Abstract
Recent progress determining the structure of the host-encoded prion protein (PrPC) and the role of auxiliary molecules in prion replication permits a more rational approach in the development of therapeutic interventions. Our objective is to identify a new class of lead compounds that mimic the dominant negative PrPC mutants, which inhibit an abnormal isoform (PrPSc) formation. A computational search was conducted on the Available Chemicals Directory for molecules that mimic both the spatial orientation and basic polymorphism of PrP residues 168, 172, 215, and 219, which confer dominant negative inhibition. The search revealed 1,000 potential candidates that were visually analyzed with respect to the structure of this four-residue epitope on PrPC. Sixty-three compounds were tested for inhibition of PrPSc formation in scrapie-infected mouse neuroblastoma cells (ScN2a). Two compounds, Cp-60 (2-amino-6-[(2-aminophenyl)thio]-4-(2-furyl)pyridine-3,5-dicarbonitrile) and Cp-62 (N′1-({5-[(4,5-dichloro-1H-imidazol-1-yl)methyl]-2-furyl}carbonyl)-4 methoxybenzene-1-sulfonohydrazide), inhibited PrPSc formation in a dose-dependent manner and demonstrated low levels of toxicity. A substructure search of the Available Chemicals Directory based on Cp-60 identified five related molecules, three of which exhibited activities comparable to Cp-60. Mimicking dominant negative inhibition in the design of drugs that inhibit prion replication may provide a more general approach to developing therapeutics for deleterious protein–protein interactions.
Prions cause fatal neurodegenerative diseases in mammals, including Creutzfeldt-Jakob disease (CJD) in humans, bovine spongiform encephalopathy (BSE), and scrapie in sheep (1). Although significant advances have been made in understanding the molecular and cellular basis of prion diseases, these efforts have not yet led to promising therapeutic interventions (2). The emergence of more than 50 cases of a new variant CJD in humans that seems to be caused by BSE prions from cattle has heightened the urgency for development of effective therapeutics (3–10). More importantly, lessons from the development of inhibitors of prion replication are likely to be relevant to the creation of therapeutics for other neurodegenerative syndromes.
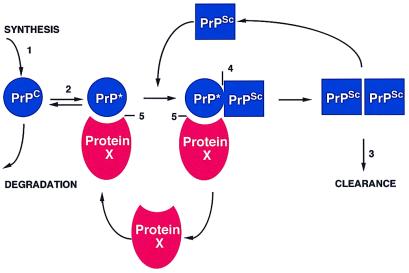
At the molecular level, the prion diseases are characterized by the accumulation of an abnormal isoform (PrPSc) of the host-encoded prion protein (PrPC) (11, 12). The recent progress in understanding the tertiary structure of PrPC and the role of auxiliary molecules e.g., protein X, in PrPSc replication permit a more rational approach in the development of therapeutic agents. From a consideration of many transgenic and cellular transfection studies, a model of the conversion of PrPC into PrPSc has emerged (Fig. 1) (13, 14). This model suggests that PrPSc accumulation can be blocked at five logical sites. One approach involves the inhibition of PrPC synthesis by antisense oligonucleotides or ribozymes targeted to PrP mRNA and is supported by work on knockout MoPrP mice demonstrating that these animals are resistant to prion disease (15–17). A second approach would be to stabilize the PrPC molecule and make the conformational change energetically less favorable (18). Alternatively, drugs that destabilize PrPSc, rendering it protease sensitive, might enhance clearance of PrPSc (Fig. 1) (8, 19–22). Compounds that bind to PrPSc and prevent it from serving as a template for the replication of nascent PrPSc could be effective. This scenario has been suggested as the mechanism of action of the amyloid-binding dye, Congo red (23). However, Congo red is effective in rodents only if it is administered close to the day of scrapie injection, and its highly anionic structure is likely to interfere with transport across the blood–brain barrier (24). Another strategy might be to prevent the formation of molecular complexes by interfering with the interactions between protein X, PrPC, and PrPSc (Fig. 1). Disrupting the interaction between PrPC and PrPSc with a small molecule is likely to be difficult, as the PrPSc binding site on the surface of the PrP(90–231) NMR structure appears to span a rather large area (25). Moreover, biochemical data suggest that this interaction is of high affinity and thus, small molecules are unlikely to disrupt PrPC/PrPSc binding. Blocking the interaction between protein X and PrPC seems to be the best approach for several reasons. The protein X binding site on PrPC maps to a small area primarily involving residues Q168, Q172, T215, and Q219 (14, 26). This region is of special interest as polymorphisms at codon 171(Q→R) in Suffolk sheep and codon 219(Q→K) in the Japanese population were reported to prevent disease naturally (27–31). When mouse Prnp genes carrying these polymorphisms were expressed in scrapie-infected neuroblastoma (ScN2a) cells, not only did they not form PrPSc, but they also blocked formation of wild-type PrPSc, presumably by sequestering protein X (14). The existence of a dominant negative phenotype argues that the protein X/PrPC interaction is the rate-limiting step in PrPSc formation. The ability of protein X to be recycled during PrPSc formation contends that this is a relatively low-affinity interaction that could be blocked by a small molecule.
Figure 1.
