Abstract
Background and aim
Known colorectal cancer syndromes, such as familial adenomatous polyposis and hereditary non‐polyposis colorectal cancer, have been identified in only a small proportion of cases with a family history of disease. In an attempt to identify loci harbouring novel predisposing genes, we have performed a genome wide linkage analysis in 18 colorectal cancer families recruited from the Department of Clinical Genetics at Karolinska Hospital, Sweden.
Methods
Multipoint parametric and non‐parametric linkage analyses were performed using two affected status criteria, stringent and less stringent. Parametric analysis was performed under the assumption of locus homogeneity and locus heterogeneity.
Results
The initial scan performed using the less stringent affected status criteria revealed regions of interest on chromosome 11 (marker D11S1314: heterogeneity logarithm of odds (HLOD) score 1.96, non‐parametric LOD (NPL) score 1.28; and marker D11S908: HLOD score 2.10, NPL score 2.16) and chromosome 14 (marker D14S258: HLOD score 2.61, NPL score 2.88). Using the stringent affected status criteria, a locus on chromosome 22 was suggested in the parametric analysis (marker D22S315: HLOD score 1.26). After finemapping of the regions on chromosomes 11 and 14, HLOD and NPL scores were reduced but still within the range of suggestive linkage. Haplotype analysis revealed overlapping regions between D11S987 and D11S4207 (proximal region), D11S4120 and D11S4090 (distal region), on chromosome 11, and between D14S1038 and D14S1069 on chromosome 14.
Conclusion
Our study provides evidence of genetic heterogeneity among Swedish colorectal cancer families. Three novel regions were suggested to be of interest in a proportion of families analysed. Further studies are needed to confirm this result.
Keywords: linkage analysis, hereditary non‐polyposis colorectal cancer, familial adenomatous polyposis, colorectal cancer, chromosome 11, chromosome 14, chromosome 22
Colorectal cancer is the third most common type of malignancy in Sweden and affects both men and women equally. The estimated lifetime colorectal cancer risk in the general population is 5%. Effects of environment and diet on the development of colorectal cancer are known,1 but epidemiological2,3 and twin4 studies have established a large genetic component in the aetiology of the disease. Hereditary syndromes have been described, of which the two best characterised are familial adenomatous polyposis (FAP) and hereditary non‐polyposis colorectal cancer (HNPCC).5 However, known syndromes have been identified in only a proportion of colorectal cancer families, leaving a majority of hereditary cases unexplained, which points to the existence of novel colorectal cancer genes. Identification of families not linked to any of the known colorectal cancer loci supports this idea.6,7,8
In Sweden, HNPCC and FAP have been identified in <3% of all colorectal cancer cases, but in a further 10% of colorectal cancer cases a hereditary component is recognised.9 However, in these families, disease causing genes have not been identified. Of these families, 1.9% are considered to be at high risk (hereditary colorectal cancer families (HCRC)). They are composed of three or more first degree relatives affected with colorectal cancer in at least two generations and are likely to segregate high risk genes transmitted in a dominant manner.9 Another 8.3% of Swedish colorectal cancer families comprise a low risk group, where two first degree relatives affected with colorectal cancer are identified (two close relatives families (TCR)). Inheritance of mildly to moderately penetrant susceptibility factors is a possible cause of the disease in these families but environmental factors cannot be excluded.9
Traditional linkage analysis has been very successful in identifying a number of disease causing genes. Selection of families with distinctive phenotypes has been shown to be of major importance in identifying cancer genes, as was done in the case of the FAP syndrome and identification of the APC gene.10,11 In contrast, due to locus heterogeneity, HNPCC causing genes were identified only after large single families were used alone in linkage analysis.12,13
Since 1990, families at risk of developing colorectal cancer (HNPCC, HCRC, and TCR families) have been included in a surveillance programme at Karolinska Hospital, where all subjects with a risk of colorectal cancer development greater than 10% have been offered regular colonoscopy and polypectomy every two years. Analysis of patients under surveillance has revealed a significantly increased risk of developing an adenoma before the age of 54 years not only in HNPCC but also in HCRC and TCR families.14 Moreover, a significant correlation between adenoma and hyperplastic polyps has been seen in all three types of families, indicating that adenomas and hyperplastic polyps can be used as markers in detecting individuals at risk due to inherited factors.15 Several other studies have confirmed these results.16,17,18
In an attempt to locate novel colorectal cancer predisposing genes, we have selected 12 high risk and six low risk colorectal cancer families (non‐FAP/non‐HNPCC) and performed a genome wide parametric and non‐parametric linkage analysis.
