it was significantly less stable at this temperature. The temperature stability of recombinant GST-mhp379 was shown to be dependent on the concentration of calcium ions. When recombinant GST-mhp379 was incubated at 65°C for 10 min in the presence of 150 mM Ca2+, approximately 30% of the nuclease activity was conserved (data not shown). However, no nuclease activity could be detected after recombinant GST-mhp379 was incubated at 50°C for 10 min in the presence of 15 mM Ca2+ (Fig. 7E).
FIG. 7.
Effects of pH (A), calcium ion concentration (B), sodium and potassium ion concentration, (C) and temperature (D) on the nuclease activity of recombinant GST-mhp379. The stability of recombinant GST-mhp379 at different temperatures was also examined (E). The nuclease activity and stability were determined by comparing the initial reaction velocities under the different experimental conditions with a real-time nuclease assay based on the enhancement of fluorescence of SYBR green I bound to dsDNA.
DISCUSSION
Nucleic acid precursors are essential for the growth of all mycoplasmas (48). The acquisition of nucleic acid precursors is thought to involve the degradation of exogenous nucleic acids by mycoplasma nucleases located at or secreted from the cell surface. This is supported by the identification of transport mechanisms for the import of exogenous nucleotides and the identification of membrane-associated nuclease activity in all mycoplasma species studied so far. In particular, the nuclease activity of most mycoplasma species appears to be largely associated with the cell membrane. No transport mechanism for the import of oligonucleotides has yet been identified, and nucleases located in the cytoplasm require stringent control of activity. Nucleases located in the cytoplasm are thought to be primarily involved in recombination and repair and in restricting foreign nucleic acids. The import of exogenous nucleotides usually precedes their dephosphorylation, and a membrane-associated protein with similarity to bacterial 5′ nucleotidase was recently shown to be expressed by M. hyopneumoniae in infected pigs (31). However, M. mycoides subspecies mycoides has a novel ability to import and utilize nucleotide 5′ monophosphates without prior dephosphorylation (38, 39). Nucleases are involved in a variety of cellular processes, and genomic sequence analyses have identified multiple genes encoding putative nucleases in all mycoplasma species studied so far.
The TNASE_3 thermonuclease domain profile was identified in the hypothetical M. hyopneumoniae protein mhp379. In order to determine whether mhp379 is a nuclease, the gene was expressed as a recombinant GST fusion protein and the nuclease activity of recombinant GST-mhp379 was examined. Agarose gel electrophoresis of the nuclease reaction products with different nucleic acid substrates showed that recombinant GST-mhp379 is a sugar-nonspecific exonuclease that preferentially cleaves nucleic acid residues from the 5′ end. Significantly greater exonuclease activity was observed against RNA and ssDNA than against dsDNA. Recombinant GST-mhp379 also displayed endonuclease activity by nicking closed circular plasmid DNA. The optimum reaction conditions for the exonuclease activity of recombinant GST-mhp379 were determined in a real-time nuclease assay based on the enhancement of fluorescence of SYBR green I bound to dsDNA. Activity was optimal in the presence of 15 mM Ca2+, and no activity could be detected in the presence of EDTA. These observations are consistent with the identification of the conserved calcium binding site of the TNASE_3 thermonuclease domain profile in the gene encoding mhp379. The activity of all mycoplasma nucleases studied so far has been strictly dependent on the presence of divalent cations (2, 32, 33, 43) and has been optimal in the presence of both magnesium and calcium ions (2, 32, 33, 43). Recombinant GST-mhp379 did not exhibit exonuclease activity in the presence of only magnesium ions, and the addition of magnesium ions did not significantly change the exonuclease activity observed at optimal pH and concentration of calcium ions. A decrease in exonuclease activity with increasing ionic strength has been reported previously for other mycoplasma nucleases (2, 32, 43). The pH optimum for recombinant GST-mhp379 was found to be approximately 9.5, and an increase in the concentration of calcium ions increased the relative exonuclease activity at pH values above and below the optimum. This is likely to be associated with the function of calcium ions in stabilizing the structure of the nuclease, as suggested by the ability of excess calcium ions to conserve some activity of recombinant GST-mhp379 at 65°C. Under optimal conditions, the exonuclease activity of recombinant GST-mhp379 was stable between 35 and 40°C. While the pH optimum of nucleases from M. penetrans and M. pulmonis ranges between 7.0 and 8.0 and between 8.0 and 9.0, respectively (2, 43), the comparatively high pH optimum for recombinant GST-mhp379 is consistent with that reported for the homologous Nuc thermonuclease of S. aureus (7). It will be important to determine whether the optimal conditions for the exonuclease activity of recombinant GST-mhp379 are correlated with the pH and concentration of calcium ions in the respiratory tract of pigs colonized with M. hyopneumoniae. It is relevant to note that the pH optimum for recombinant GST-mhp379 is similar to that reported for the lipase activity of a cell surface-exposed lipoprotein of M. hyopneumoniae (53). The biochemical properties of mycoplasma nucleases have been reported previously only for the calcium and magnesium ion-dependent endonucleases of a number of mycoplasma species (2, 32, 43). This is the first study to characterize the biochemical properties of a mycoplasma exonuclease.