Model of PrPSc formation. Five potential strategies for drug discovery are suggested by this model: 1) block PrPC synthesis, 2) stabilize PrPC, 3) enhance PrPSc clearance, 4) interfere with binding of PrPC to PrPSc, and 5) prevent binding of protein X to PrPC.
We present a structure-based approach to the identification of small heterocyclic molecules that were designed to mimic the dominant negative inhibition of prion replication by polymorphic variants of PrP. Although most structure-based drug design focuses on filling cavities on the surface of a molecule to inhibit an interaction or on optimizing the efficiency of small molecule whose detailed interactions with its molecular target are visualized crystallographically, we followed a distinctly different route. We sought to mimic the surface of a macromolecule by using a small molecule. To the extent that our computational algorithm identifies appropriate mimetics of the PrPC epitope that specifies the dominant negative phenotype, PrPSc formation will be blocked. Our studies demonstrate that it is possible to identify plausible mimetics of a localized epitope on the surface of PrPC and that a subset of these molecules inhibits prion replication in cultured cells.
Materials and Methods
Chemicals.
Compounds selected from our computational pharmacophore search strategy were purchased from Aldrich, Bionet (Cornwall, U.K.), ChemService (West Chester), Nova Biochem, Parish (Vineyard, UT), Research Biochemicals, Sigma, TCI America (Portland, OR), and Wako. Compounds that have an alpha-numeric name were provided by Maybridge (Cornwall, U.K.) (see Table 1, which is published as supplementary material on the PNAS web site, www.pnas.org). Cp-60 is 2-amino-6-[(2-aminophenyl)thio]-4-(2-furyl)pyridine-3,5-dicarbonitrile. Cp-62 is N′1-({5-[(4,5-dichloro-1H-imidazol-1-yl)methyl]-2-furyl}carbonyl)-4 methoxybenzene-1-sulfonohydrazide. Stock solutions at 1 mM concentration were prepared according to the solubility and solvent recommended by the companies. Analogs of Cp-60 also were purchased from Maybridge. Their alpha-numeric names are A3-KM00566, A4-KM00567, and A5-KM00563. Stock solutions at 5 mM concentration were prepared in DMSO.
Cells and Antibodies.
Mouse neuroblastoma cells (N2a) were obtained from the American Tissue Culture Collection. Scrapie-infected mouse neuroblastoma (ScN2a) cells have been described (32). Cells were grown and maintained at 37°C in DMEM supplemented with 10% FBS or 1% lipid reduced serum for the mevastatin samples, 1% glutamax, and 1% penicillin/streptomicin. The mAb 3F4 was raised against Syrian hamster (SHa) PrP 27–30 (33). To distinguish the products of a mouse-hamster chimeric (MHM2)PrP construct from endogeneous mouse PrP (MoPrP), the 3F4 mAb was used. This antibody recognizes SHa as well as MHM2PrP (14) but not MoPrP, because of sequence differences at residues 109 and 112. The polyclonal RO73 antiserum was raised in a rabbit against SDS/PAGE-purified SHaPrP 27–30 that reacts with SHaPrP, MoPrP, and MHM2PrP (34).
Screening Procedure.
Identically seeded ScN2a cells (60-mm plate) at 25% confluent density were transiently transfected with 20 μg of pSPOX vector carrying the MHM2 construct by using the DOTAP (N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate) liposomal transfection reagent (Boehringer Mannheim). Compounds at a final concentration of 10 μM were applied 24 h after the transfection directly in the cell culture medium over 3 days. Confluent cells then were lysed by addition of 500 μl of lysis buffer (10 mM Tris⋅HCl, pH 8/100 mM NaCl/0.5% NP-40/0.5% deoxycholate). The protein concentration of each sample was measured with the BCA reagent (Pierce). Samples of equal protein amounts and volumes then were digested with 20 μg/ml proteinase K at a ratio of 1:25 protease to protein for 1 h at 37°C. Digestions were stopped by 2 mM PMSF, and the samples were ultracentrifuged. Pellets then were resuspended in 20 μl of lysis buffer and 20 μl of SDS loading buffer and boiled 5 min before loading on 12% SDS/PAGE precast Novex gel. Immunoblot analysis was performed according to a protocol described elsewhere (35). The 3F4 mAb was used to detect the MHM2PrP protein on the membrane.
Quantification of PrPSc by Immunoassay Using Time-Resolved Fluorescence (TRF) Spectroscopy.
Different concentrations of Cp-60 were applied to transiently transfected ScN2a cells according to the same protocol described in the screening procedure. Cells were trypsinized, collected in tubes, and washed twice with 5 ml of 1× PBS solution. Each sample was lysed with 1 ml of PBS containing 2% sarkosyl. The crude cell extract was sheared with a syringe and stored overnight at −80°C. Protein concentration was measured with the BCA reagent and samples were digested with proteinase K as described (21). The reaction was stopped by a mixture of protease inhibitors (5 mM PMSF, aprotinin, and leupeptin at 4 μg/ml each). Samples were precipitated with a solution of 4% sodium phosphotungstate (NaPTA) and 34 mM MgCl2 to obtain a final concentration of 0.3% NaPTA. After centrifugation at 20,000 g, the pellets were resuspended in 25 μl of distilled water and mixed with guanidine-HCl to a final concentration of 4 M, then denatured by heating for 5 min at 80°C. The immunoassay was performed according to a protocol described elsewhere (21). The remaining protein was quantified by immunoassay using TRF. The europium-labeled mAb 3F4 was used to detect MHM2PrP proteins.