Material and methods
Families
Eighteen colorectal cancer families (190 subjects) were used in a genome wide screen. All families were recruited from the Department of Clinical Genetics at Karolinska Hospital, Sweden. Family history was obtained and all diagnoses were confirmed through medical records or death certificates. Informed consent was obtained from each family member in accordance with Swedish law concerning ethics approval of research on human subjects (2000:291). None of the families included in this study had classical or attenuated polyposis.
As it has been shown that microsatellite instability (MSI) is a powerful predictor of mutations in mismatch repair genes,7 we used the same approach in this study to eliminate HNPCC families. Available tumours from all 18 families were previously analysed for MSI and all were shown to be microsatellite stable.7 In addition, all families fulfilling the Amsterdam criteria (families 53, 155, 181) were tested for mutations in the hMLH1 and hMSH2 genes, and no mutations were detected.8 Mutation screening of the hMSH6 and hPMS2 genes was also performed in families 53, 68, 70, 100 101, 134 and 155, while family 161 was screened for the hMSH6 gene only. No mutations were found (unpublished data). Characteristics of the families included in this study are presented in table 1.
Table 1 Families included in the linkage analysis.
| Family ID | No of CRC cases | No affected (criteria 1) | No affected (criteria 2) | Other cancers | Family type |
|---|---|---|---|---|---|
| 29 | 3 | 3 | 7 | Endo | HCRC |
| 53 | 3 | 3 | 6 | Endo | HCRC |
| 68 | 3 | 6 | 7 | Endo, Br | HCRC |
| 70 | 2 | 2 | 5 | — | TCR |
| 100 | 2 | 4 | 9 | GB | TCR |
| 101 | 3 | 4 | 7 | Br | HCRC |
| 134 | 3 | 3 | 5 | Ga | HCRC |
| 155 | 6 | 10 | 13 | Ga, Ki | HCRC |
| 161 | 3 | 3 | 6 | Ov, Le, Ce, Pr | HCRC |
| 181 | 3 | 4 | 4 | Pa | HCRC |
| 191 | 3 | 4 | 5 | — | HCRC |
| 197 | 2 | 2 | 5 | Lung, Br | TCR |
| 201 | 2 | 2 | 5 | Ov | TCR |
| 202 | 2 | 4 | 9 | — | TCR |
| 216 | 3 | 3 | 6 | Ga, Br | HCRC |
| 242 | 6 | 9 | 16 | Ga | HCRC |
| 244 | 1 | 1 | 6 | — | TCR |
| 309 | 4 | 4 | 6 | Br | HCRC |
CRC, colorectal cancer; Endo, endometrial; Br, breast; GB, gall bladder; Ga, gastric; Ki, kidney; Ov, ovarian; Le, leukaemia; Ce, cervical; Pr, prostate; Pa, pancreas; HCRC, hereditary colorectal cancer families; TCR, two close relatives families
Criteria 1, patients with CRC or adenomas with high grade dysplasia.
Criteria 2, patients with CRC, adenomas with high or low grade dysplasia, two or more hyperplastic polyps, or one polyp >10 mm.
Genotyping
Peripheral blood lymphocytes were collected from available family members and DNA was extracted by standard methods.
A genome wide linkage scan was carried out using the ABI Prism Linkage Mapping Set version 2.5 (Applied Biosystems, Foster City, California, USA) which consists of 400 fluorescently labelled microsatellite markers distributed across the genome with an average spacing of 10 cM and an average heterozygosity of 0.76. Each marker was amplified independently according to the manufacturer's protocol. An additional 11 markers on chromosome 11 (D11S4136, D11S4206, D11S1362, D11S4135, D11S1887, D11S1332, D11S4120, D11S4206, D11S4090, D11S4142, D11S4129) and 19 markers on chromosome 14 (D14S1027, D14S269, D14S1018, D14S285, D14S1038, D14S997, D14S290, D14S63, D14S981, D14S1069, D14S1011, D14S1002, D14S277, D14S268, D14S1028, D14S1047, D14S61, D14S287, D14S1037) were selected for finemapping. Their position, relative to the position of the first marker set, was estimated from the Généthon human linkage map (polymerase chain reaction (PCR) conditions are available on request). PCR products were pooled and separated on an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster city, California, USA) together with Genescan 400HD ROX size standard (Applied Biosystems). Electrophoretic data were analysed using Genescan3.1 and Genotyper2.0 software programs (Applied Biosystems).