The mycoplasma membrane is rich in essential enzymes, and most of the nuclease activity of all mycoplasma species studied so far is either located at or secreted from the cell surface. The identification of an amino-terminal signal sequence and prokaryotic lipoprotein cleavage site indicates that mhp379 is processed for attachment to the cell membrane through the addition of a lipid moiety. The expression of mhp379 was examined by using antiserum raised against recombinant mhp379 in Western immunoblots of TX-114-fractionated proteins and trypsin-treated M. hyopneumoniae cells. These results indicate that mhp379 is a 33-kDa membrane-associated protein exposed on the cell surface. While rabbit antiserum against M. hyopneumoniae whole cells and antiserum from a pig experimentally infected with M. hyopneumoniae did not bind the recombinant mhp379 protein in Western immunoblots, it is possible that antibodies were raised against conformational epitopes of mhp379 that were subsequently lost when the recombinant mhp379 protein was reduced and denatured for separation by SDS-PAGE. This observation may alternatively indicate that the expression of mhp379 during the growth of M. hyopneumoniae both in vitro and in vivo is not sufficient to elicit an immune response in rabbits and pigs. Three cell-associated nucleases, of approximately 15, 29, and 40 kDa, in M. hyopneumoniae have been identified previously (33). While the homologous Nuc thermonuclease of S. aureus is an extracellular protein, mhp379 could not be detected in the supernatants of M. hyopneumoniae cultures by antibody affinity purification and/or Western immunoblotting. Furthermore, no extracellular nuclease activity was identified in M. hyopneumoniae in a real-time nuclease assay of culture supernatants. Minion et al. (33) also failed to identify extracellular nuclease activity in M. hyopneumoniae under the experimental conditions used. However, it is relevant to note that the 40-kDa extracellular endonuclease of M. penetrans is thought to be derived from the proteolytic cleavage of a 50-kDa precursor lipoprotein (2). Furthermore, although the membrane nuclease of M. pulmonis contains a prokaryotic lipoprotein cleavage site, no lipid-modified nuclease could be identified (33). The proteolytic cleavage of a mycoplasma lipoprotein has been previously reported for the macrophage-activating lipoprotein of Mycoplasma fermentans (9). The possibility that mhp379 may be proteolytically processed for extracellular release cannot be definitively excluded. In particular, the extracellular release of mhp379 would allow the activity of the protein to be supplied in trans to other cells; this may explain the survival of M. genitalium transposon mutants that contained a disruption in the mhp379 homolog when cultured in mixed populations of mutants (16). The inability to obtain pure clonal populations of the M. genitalium mhp379 homolog mutants suggests that mhp379 performs an essential biological function.