Cellular Viability Measure.
The reagent 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide, thiazolyl blue (MTT) was used to determined the cellular viability of ScN2a cells, in the presence of Cp-60 or methanol. One-tenth of the volume of the original culture volume of a filtered stock solution of MTT (5 mg/ml) was added to each culture being assayed and incubated for 4 h. The medium was removed, and the converted dye was solubilized with acidic isopropanol (0.1 M HCl). Absorbance of the converted dye was measured at a wavelength of 562 nm with a background subtraction at 650 nm.
Nomenclature.
Residue 171 in sheep PrP corresponds to codon 168 in human PrP and codon 167 in MoPrP.
Results
Selection of Compounds by a Structure-Based Drug Design Strategy.
We assumed that the most suitable therapeutic target within the prion replication cycle is the dominant negative epitope localized on the surface of PrPC. For convenience we refer to this as the protein X/PrPC molecular complex to reflect the fact that we have not yet identified the binding partner or partners that mediate this effect (14, 26). Theoretically, three types of ligands might interrupt the binding between these two entities: (i) a small molecule that alters the protein X binding site on the surface of PrPC by binding PrPC, (ii) a compound that mimics the protein X binding site on the surface of PrPC, and (iii) a compound that mimics the PrPC binding site on the surface of the protein X. Because the protein X binding site on the surface of PrP lacks suitable cavities, the dock algorithm of Shoichet and Kuntz (36) proved ineffective. As the identity and structure of protein X is unknown, we focused on identifying a mimic of the protein X binding site on the surface of PrPC.
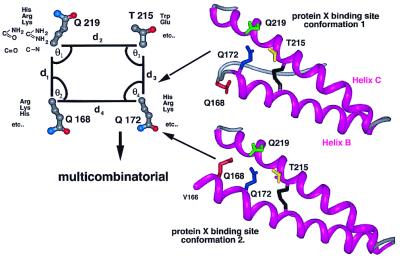
Disruption of protein–protein interactions by using a structure-based drug design approach has proved difficult; however, this task should be easier if a small subset of residues contribute to a significant fraction of the binding energy and occupy only a small percentage of the overall protein surface (37–39). Kaneko et al. (14) showed that residues Q168, Q172, T215, and Q219 on the surface of the PrPC molecule contribute most prominently to the stability of the molecular complex between PrPC and protein X. Their side-chain coordinates from the PrP(90–231) NMR structure define a plausible pharmacophore target for mimetic design (14, 40). In the PrP(90–231) NMR structure, residue Q168 is part of a nonhelical loop that is not in close contact with the three other residues involved in the protein X binding site. The apparent discontinuity of the protein X binding site side chain owes to nuclear Overhauser effect restraints between the side chain of V166 and residues Y218, E221, S222, and Y225 in uncomplexed recombinant PrP(90–231). However, it would be quite feasible for Q168 to move adjacent to Q172, T215, and Q219 upon binding to protein X by extending helix B one turn. We therefore placed Q168 in two positions, first as it is found in the PrP(90–231) NMR structure and second, with helix B extended from residue Q172 to V166 (Fig. 2). As the substitution of basic residues at Q168, Q172, and Q219, and acidic or hydrophobic residues at T215 appear to inhibit PrPSc replication by increasing the affinity of protein X for PrPC, we modeled Arg, Lys, His, Asp, Glu, and Trp onto the relevant side-chain positions of the PrP(90–231) NMR structure by using the program scwrl (41).
Figure 2.
Possible conformations of the protein X binding site used to create a data set of pharmacophores. In the first conformation, residue 168 is positioned as found in the NMR structure of PrP(90–231) whereas in the second one, helix B is extended to residue V166 (25). In this case, residue Q168 forms a contiguous structural epitope with the other three protein X binding site residues, Q172, T215, and Q219.
Using these amino acid side-chain coordinates, we generated a data set of three-dimensional pharmacophores such that all possible combinations of amino acid substitutions at the protein X binding site were represented (Fig. 2). Emphasis was placed on mimicking the position of the functional atoms in each side chain as it is unlikely that any small molecules will contain all of the atoms of each of the side chains that define the dominant negative epitope. Pharmacophores with more than one basic residue were eliminated as substitution of more than one of the basic residues at the protein X binding site interferes with the dominant negative inhibition of prion replication (26). This analysis resulted in 1,000 potential templates, which were compared with the 210,000 compounds present in the Available Chemicals Database (ACD) for compounds that mimic both the spatial orientation and chemical properties present in the data set of pharmacophores. The computer program concord was used to generate the three-dimensional structure of each of the ACD compounds (42). Our algorithm, called genx, has two main stages (43). First, the covalent connectivity of the pharmacophore functional atoms is compared with the compounds. Then, if at least one match for each of these groups of functional atoms is located within the compound, the mean coordinate position of each group is calculated. This generates a single pseudoatom representative for each match corresponding to a functional atom cluster. The interatomic distances and bond angles defined by these pseudoatoms then are compared with the distances and angles previously specified for each of the pharmacophores from an analysis of the relevant side-chain centroids. For each possible match, the small molecule is superimposed on the optimal collection of side-chain atoms and the rms distance is calculated. A rms distance of less than 2 Å is classified as a match. genx identified 5,000 compounds from the ACD, which were further clustered according to their functional groups and screened visually by using midasplus. Of the 80 compounds that were identified for experimental characterization, 63 were available for purchase.
