Genetic predisposition probably plays an important part in the pathogenesis of systemic lupus erythematosus (SLE). Several loci have been suggested to be related to SLE, among which is the 1q41–42 region on chromosome 1.1 This region is homologous to the Sle1 region in an SLE mouse model.2 One of the genes on this region is the poly(ADP‐ribose) polymerase 1 (PARP1) gene, which has been associated with SLE in several studies. This gene encodes a nuclear enzyme involved in DNA repair and apoptosis, and its activity is reduced in patients with SLE,3 thereby possibly facilitating autoimmunity and inflammation. However, conflicting results with regard to this association have been reported.4,5,6 Tsao et al4 reported preferential transmission of PARP alleles to patients with SLE, whereas Criswell et al5 and Delrieu et al6 could not confirm these results. The first two studies were performed in an American, multiethnic population, and the third used a sample of French Caucasian patients. In a comment on the negative finding of Delrieu et al, Tan et al7 performed additional analyses showing that PARP1 is a candidate gene for SLE, at least in the African–American population. Another locus on this region, LOC127086, has not been studied before.
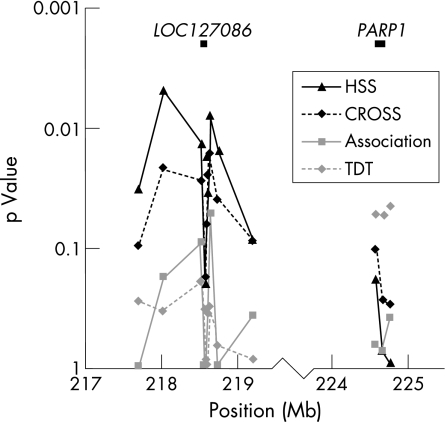
In this study, we analysed the involvement of the 1q41–42 region, in particular the PARP1 gene and LOC127086, in a Dutch Caucasian population of 103 patients with SLE (table 1). DNA from family members was also obtained, thereby constituting 103 trios. Microsatellite markers covering this region were genotyped. Strong linkage disequilibrium was observed between markers close to PARP1 and close to LOC127086. Haplotype sets of patients and controls were compared using the Haplotype Sharing Statistic (HSS) and the CROSS test. The HSS compares the length of haplotypes between patients and controls, and the CROSS test assumes that the haplotypes spanning the disease locus differ more between patients and controls than between patients themselves. These methods have proved to be valid in other studies.8,9,10 HSS and CROSS showed significant differences between patients and controls at markers close to LOC127086 (p = 0.005; fig 1). This result was confirmed by single‐locus association analysis, although just not significant (p = 0.051; fig 1). The Transmission/Disequilibrium Test was performed on trios consisting of patients and their parents. This showed significantly lower transmission of alleles surrounding PARP1 (p = 0.045; fig 1), suggesting a protective effect of these alleles.
Table 1 Basic characteristics of 103 patients with systemic lupus erythematosus.
| Median age (years; min–max) | 43 (23–78) |
| Sex: female, n (%) | 89 (86) |
| Median age of onset (years; min–max) | 31 (8–73) |
| Median duration of disease (months; min–max) | 131 (21–516) |
| Race, n (%) | |
| Caucasian | 103 (100) |
| ACR criteria, n (%) | |
| Malar rash | 37 (36) |
| Discoid rash | 31 (30) |
| Photosensitivity | 52 (50) |
| Oral ulcers | 13 (13) |
| Arthritis | 67 (65) |
| Serositis | 39 (38) |
| Renal disorder | 42 (41) |
| Neurological disorder | 7 (7) |
| Haematological disorder | 75 (73) |
| Immunological disorder | 91 (88) |
| Anti‐dsDNA | 81 (79) |
| Anti‐Sm | 13 (13) |
| Anti‐phospholipid antibodies | 21 (20) |
| Antinuclear antibody | 103 (100) |
ACR, American College of Rheumatology.
Figure 1 Results of the Haplotype Sharing Statistic (HSS), CROSS test, association analysis and the Transmission/Disequilibrium Test (TDT).
Our study shows that PARP1 is a candidate gene for SLE in a Dutch Caucasian population. As this gene encodes a nuclear enzyme that is involved in DNA repair and apoptosis, it is a logical candidate gene for SLE. We also found a novel candidate locus, LOC127086, which is a pseudogene similar to the XRCC6 gene on chromosome 22q11–13. This gene encodes subunit p70 of the lupus Ku p70/80 autoantigen, an SLE‐related gene. LOC127086 is a pseudogene without a transcription product, hence its exact function remains unclear. However, the fact that it is a copy of an SLE‐related gene is noteworthy.
Footnotes
Competing interests: None declared.
References
- 1.Tsao B P. The genetics of human systemic lupus erythematosus. Trends Immunol 200324595–602. [DOI] [PubMed] [Google Scholar]
- 2.Kono D H, Burlingame R W, Owens D G, Kuramochi A, Balderas R S, Balomenos D.et al Lupus susceptibility loci in New Zealand mice. Proc Natl Acad Sci U S A 19949110168–10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sibley J T, Haug B L, Lee J S. Altered metabolism of poly(ADP‐ribose) in the peripheral blood lymphocytes of patients with systemic lupus erythematosus. Arthritis Rheum 1989321045–1049. [DOI] [PubMed] [Google Scholar]
- 4.Tsao B P, Cantor R M, Grossman J M, Shen N, Teophilov N T, Wallace D J.et al PARP alleles within the linked chromosomal region are associated with systemic lupus erythematosus. J Clin Invest 19991031135–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Criswell L A, Moser K L, Gaffney P M, Inda S, Ortmann W A, Lin D.et al PARP alleles and SLE: failure to confirm association with disease susceptibility. J Clin Invest 20001051501–1502. [PubMed] [Google Scholar]
- 6.Delrieu O, Michel M, Frances C, Meyer O, Michel C, Wittke F.et al Poly(ADP‐ribose) polymerase alleles in French Caucasians are associated neither with lupus nor with primary antiphospholipid syndrome. GRAID Research Group. Group for Research on Auto‐Immune Disorders. Arthritis Rheum 1999422194–2197. [DOI] [PubMed] [Google Scholar]
- 7.Tan F K, Reveille J D, Arnett F C, Stivers D N, Tsao B P. Poly(ADP)‐ribose polymerase and susceptibility to systemic lupus erythematosus and primary antiphospholipid syndrome: comment on the article by Delrieu et al. Arthritis Rheum 2000431421–1423. [DOI] [PubMed] [Google Scholar]
- 8.Beckmann L, Fischer C, Deck K G, Nolte I M, te Meerman G, Chang‐Claude J. Exploring haplotype sharing methods in general and isolated populations to detect gene(s) of a complex genetic trait. Genet Epidemiol 200121(Suppl 1)S554–S559. [DOI] [PubMed] [Google Scholar]
- 9.Boon M, Nolte I M, Bruinenberg M, Spijker G T, Terpstra P, Raelson J.et al Mapping of a susceptibility gene for multiple sclerosis to the 51 kb interval between G511525 and D6S1666 using a new method of haplotype sharing analysis. Neurogenetics 20013221–230. [DOI] [PubMed] [Google Scholar]
- 10.Levinson D F, Kirby A, Slepner S, Nolte I, Spijker G T, te Meerman G. Simulation studies of detection of a complex disease in a partially isolated population. Am J Med Genet 200110565–70. [PubMed] [Google Scholar]