The organization of mhp379 in a putative ABC transport operon and its exposure on the cell surface suggests that the nuclease activity of mhp379 is involved in the ABC import of nucleic acid precursors. While mhp379 is not predicted to play a role in pathogenesis, the indirect role of cell surface-exposed enzymes in mycoplasma-host interactions is a recurring theme in the functional analyses of mycoplasma proteins. Indeed, the potential pathogenetic role of mycoplasma nucleases has been previously reported for the extracellular endonucleases of M. pulmonis (3) and M. hyorhinis (40, 41). The role of mhp379 in an active ABC transport operon is supported by the identification of overlapping transcripts between the transcriptional initiation site of mhp377 and the transcriptional termination site of mhp382 in preliminary reverse transcription-PCR analyses (data not shown). Homologs of mhp379 immediately upstream of the putative homologous ABC transport system in a number of phylogenically distant mycoplasma species have been identified, which suggests that mhp379 is organized in a conserved putative operon and that the functions of both mhp379 and the putative operon are likely to be conserved. However, the mhp379 homolog of M. synoviae is not located immediately upstream of the homologous putative ABC transport operon. In most mycoplasma species, homologs of M. pneumoniae lipoprotein multigene family 2 could be identified immediately upstream of the gene encoding the homolog of mhp379. Transposon mutagenesis of this homolog in M. genitalium was associated with the inability of the mycoplasma to adhere to plastic (16). Lipoproteins upstream of mycoplasma ABC import systems are predicted to function as substrate-binding proteins (52). Several binding protein-dependent ABC transport systems have also been shown to function in both import and export (22).
This is the first study to identify and characterize an exonuclease in a mycoplasma. M. hyopneumoniae mhp379 is a cell surface-exposed exonuclease that is located in a putative ABC transport operon and is conserved in a number of mycoplasma species. The exonuclease activity of mhp379 is predicted to be important in the import of nucleic acid precursors. However, the function of mhp379 in infected pigs remains to be determined. Despite the importance of genomic sequence analyses in predicting the function of hypothetical proteins, definitive characterization of the functions of mycoplasma proteins, based on biochemical analyses, is fundamental to our understanding of the life cycle of these minimalist prokaryotes. Further understanding of the functional role of mycoplasma proteins is likely to be essential for improving the control of mycoplasmoses.
Acknowledgments
This work was supported by funding from the Australian Research Council and Bioproperties Australia Pty. Ltd.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bendjennat, M., A. Blanchard, M. Loutfi, L. Montagnier, and E. Bahraoui. 1997. Purification and characterization of Mycoplasma penetrans Ca2+/Mg2+-dependent endonuclease. J. Bacteriol. 179:2210-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendjennat, M., A. Blanchard, M. Loutfi, L. Montagnier, and E. Bahraoui. 1999. Role of Mycoplasma penetrans endonuclease P40 as a potential pathogenic determinant. Infect. Immun. 67:4456-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Counis, M. F., and A. Torriglia. 2000. DNases and apoptosis. Biochem. Cell Biol. 78:405-414. [PubMed] [Google Scholar]
- 7.Cuatrecasas, P., S. Fuchs, and C. B. Anfinsen. 1967. Catalytic properties and specificity of the extracellular nuclease of Staphylococcus aureus. J. Biol. Chem. 242:1541-1547. [PubMed] [Google Scholar]
- 8.Czaja, T., A. Kanci, L. C. Lloyd, P. F. Markham, K. G. Whithear, and G. F. Browning. 2002. Induction of enzootic pneumonia in pigs by the administration of an aerosol of in vitro-cultured Mycoplasma hyopneumoniae. Vet. Rec. 150:9-11. [DOI] [PubMed] [Google Scholar]