Drug Screening.
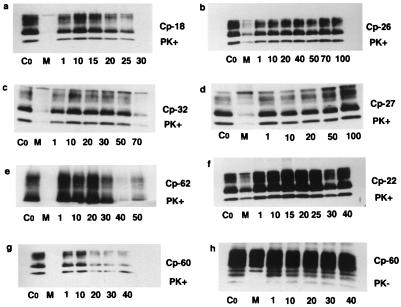
Compounds were screened in a cellular assay for prion replication. To analyze the effect of the compounds on newly synthesized PrPSc, ScN2a cells were transiently transfected. The SHa/MoPrP chimeric construct designated MHM2PrP, which contains an epitope for the anti-SHaPrP 3F4 mAb, was used (33, 34, 44). In this assay, MoPrPSc is immunologically silent. Twenty-four hours after the transfection, each of the 63 compounds was added to the medium and incubated for 3 days. After 72 h, the cells were lysed and the crude extract was analyzed on an immunoblot before and after proteinase K digestion (Fig. 3). Initial screening of compounds at 10 μM identified 10 potential inhibitors of PrPSc formation. The inhibition observed for five of these candidates was not reproducible. Complete dose-response curves confirmed the activity of four of the 10 compounds: Cp-18 (purine riboside), Cp-32 (2′, 3′-di-O-acetyladenosine), Cp-60, and Cp-62 indicated in Fig. 3. Unlike these four compounds, Cp-7 (1,N-6-ethenoadenosine-5′-monophosphate) failed to exhibit a classical dose-response curve. As Cp-60 and Cp-62 were solubilized at 1 mM in pure methanol, we verified that the inhibition was not caused by the solvent. Immunoblots revealed that 4% methanol, which corresponds to the volume of solvent used with 40 μM of Cp-60, did not inhibit PrPSc formation. For all of the compounds screened, we made immunoblots of the samples not treated with proteinase K. This provided a control for the transfection efficiency and shows that each of the samples were equally transfected (data shown only for Cp-60, see Fig. 3). The compound concentrations at which 50% of PrPSc replication was inhibited (IC50) were determined by densitometry as a ratio of PrPSc/total PrP. The compounds, in descending order of apparent potency are Cp-60 (18 μM), Cp-18 (20 μM), Cp-62 (30 μM), and Cp-32 (60 μM) (Fig. 3). Cytotoxicity was evaluated for each of the 63 compounds and carefully analyzed for the four active compounds. The cellular toxicity of the compounds was determined by the quantification of the total protein amounts in the lysates and compared with the untreated control. At concentrations near the IC50s, Cp-18 and Cp-32 showed toxic effects on ScN2a cells. At 30 μM of Cp-18, 5 ± 0.1% of the protein remained and 47 ± 3% of the protein was found after the cells were exposed to 70 μM of Cp-32. Experiments with Cp-18 and Cp-32 were not pursued. In contrast, 95 ± 4% of the protein was found after exposure to 20 μM Cp-60, and 80 ± 3% was recovered after exposure to 40 μM Cp-62. Among the 63 compounds screened, three inhibited PrPSc formation without significant cellular toxicity (Cp-7, Cp-60, and Cp-62). Fifty-eight compounds did not inhibit PrPSc formation; among these, five were toxic to ScN2a cells [Cp-25, amidinophenyl(6amidino-2-indolyl)phenyl ether; Cp-38, allantoic acid; Cp-40, 2′-O-anthraniloyladenosine 3′, 5′-cyclic monophosphate; Cp-48,n-α-benzoyl-l-histidinol; and Cp-65, 4-nitro-phenyl-methoxy-benzoyl]. As Cp-60 was not overtly toxic to cells and exhibited the lowest IC50, we studied its impact on cellular viability by using the MTT reagent. At 20 μM Cp-60, 82 ± 0.3% of ScN2a cells were alive; at 40 μM 66 ± 0.1% were viable. To assess the toxicity of the methanol used to solubilize Cp-60, the viability of the cells was determined with solvent alone. Methanol was responsible for 10% and 20% of the cell death at 20 μM and 40 μM of Cp-60, respectively.
Figure 3.
Inhibition of MHM2PrPSc formation in ScN2a cells. Control without compound is designated Co. Positive control with mevastatin is denoted M. Immunoblot analysis of the lysates were performed before (PK−) and after (PK+) proteinase K digestion. (a) Cp-18 is purine riboside. (b) Cp-26 is hydroxy-guanidino purine riboside. (c) Cp-32 is 2′3′-di-O-acetyladenosine. (d) Cp-27 is adenylyl (3′5′)cytidine. (e) Cp-62 is KM-06274 (see Materials and Methods for complete chemical name). (f) Cp-22 is dibekacin. (g and h) Cp-60 is KM-00561 (see Materials and Methods for complete chemical name). (a, c, e, g, and h) Shown are the four drugs screened that inhibit MHM2PrPSc formation. (b, d, and f) Examples of compounds without inhibitory effect. The numbers below the individual gels correspond to various concentrations of compounds in μM.