Statistical analysis
Genotyping data were checked for Mendelian inconsistencies through all levels of PedCheck19 and errors were either resolved unambiguously or families with errors were discarded from the analysis for the marker in question. The mistyping analysis option of the SimWalk2 program version 2.8320 was used in order to detect possible non‐Mendelian genotyping errors. Under this analysis option, SimWalk2 reports an overall probability of mistyping at each observed genotype. Marker genotypes with significant probability of mistyping were deleted. Marker allele frequencies were calculated from all genotyped family members using the PedCheck program.
Linkage analysis was performed using two different criteria for the affected status. In the first more stringent affected status criteria, all patients with colorectal cancer or adenomas with high grade dysplasia of any size were coded as affected, regardless of age of onset, while patients with adenomas with low grade dysplasia or with hyperplastic polyps were coded as unknown. In the second less stringent affected status criteria, all patients with colorectal cancer, adenomas of any size (with high or low grade dysplasia), two or more hyperplastic polyps, or one hyperplastic polyp larger than 10 mm were coded as affected, regardless of age of onset.
Both multipoint parametric and non‐parametric linkage analysis of all autosomes was carried out using the SimWalk2 program version 2.83. This program uses the Markov chain Monte Carlo and simulated annealing algorithms to compute location scores which are directly comparable with multipoint logarithm of odds (LOD) scores and are presented in log10 units. Markov chain sampling of descent graphs also permits evaluation of marker allele sharing among affected pedigree members.
Parametric linkage analysis using either of the two affected status criteria was performed assuming both locus homogeneity and locus heterogeneity. Under the assumption of locus heterogeneity, location scores were calculated by varying a proportion of linked families (α value) until the highest heterogeneity LOD (HLOD) scores were obtained. An autosomal dominant mode of inheritance was assumed with a disease allele frequency of 0.0001 and an equal female and male recombination rate. Penetrance for homozygous normal, heterozygous, and homozygous affected was 0.05, 0.80, and 0.80, respectively.
The SimWalk2 program was also used for generating haplotypes. The X chromosome was analysed by computing single point LOD scores using the FASTLINK program.21
Results
Linkage analysis using the stringent affected status criteria
Using the stringent affected status criteria, parametric and non‐parametric linkage analysis was performed in 18 families. Based on these criteria, chromosome 22 was suggested to be of interest, but only in the parametric linkage analysis performed under the assumption of locus heterogeneity. Suggestive linkage was observed for the region around marker D22S315 where a HLOD score of 1.26 was obtained for α = 0.60 (table 2). Non‐parametric analysis using the same stringent criteria showed no evidence of linkage to any of the autosomes.
Table 2 Markers showing the highest non‐parametric logarithm of odds (LOD) scores (STAT E) and parametric heterogeneity LOD (HLOD) scores after the first scan.
| Chromosome | Marker name | cM† | NPL score (STAT E) | p Value‡ | Parametric HLOD score | α |
|---|---|---|---|---|---|---|
| 11 | D11S1314 | 77.5 | 1.28 | 0.05 | 1.96 | 0.25 |
| 11 | D11S908 | 112.5 | 2.16 | 0.006 | 2.10 | 0.35 |
| 14 | D14S258 | 65.8 | 2.88 | 0.001 | 2.61 | 0.25 |
| 22 | D22S315* | 16.2 | 0.96 | 0.10 | 1.26 | 0.60 |
*Result obtained using stringent diagnostic criteria.
†Based on the Généthon map.
‡p values calculated for non‐parametric LOD scores.
α, Proportion of linked families.
Linkage analysis using the less stringent affected status criteria
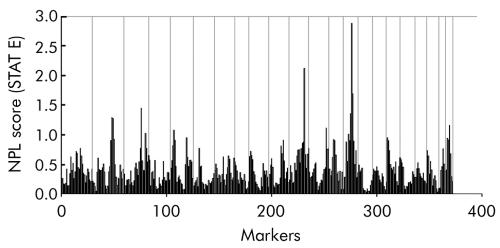
When the less stringent affected status criteria were used, no evidence of linkage was found by multipoint parametric linkage analysis, assuming locus homogeneity (data not shown). However, analysis under the assumption of locus heterogeneity with the same criteria revealed three regions of interest. Suggestive linkage was observed for chromosome 14 and chromosome 11 (table 2). Parametric analysis of chromosome 14 resulted in one peak with the highest HLOD score of 2.61 (α = 0.25) for the marker D14S258 (table 2). Two peaks were observed on chromosome 11: a HLOD score of 2.10 (α = 0.35) was calculated for the marker D11S908 and a HLOD score of 1.96 (α = 0.25) for the marker D11S1314, located ∼34 cM proximal to D11S908 (table 2). Results from the non‐parametric multipoint linkage analysis with the less stringent criteria over all autosomes, summarised for all pedigrees, are shown in fig 1. An NPL score of 2 or more, which indicated significance in this analysis, was reached twice. The strongest linkage was found at chromosome 14, an NPL score (STAT E as implemented in the SimWalk 2 program) of 2.88 (p = 0.001) was obtained for marker D14S258 (table 2). In addition, linkage was observed for the marker D11S908 on chromosome 11 (NPL score of 2.16, p = 0.006).