- 9.Davis, K. L., and K. S. Wise. 2002. Site-specific proteolysis of the MALP-404 lipoprotein determines the release of a soluble selective lipoprotein-associated motif-containing fragment and alteration of the surface phenotype of Mycoplasma fermentans. Infect. Immun. 70:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duffy, M. F., A. H. Noormohammadi, N. Baseggio, G. F. Browning, and P. F. Markham. 1998. Immunological and biochemical characterisation of membrane proteins, p. 267-278. In R. J. Miles and R. A. J. Nicholas (ed.), Methods in molecular biology: mycoplasma protocols. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 11.Duffy, M. F., A. H. Noormohammadi, N. Baseggio, G. F. Browning, and P. F. Markham. 1998. Polyacrylamide gel-electrophoresis separation of whole-cell proteins, p. 279-298. In R. J. Miles and R. A. J. Nicholas (ed.), Methods in molecular biology: mycoplasma protocols. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 12.Dybvig, K., R. Sitaraman, and C. T. French. 1998. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc. Natl. Acad. Sci. USA 95:13923-13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Etheridge, J. R., G. S. Cottew, and L. C. Lloyd. 1979. Isolation of Mycoplasma hyopneumoniae from lesions in experimentally infected pigs. Aust. Vet. J. 55:356-359. [PubMed] [Google Scholar]
- 14.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friis, N. F. 1975. Some recommendations concerning primary isolation of Mycoplasma suipneumoniae and Mycoplasma flocculare: a survey. Nord. Vet. Med. 27:337-339. [PubMed] [Google Scholar]
- 16.Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gumulak-Smith, J., A. Teachman, A. H. Tu, J. W. Simecka, J. R. Lindsey, and K. Dybvig. 2001. Variations in the surface proteins and restriction enzyme systems of Mycoplasma pulmonis in the respiratory tract of infected rats. Mol. Microbiol. 40:1037-1044. [DOI] [PubMed] [Google Scholar]
- 18.Halden, N. F., J. B. Wolf, and W. J. Leonard. 1989. Identification of a novel site specific endonuclease produced by Mycoplasma fermentans: discovery while characterizing DNA binding proteins in T lymphocyte cell lines. Nucleic Acids Res. 17:3491-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallamaa, K. M., G. F. Browning, and S. L. Tang. 2006. Lipoprotein multigene families in Mycoplasma pneumoniae. J. Bacteriol. 188:5393-5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harlow, E., and D. P. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosie, A. H., D. Allaway, M. A. Jones, D. L. Walshaw, A. W. Johnston, and P. S. Poole. 2001. Solute-binding protein-dependent ABC transporters are responsible for solute efflux in addition to solute uptake. Mol. Microbiol. 40:1449-1459. [DOI] [PubMed] [Google Scholar]
- 23.Hynes, T. R., and R. O. Fox. 1991. The crystal structure of staphylococcal nuclease refined at 1.7 A resolution. Proteins 10:92-105. [DOI] [PubMed] [Google Scholar]
- 24.Jaffe, J. D., N. Stange-Thomann, C. Smith, D. DeCaprio, S. Fisher, J. Butler, S. Calvo, T. Elkins, M. G. FitzGerald, N. Hafez, C. D. Kodira, J. Major, S. Wang, J. Wilkinson, R. Nicol, C. Nusbaum, B. Birren, H. C. Berg, and G. M. Church. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarvill-Taylor, K. J., C. VanDyk, and F. C. Minion. 1999. Cloning of mnuA, a membrane nuclease gene of Mycoplasma pulmonis, and analysis of its expression in Escherichia coli. J. Bacteriol. 181:1853-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunitz, M. 1950. Crystalline desoxyribonuclease; digestion of thymus nucleic acid; the kinetics of the reaction. J. Gen. Physiol. 33:363-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd, L. C., and J. R. Etheridge. 1981. The pathological and serological response induced in pigs by parenteral inoculation of Mycoplasma hyopneumoniae. J. Comp. Pathol. 91:77-83. [DOI] [PubMed] [Google Scholar]