Quantification of PrPSc by TRF Spectroscopy.
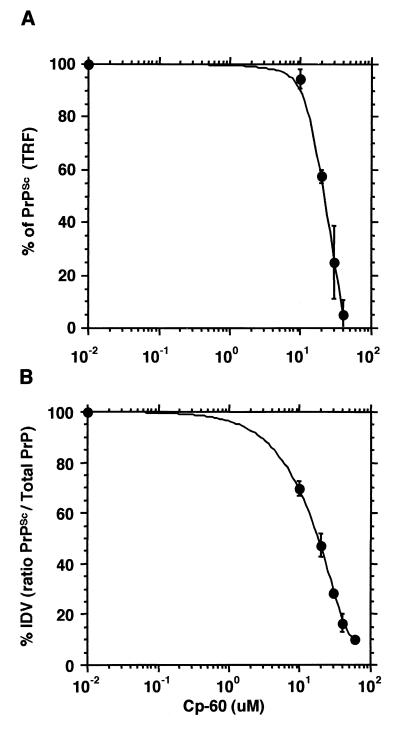
To obtain an independent verification of the IC50 of the most active compound, we used a more sensitive and quantitative immunoassay using TRF (21). Transiently transfected ScN2a cells were incubated with various concentrations of Cp-60 over 3 days. The cell lysates were digested with proteinase K and the PrPSc was measured by immunoassay using a europium-labeled mAb (Eu-mAb 3F4) (Fig. 4A). The IC50 for Cp-60 obtained by the TRF technique was 21.5 ± 0.5 μM. The result of this approach agreed well with the dose-response curve determined by immunoblotting where the IC50 for Cp-60 was 18.0 ± 1.5 μM (Fig. 4B).
Figure 4.
Cp-60 inhibition on MHM2PrPSc formation. (A) Immunoassay using TRF. The data represent the mean ± SE from three independent experiments conducted in duplicate. (B) Immunoblots quantified by densitometry. Treatment of transfected ScN2a cells with Cp-60 and immunoblot analysis were described in Fig. 3. Densitometry was conducted with four immunoblot films from independent experiments. Values represent the mean ± SE.
Treatment of ScN2a Cells with Cp-60.
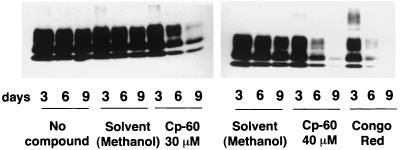
Finally, we studied the ability of Cp-60 to cure the ScN2a cells. Cells were incubated with solvent alone, Cp-60, or Congo red as a positive control. These cells were serially passaged every 3 days for 9 days. An immunoblot analysis of the samples showed that after 6 days of incubation with 40 μM of Cp-60, half of the PrPSc molecules disappeared. After 9 days, the cells exhibited little detectable PrPSc. The magnitude of this effect is comparable to Congo red (1 μM), a reagent previously reported to block the PrPSc formation in cells (Fig. 5).
Figure 5.
Inhibition of MoPrPSc formation in ScN2a cells after treatment with Cp-60. ScN2a cells were incubated with various compounds and serially passaged for 3, 6, and 9 days. Two sets of four dishes (60-mm plate) containing identically seeded ScN2a cells at 25% confluency were independently treated with 1 μM of Congo red, 30 μM of Cp-60, or 40 μM of Cp-60, and a control plate had no compound. Cells were grown to confluence within 3 days. One set of four dishes was lysed, digested by proteinase K (PK), and analyzed by immunoblot according to the same protocol described in Fig. 3. For the other set, each plate was split into duplicates and incubated with the relevant compounds for another 3 days, until the cells reached confluence. After 6 days of incubation, half of the plates were lysed and analyzed for PK resistance; the other half was serially passaged again by the same procedure. The experiment also was done with pure methanol at final concentrations of 3% and 4%, which correspond to the volume of solvent used for 30 μM and 40 μM of Cp-60. Immunoblot detection of PrPSc was performed in the PK-digested lysates from crude cellular extracts by using the polyclonal antiserum RO73. Experiments were repeated independently.
Discussion
Prion replication is a complex cellular event that does not easily lend itself to the high throughput screening efforts that are a standard element of most modern drug discovery efforts. Although PrPC and PrPSc have been purified, some critical elements required for prion replication remain unknown. Two lines of evidence support this view. Although in vitro biochemical methods have succeeded in creating a protease-resistant form of PrP, they have failed to demonstrate the conversion of PrPC into infectious PrPSc. The existence of dominant negative variants of PrP implies the sequestration of one or more critical factors in PrPSc formation (e.g., protein X). By contrast, cell culture-based systems that support PrPSc formation correlate well with animal-based assays of prion infectivity (32, 45). Unfortunately, these assays are difficult to automate. Thus, the identification of molecules that inhibit PrPSc formation traditionally has relied on empirical or serendipitous observations (8). For most of these compounds, the mechanism of action is largely unknown. This complicates medicinal chemistry efforts to optimize compound potency. Our goal was to identify lead molecules that are able to inhibit PrPSc formation by using a structure-based strategy.