Figure 1 Non‐parametric linkage analysis for the colorectal cancer susceptibility loci in 18 families. Non‐parametric logarithm of odds (NPL) scores are shown across all autosomes, from pter to qter. Vertical lines indicate chromosome boundaries.
No other chromosomes, apart from chromosomes 11 and 14, showed evidence of linkage in the parametric analysis using the less stringent criteria for the affected status. An indication of linkage was, however, detected on several chromosomes in the non‐parametric linkage analysis. NPL scores (STAT E) reaching values over 1 were seen on chromosome 2 (NPL score of 1.29 for D2S142), chromosome 3 (NPL score of 1.45 for D3S1279), chromosome 5 (NPL score of 1.07 for D5S16), chromosome 12 (NPL score 1.11 for D12S324), and chromosome 22 (NPL score of 1.15 for D22S280).
Two point parametric linkage analysis of the X chromosome showed no evidence of linkage to any of the markers (data not shown).
Not all families were tested for mutations in the hMSH6 gene, which has been shown to be associated with MSI negative tumours. However, no evidence of linkage was found to the region on chromosome 2 harbouring this gene in either the entire family group or in any individual family.
Finemapping
As chromosomes 11 and 14 showed the most significant evidence of linkage in this study, additional markers within these regions were selected for analysis. Nineteen markers spanning a region of ∼42 cM on chromosome 14 and 11 markers spanning a region of ∼45 cM on chromosome 11 were genotyped for the original 18 families, and parametric and non‐parametric linkage analysis were performed using both affected status criteria.
After finemapping, HLOD scores and NPL scores decreased across the region on chromosome 14 (table 3). The highest HLOD score was 0.62 (α = 0.20) and was obtained for the novel marker D14S63, located 6.8 cM proximal to D14S258. The highest NPL score of 1.40 (p = 0.03) was observed for the same marker. For the proximal region on chromosome 11, the HLOD score decreased to 0.87 (α = 0.25) for the marker D11S1314 (table 3) but the highest NPL score of 1.57 (p = 0.02) in this region was calculated for D11S4207, located 2.7 cM distal to D11S1314. For this marker the parametric HLOD score was 0.44 for an α value of 0.20. HLOD and NPL scores decreased, after finemapping, in the distal region on chromosome 11 (table 3). The highest values were obtained for D11S4090, located 2.6 cM proximal to D11S908 (HLOD score of 0.25, α = 0.15 and NPL score of 1.55, p = 0.02).
Table 3 Finemapping results for markers on chromosomes 11 and 14.
| Chromosome | Marker name | cM* | NPL score (STAT E) | p Value† | Parametric HLOD score | α |
|---|---|---|---|---|---|---|
| 11 | D11S1314 | 77.5 | 1.31 | 0.04 | 0.87 | 0.25 |
| 11 | D11S908 | 112.5 | 1.46 | 0.03 | 0.20 | 0.10 |
| 14 | D14S258 | 65.8 | 1.31 | 0.04 | 0.22 | 0.15 |
LOD, logarithm of odds; HLOD, heterogeneity LOD; NPL, non‐parametric LOD. *Based on the Généthon map.
†p values calculated for NPL scores.
α, proportion of linked families.
Although HLOD and NPL scores failed to reach statistical significance after finemapping of chromosomes 11 and 14, their values were still within the range of suggestive linkage. Moreover, haplotype analysis revealed several families with linkage to these two chromosomes. Families 70, 100, 161, 197, 201, and 244 showed linkage to chromosome 11, and haplotype analysis revealed two regions of linkage: the first between D11S987 and D11S4207 (11q13.2–11q13.4) and the second between D11S4120 and D11S4090 (11q22.1–11q23.1). Families 53, 70, 101, 161, 201, and 202 were linked to chromosome 14, where an overlapping region between markers D14S1038 and D14S1069 (14q23.1–14q24.1) was identified.