- 30.Markham, P. F., M. D. Glew, K. G. Whithear, and I. D. Walker. 1993. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect. Immun. 61:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meens, J., M. Selke, and G. F. Gerlach. 2006. Identification and immunological characterization of conserved Mycoplasma hyopneumoniae lipoproteins Mhp378 and Mhp651. Vet. Microbiol. 116:85-95. [DOI] [PubMed] [Google Scholar]
- 32.Minion, F. C., and J. D. Goguen. 1986. Identification and preliminary characterization of external membrane-bound nuclease activities in Mycoplasma pulmonis. Infect. Immun. 51:352-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minion, F. C., K. J. Jarvill-Taylor, D. E. Billings, and E. Tigges. 1993. Membrane-associated nuclease activities in mycoplasmas. J. Bacteriol. 175:7842-7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minion, F. C., E. J. Lefkowitz, M. L. Madsen, B. J. Cleary, S. M. Swartzell, and G. G. Mahairas. 2004. The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J. Bacteriol. 186:7123-7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell, A., and L. R. Finch. 1979. Enzymes of pyrimidine metabolism in Mycoplasma mycoides subsp. mycoides. J. Bacteriol. 137:1073-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell, A., and L. R. Finch. 1977. Pathways of nucleotide biosynthesis in Mycoplasma mycoides subsp. mycoides. J. Bacteriol. 130:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell, A., I. L. Sin, and L. R. Finch. 1978. Enzymes of purine metabolism in Mycoplasma mycoides subsp. mycoides. J. Bacteriol. 134:706-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neale, G. A., A. Mitchell, and L. R. Finch. 1983. Pathways of pyrimidine deoxyribonucleotide biosynthesis in Mycoplasma mycoides subsp. mycoides. J. Bacteriol. 154:17-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neale, G. A., A. Mitchell, and L. R. Finch. 1984. Uptake and utilization of deoxynucleoside 5′-monophosphates by Mycoplasma mycoides subsp. mycoides. J. Bacteriol. 158:943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paddenberg, R., A. Weber, S. Wulf, and H. G. Mannherz. 1998. Mycoplasma nucleases able to induce internucleosomal DNA degradation in cultured cells possess many characteristics of eukaryotic apoptotic nucleases. Cell Death Differ. 5:517-528. [DOI] [PubMed] [Google Scholar]
- 41.Paddenberg, R., S. Wulf, A. Weber, P. Heimann, L. A. Beck, and H. G. Mannherz. 1996. Internucleosomal DNA fragmentation in cultured cells under conditions reported to induce apoptosis may be caused by mycoplasma endonucleases. Eur. J. Cell Biol. 71:105-119. [PubMed] [Google Scholar]
- 42.Papazisi, L., T. S. Gorton, G. Kutish, P. F. Markham, G. F. Browning, D. K. Nguyen, S. Swartzell, A. Madan, G. Mahairas, and S. J. Geary. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low). Microbiology 149:2307-2316. [DOI] [PubMed] [Google Scholar]
- 43.Pollack, J. D., and P. J. Hoffmann. 1982. Properties of the nucleases of mollicutes. J. Bacteriol. 152:538-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pollack, J. D., M. A. Myers, T. Dandekar, and R. Herrmann. 2002. Suspected utility of enzymes with multiple activities in the small genome Mycoplasma species: the replacement of the missing “household” nucleoside diphosphate kinase gene and activity by glycolytic kinases. OMICS 6:247-258. [DOI] [PubMed] [Google Scholar]
- 45.Pollack, J. D., S. Razin, and R. C. Cleverdon. 1965. Localization of enzymes in Mycoplasma. J. Bacteriol. 90:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollack, J. D., M. V. Williams, and R. N. McElhaney. 1997. The comparative metabolism of the mollicutes (mycoplasmas): the utility for taxonomic classification and the relationship of putative gene annotation and phylogeny to enzymatic function in the smallest free-living cells. Crit. Rev. Microbiol. 23:269-354. [DOI] [PubMed] [Google Scholar]