The tertiary and quaternary structure of PrPSc and the identity of the protein X are still unknown. Therefore, we focused our attention on the NMR structure of PrPC and the observation that specific polymorphic variants of PrPC are not substrates for PrPSc formation and block PrPSc replication when coexpressed with wild-type PrP. A number of in vivo studies demonstrated that PrPSc formation and the development of disease is inhibited by PrP alleles carrying basic residues at position 171 in sheep (which corresponds to human PrP codon 168 and MoPrP codon 167) and 219 in humans (which corresponds to MoPrP codon 218). This dominant negative inhibition effect has been reproduced in cultured cells (14, 26). Furthermore, Tg(MoPrP Q167R) Prnp0/0 mice expressing mutant MoPrP at 1–2 times the level of wild-type MoPrPC remain well more than 400 days after inoculation with RML prions (V.P., K.K., S.B.P., and F.E.C., unpublished data). By contrast, non-Tg FVB mice expressing wild-type MoPrPC developed disease at ≈120 days after inoculation with RML prions. Although these studies are incomplete, the results serve to reinforce the assertion that the Q167 polymorphism delays or prevents the development of prion disease.
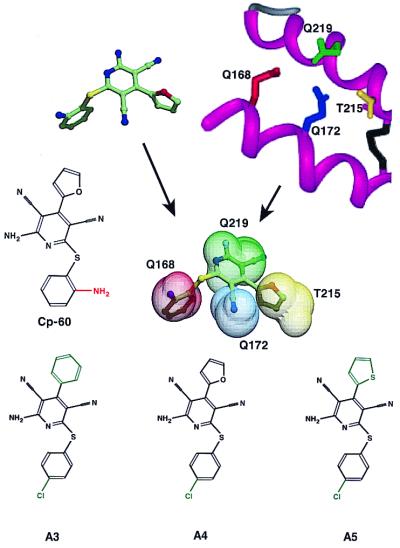
In view of the foregoing results, we focused our efforts on the search for mimetics of the epitope defined by the side chains of residues 168, 172, 215, and 219. High throughput screening of more than 200,000 compounds was conducted in compuo and a more defined subset was characterized experimentally in a cultured cell-based assay. Of the initial list of computationally sensible compounds, about 6% inhibit PrPSc formation at reasonable concentrations. The best of these compounds, Cp-60, was used to search the Available Chemical Directory for related molecules. To date, five related compounds have been analyzed (Fig. 6), and three exhibit activity that is comparable to Cp-60. In our cellular assay, the IC50 for A3 is 35 ± 3.0 μM, for A4 is 18.6 ± 3.1 μM, and for A5 is 15.5 ± 0.5 μM. Our enthusiasm for the initial leads described here is tempered by the recognition that existing animal models of scrapie replication are quite time consuming, and existing cellular data suggests that daily dosing schedules will be required.
Figure 6.
Comparison of compound Cp-60 with the protein X binding site in conformation two. The B and C helices of PrPC modeled with helix B extended by one turn are shown together with the side chain of the dominant negative residues 168 (red), 172 (blue), 215 (yellow), and 219 (green). Cp-60 is shown in the conformation predicted by the concord algorithm. Cp-60 then is superimposed on the dominant negative pharmacophore when the relevant side-chain atoms are represented by a van der Waals sphere centered on side-chain atoms' positions. The color coding is by residue in the sphere and by atom type in Cp-60: carbon (green), oxygen (red), nitrogen (blue), and sulfur (yellow). The chemical structures of the three analogs of Cp-60 (A3, A4, and A5) that inhibit PrPSc formation are shown.
Our results show that Cp-60 is able to inhibit the formation of newly synthesized PrPSc and cure infected cells. However, additional bioassay studies will be required to confirm that prion infectivity has been eliminated. We also lack direct evidence that the mechanism of action of Cp-60 is indeed to block the PrPC/protein X interaction by mimicking the dominant negative epitope. Although protein X has yet to be isolated, derivatives of Cp-60 carrying a photoactivatable crosslinking moiety have been synthesized and observed to be more potent than the original compound. Thus derivatives of Cp-60 may provide a more direct route to the purification of protein X.
The mechanism of inhibition of PrPSc formation for Cp-60 seems to be radically different from that of Congo red. Cp-60 exhibits a classical dose-response curve (Fig. 4B), whereas low concentrations of Congo red increase the production of PrPSc approximately 3-fold and high concentrations block PrPSc formation (data not shown) (46). These observations are consistent with the direct binding of PrPSc to Congo red in a mode that initially stabilizes the molecular complex but ultimately occludes molecular surfaces that are required for PrPSc to act as a replication template.
In general, many protein–protein interactions that play essential roles in disease processes have been evaluated by the pharmaceutical industry. While some, like calcineurin-immunophilin (FKPB) (47, 48) have proved to be fruitful targets, most have been difficult to influence with small molecules. We suggest that the subset of protein–protein interactions where single-residue mutations at the protein–protein interface give rise to a dominant negative phenotype may select for a subset of pharmaceutically tractable interactions. Although most protein–protein interfaces cover a large fraction of the total protein surface, the existence of these single residue dominant negatives suggest that a large component of the energetics of this interaction can be focused in a very small area that is comparable in size to small heterocyclic molecules. Clearly, small molecules like morphine are capable of mimicking the action of larger polypeptide hormones like the dynorphins at the opioid receptors. However, the search for small molecule analogs of erythropoietin and insulin has been more challenging (49, 50). The task of exchanging polypeptide scaffolds for small heterocyclic structures without loss of biological activity remains difficult. The utility of the genx approach to protein–protein interactions where domimant negative variants have been identified needs to be explored more broadly. Particular attention will be required in those cases where point mutants confer the desired phenotype. Our efforts demonstrate how the tools of structural biology and computational chemistry can be used to identify molecules that may lead to therapeutic agents for the treatment of prion diseases. Whether this approach will provide a more general method for developing novel molecules to treat the more common neurodegenerative disorders including Alzheimer's and Parkinson's diseases, as well as amyotrophic lateral sclerosis, remains to be established.