- 47.Razin, S. 1978. The mycoplasmas. Microbiol. Rev. 42:414-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Razin, S., and B. C. Knight. 1960. The effects of ribonucleic acid and deoxyribonucleic acid on the growth of Mycoplasma. J. Gen. Microbiol. 22:504-519. [DOI] [PubMed] [Google Scholar]
- 49.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Sasaki, Y., J. Ishikawa, A. Yamashita, K. Oshima, T. Kenri, K. Furuya, C. Yoshino, A. Horino, T. Shiba, T. Sasaki, and M. Hattori. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 30:5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saurin, W., and E. Dassa. 1996. In the search of Mycoplasma genitalium lost substrate-binding proteins: sequence divergence could be the result of a broader substrate specificity. Mol. Microbiol. 22:389-390. [PubMed] [Google Scholar]
- 53.Schmidt, J. A., G. F. Browning, and P. F. Markham. 2004. Mycoplasma hyopneumoniae p65 surface lipoprotein is a lipolytic enzyme with a preference for shorter-chain fatty acids. J. Bacteriol. 186:5790-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sitaraman, R., and K. Dybvig. 1997. The hsd loci of Mycoplasma pulmonis: organization, rearrangements and expression of genes. Mol. Microbiol. 26:109-120. [DOI] [PubMed] [Google Scholar]
- 55.Taniuchi, H., C. B. Anfinsen, and A. Sodja. 1967. The amino acid sequence of an extracellular nuclease of Staphylococcus aureus. 3. Complete amino acid sequence. J. Biol. Chem. 242:4752-4758. [PubMed] [Google Scholar]
- 56.Tolun, G., and R. S. Myers. 2003. A real-time DNase assay (ReDA) based on PicoGreen fluorescence. Nucleic Acids Res. 31:e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasconcelos, A. T., H. B. Ferreira, C. V. Bizarro, S. L. Bonatto, M. O. Carvalho, P. M. Pinto, D. F. Almeida, L. G. Almeida, R. Almeida, L. Alves-Filho, E. N. Assuncao, V. A. Azevedo, M. R. Bogo, M. M. Brigido, M. Brocchi, H. A. Burity, A. A. Camargo, S. S. Camargo, M. S. Carepo, D. M. Carraro, J. C. de Mattos Cascardo, L. A. Castro, G. Cavalcanti, G. Chemale, R. G. Collevatti, C. W. Cunha, B. Dallagiovanna, B. P. Dambros, O. A. Dellagostin, C. Falcao, F. Fantinatti-Garboggini, M. S. Felipe, L. Fiorentin, G. R. Franco, N. S. Freitas, D. Frias, T. B. Grangeiro, E. C. Grisard, C. T. Guimaraes, M. Hungria, S. N. Jardim, M. A. Krieger, J. P. Laurino, L. F. Lima, M. I. Lopes, E. L. Loreto, H. M. Madeira, G. P. Manfio, A. Q. Maranhao, C. T. Martinkovics, S. R. Medeiros, M. A. Moreira, M. Neiva, C. E. Ramalho-Neto, M. F. Nicolas, S. C. Oliveira, R. F. Paixao, F. O. Pedrosa, S. D. Pena, M. Pereira, L. Pereira-Ferrari, I. Piffer, L. S. Pinto, D. P. Potrich, A. C. Salim, F. R. Santos, R. Schmitt, M. P. Schneider, A. Schrank, I. S. Schrank, A. F. Schuck, H. N. Seuanez, D. W. Silva, R. Silva, S. C. Silva, C. M. Soares, K. R. Souza, R. C. Souza, C. C. Staats, M. B. Steffens, S. M. Teixeira, T. P. Urmenyi, M. H. Vainstein, L. W. Zuccherato, A. J. Simpson, and A. Zaha. 2005. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J. Bacteriol. 187:5568-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, L., J. Westberg, G. Bolske, and S. Eriksson. 2001. Novel deoxynucleoside-phosphorylating enzymes in mycoplasmas: evidence for efficient utilization of deoxynucleosides. Mol. Microbiol. 42:1065-1073. [DOI] [PubMed] [Google Scholar]
- 59.Westberg, J., A. Persson, A. Holmberg, A. Goesmann, J. Lundeberg, K. E. Johansson, B. Pettersson, and M. Uhlen. 2004. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of contagious bovine pleuropneumonia (CBPP). Genome Res. 14:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Youil, R., and L. R. Finch. 1988. Isolation and characterization of Mycoplasma mycoides subsp. mycoides mutants deficient in nucleoside monophosphate transport. J. Bacteriol. 170:5922-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]