Supplementary Material
Acknowledgments
This work was supported by Grants BS14069, AG02132, and AG10770 from the National Institutes of Health to the laboratories of S.B.P. and F.E.C. Support for V.P. is provided by fellowships from the Fondation pour la Recherche Médicale (France) and the John Douglas French Alzheimer's foundation.
Abbreviations
- PrP
prion protein
- PrPC
host-encoded PrP
- PrPSc
abnormal isoform of PrP
- SHa
Syrian hamster
- N2a
mouse neuroblastoma cells
- ScN2a
scrapie-infected mouse neuroblastoma cells
- TRF
time-resolved fluorescence
- MoPrP
mouse PrP
- Cp-60
2-amino-6-[(2-aminophenyl)thio]-4-(2-furyl)pyridine-3,5-dicarbonitrile
- Cp-62
N′1-({5-[(4,5-dichloro-1H-imidazol-1-yl)methyl]-2-furyl}carbonyl)-4 methoxybenzene-1-sulfonohydrazide
- Cp-18
purine riboside
- Cp-32
2′3′-di-O-acetyladenosine
References
- 1.Prusiner S B. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 2.Aguzzi A, Weissmann C. Nature (London) 1997;389:795–798. doi: 10.1038/39758. [DOI] [PubMed] [Google Scholar]
- 3.Anderson R M, Donnelly C A, Ferguson N M, Woolhouse M E J, Watt C J, Udy H J, MaWhinney S, Dunstan S P, Southwood T R E, Wilesmith J W, et al. Nature (London) 1996;382:779–788. doi: 10.1038/382779a0. [DOI] [PubMed] [Google Scholar]
- 4.Collinge J, Sidle K C L, Meads J, Ironside J, Hill A F. Nature (London) 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 5.Will R G, Ironside J W, Zeidler M, Cousens S N, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofman A, Smith P G. Lancet. 1996;347:921–925. doi: 10.1016/s0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 6.Cousens S N, Vynnycky E, Zeidler M, Will R G, Smith P G. Nature (London) 1997;385:197–198. doi: 10.1038/385197a0. [DOI] [PubMed] [Google Scholar]
- 7.Ghani A C, Ferguson N M, Donnelly C A, Hagenaars T J, Anderson R M. Lancet. 1998;352:1353–1354. doi: 10.1016/s0140-6736(05)60744-1. [DOI] [PubMed] [Google Scholar]
- 8.Supattapone S, Nguyen H-O B, Cohen F E, Prusiner S B, Scott M R. Proc Natl Acad Sci USA. 1999;96:14529–14534. doi: 10.1073/pnas.96.25.14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Will R G, Cousens S N, Farrington C P, Smith P G, Knight R S G, Ironside J W. Lancet. 1999;353:979. doi: 10.1016/s0140-6736(99)01160-5. [DOI] [PubMed] [Google Scholar]
- 10.Scott M R, Will R, Ironside J, Nguyen H-O B, Tremblay P, DeArmond S J, Prusiner S B. Proc Natl Acad Sci USA. 1999;26:15137–15142. doi: 10.1073/pnas.96.26.15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, et al. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen F E, Pan K-M, Huang Z, Baldwin M, Fletterick R J, Prusiner S B. Science. 1994;264:530–531. doi: 10.1126/science.7909169. [DOI] [PubMed] [Google Scholar]
- 13.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 14.Kaneko K, Zulianello L, Scott M, Cooper C M, Wallace A C, James T L, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Büeler H, Fisher M, Lang Y, Bluethmann H, Lipp H-P, DeArmond S J, Prusiner S B, Aguet M, Weissmann C. Nature (London) 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 16.Prusiner S B, Groth D, Serban A, Koehler R, Foster D, Torchia M, Burton D, Yang S-L, DeArmond S J. Proc Natl Acad Sci USA. 1993;90:10608–10612. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Büeler H, Aguzzi A, Sailer A, Greiner R A, Autenried P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 18.Tatzelt J, Prusiner S B, Welch W J. EMBO J. 1996;15:6363–6373. [PMC free article] [PubMed] [Google Scholar]
- 19.Prusiner S B. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 20.Prusiner S B, Groth D, Serban A, Stahl N, Gabizon R. Proc Natl Acad Sci USA. 1993;90:2793–2797. doi: 10.1073/pnas.90.7.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen F E, Prusiner S B. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 22.Soto C, Kascsak R J, Saborio G P, Aucouturier P, Wisniewski T, Prelli F, Kascsak R, Mendez E, Harris D A, Ironside J, et al. Lancet. 2000;355:192–197. doi: 10.1016/s0140-6736(99)11419-3. [DOI] [PubMed] [Google Scholar]
- 23.Caspi S, Sasson S B, Taraboulos A, Gabizon R. J Biol Chem. 1998;273:3484–3489. doi: 10.1074/jbc.273.6.3484. [DOI] [PubMed] [Google Scholar]
- 24.Ingrosso L, Ladogana A, Pocchiari M. J Virol. 1995;69:506–508. doi: 10.1128/jvi.69.1.506-508.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Farr-Jones S, Ulyanov N B, Llinas M, Marqusee S, Groth D, Cohen F E, Prusiner S B, James T L. Biochemistry. 1999;38:5362–5377. doi: 10.1021/bi982878x. [DOI] [PubMed] [Google Scholar]
- 26.Zulianello L, Kaneko K, Scott M, Erpel S, Han D, Cohen F E, Prusiner S B. J Virol. 2000;74:4351–4360. doi: 10.1128/jvi.74.9.4351-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitamoto T, Tateishi J. Philos Trans R Soc London B. 1994;343:391–398. doi: 10.1098/rstb.1994.0034. [DOI] [PubMed] [Google Scholar]
- 28.Westaway D, Zuliani V, Cooper C M, Da Costa M, Neuman S, Jenny A L, Detwiler L, Prusiner S B. Genes Dev. 1994;8:959–969. doi: 10.1101/gad.8.8.959. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda T, Horiuchi M, Ishiguro N, Muramatsu Y, Kai-Uwe G D, Shinagawa M. J Gen Virol. 1995;76:2577–2581. doi: 10.1099/0022-1317-76-10-2577. [DOI] [PubMed] [Google Scholar]
- 30.Hunter N, Moore L, Hosie B D, Dingwall W S, Greig A. Vet Rec. 1997;140:59–63. doi: 10.1136/vr.140.3.59. [DOI] [PubMed] [Google Scholar]
- 31.Shibuya S, Higuchi J, Shin R-W, Tateishi J, Kitamoto T. Lancet. 1998;351:419. doi: 10.1016/S0140-6736(05)78358-6. [DOI] [PubMed] [Google Scholar]
- 32.Butler D A, Scott M R D, Bockman J M, Borchelt D R, Taraboulos A, Hsiao K K, Kingsbury D T, Prusiner S B. J Virol. 1988;62:1558–1564. doi: 10.1128/jvi.62.5.1558-1564.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kascsak R J, Rubenstein R, Merz P A, Tonna-DeMasi M, Fersko R, Carp R I, Wisniewski H M, Diringer H. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serban D, Taraboulos A, DeArmond S J, Prusiner S B. Neurology. 1990;40:110–117. doi: 10.1212/wnl.40.1.110. [DOI] [PubMed] [Google Scholar]
- 35.Scott M R, Köhler R, Foster D, Prusiner S B. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shoichet B K, Kuntz I D. J Mol Biol. 1991;221:327–346. doi: 10.1016/0022-2836(91)80222-g. [DOI] [PubMed] [Google Scholar]
- 37.Tilley J W, Chen L, Fry D C, Emerson S D, Powers G D, Biondi D, Varnell T, Trilles R, Guthrie R, Mennona F, et al. J Am Chem Soc. 1997;119:7589–7590. [Google Scholar]
- 38.Clackson T, Wells J A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 39.Bogan A A, Thorn K S. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 40.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, et al. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bower M J, Cohen F E, Dunbrack R L J. J Mol Biol. 1997;267:1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 42.Ho C M, Marshall G R. J Comput Aided Mol Des. 1995;9:65–86. doi: 10.1007/BF00117279. [DOI] [PubMed] [Google Scholar]
- 43.Ullman J R. J Assoc Comput Machinery. 1976;23:31–42. [Google Scholar]
- 44.Rogers M, Serban D, Gyuris T, Scott M, Torchia T, Prusiner S B. J Immunol. 1991;147:3568–3574. [PubMed] [Google Scholar]
- 45.Race R E, Fadness L H, Chesebro B. J Gen Virol. 1987;68:1391–1399. doi: 10.1099/0022-1317-68-5-1391. [DOI] [PubMed] [Google Scholar]
- 46.Demaimay R, Harper J, Gordon H, Weaver D, Chesebro B, Caughey B. J Neurochem. 1998;71:2534–2541. doi: 10.1046/j.1471-4159.1998.71062534.x. [DOI] [PubMed] [Google Scholar]
- 47.Bierer B, Somers P K, Wandless T J, Burakoff S J, Schreiber S L. Science. 1990;250:556–559. doi: 10.1126/science.1700475. [DOI] [PubMed] [Google Scholar]
- 48.Clipstone N, Fiorentino D F, Crabtree G R. J Biol Chem. 1994;269:26431–26437. [PubMed] [Google Scholar]
- 49.Livnah O, Stura E A, Johnson D L, Middleton S A, Mulcahy L S, Wrighton N C, Dower W J, Jolliffe L K, Wilson I A. Science. 1996;273:464–471. doi: 10.1126/science.273.5274.464. [DOI] [PubMed] [Google Scholar]
- 50.Zhang B, Salituro G, Szalkowski D, Li Z, Zhang Y, Royo I, Vilella D, Diez M T, Pelaez F, Ruby C, et al. Science. 1999;284:974–977. doi: 10.1126/science.284.5416.974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.