Discussion
In an attempt to identify novel colorectal cancer predisposing genes, we performed a genome wide linkage analysis in 18 colorectal cancer families from Sweden. All families were included in a surveillance programme at Karolinska Hospital because of a family history of the disease. Linkage analysis was performed using two affected status criteria, one stringent and the second less stringent. Using the stringent criteria, only patients with colorectal cancer or adenomas with high grade dysplasia of any size were coded as affected. Due to the successful prevention programme, the number of new cancer cases in these families was very low and the size of adenomas rarely exceed 5 mm. Bearing in mind that adenomas and hyperplastic polyps can be used as markers for individuals at risk of colorectal cancer,15 in the less stringent affected status criteria we also considered patients with adenomas of any size (with high or low grade dysplasia), patients with two or more hyperplastic polyps, or one hyperplastic polyp larger than 10 mm as affected.
The families included in this study were classified as high risk or low risk, based on the number of family members affected with colorectal cancer. However, it is difficult to know whether different genetic factors contributed to the disease in these two groups of families.22 For this reason linkage analysis was performed for the two groups together. As the location scores and haplotypes were generated for each family, it was possible to analyse families individually and no evidence for involvement of a single locus in either of the two family groups was found. Neither did the current study identify one major locus for the entire group of families examined, regardless of the statistical analysis used. The data support the hypothesis that genetic heterogeneity plays a role in the aetiology of colorectal cancer. Further support for the role of genetic heterogeneity is provided by the overall results of this genome wide scan where three chromosomes were suggested in a proportion of families analysed.
Recently, two novel colorectal cancer candidate loci have been identified. Linkage to chromosome 15q13‐q14 has been found in several Ashkenazi families, but this locus has been shown to be associated with a specific phenotype; the hereditary mixed polyposis syndrome.23 A sibling pair analysis on 53 kindreds affected by colon cancer or advanced adenomas has led to identification of a second candidate colorectal cancer locus on chromosome 9q22.2–q31.2.24 In our analysis we found no evidence for linkage to the suggested regions on 9q and 15q using either of the two affected status criteria. Similarly, none of the regions described here were detected in the study by Wiesner and colleagues24 or Jaeger and colleagues.23 These findings further support evidence of genetic heterogeneity in the aetiology of colorectal cancer.
In the current analysis performed using the stringent affected status criteria, suggestive linkage was observed for the region on chromosome 22, around marker D22S315, but only in the parametric analysis under the assumption of locus heterogeneity. This locus was also supported by the result of non‐parametric analysis using the less stringent affected status criteria where the NPL score of 1.15 was detected for the marker D22S280 (located 10.4 cM distal to D22S315).
Two regions on chromosome 11 and one region on chromosome 14 were identified after the initial parametric and non‐parametric linkage analysis using the less stringent affected status criteria. Following finemapping of the three loci, HLOD and NPL scores were reduced across all three regions but remained within the range of suggestive linkage. Haplotype analysis identified families linked to these regions and revealed overlapping regions of 8.7 and 9.4 cM on chromosome 11 and 5.5 cM on chromosome 14.
The regions on chromosomes 11 and 14 were not detected in the analysis performed using the stringent affected status criteria. Performing linkage analysis in colorectal cancer families is difficult due to incomplete and age dependent penetrance and the effect of environmental factors. An estimate of the phenocopy rate was included in the statistical analysis. However, it is possible that this estimate was too low, especially when performing the analysis using the less stringent affected status criteria. This was also suggested by haplotype analysis of the linked families after finemapping. Incomplete segregation of the disease haplotype and the affected status was seen, which can explain the reduction in LOD scores.
The difficulty in identifying a major colorectal cancer predisposing gene may be due not only to the expected heterogeneity but also to the existence of a number of genes with reduced penetrance, each of them responsible for only a small fraction of familial clustering,25 or the effect of a number of additive or modifying low risk genes.22 Analyses of the suggested regions on chromosomes 11q and 14q found that some families were linked to both chromosomes (families 70, 161, 201) which could support the idea of different genes contributing to disease within the same family.
In summary, a genome wide linkage analysis in 18 colorectal cancer families from Sweden did not reveal a single predisposing locus using either stringent or less stringent affected status criteria. An indication for linkage was found to chromosomes 11q, 14q, and 22q. Although the results seen here failed to reach statistical significance, it would be worthwhile to further investigate suggested regions in an independent set of families in order to determine the possible role of these loci in colorectal cancer predisposition.
Acknowledgements
We are indebted to the families for their cooperation. We also thank Katariina Lunnas for help with genotyping. This work was funded by the Swedish Cancer Foundation, the Swedish Cancer Society (03‐0300, 3557), the Stockholm Cancer Society (03:316), and the Nilsson‐Ehle Foundation.
Abbreviations
HNPCC - hereditary non‐polyposis colorectal cancer
FAP - familial adenomatous polyposis
HCRC - hereditary colorectal cancer
TCR - two close relatives
MSI - microsatellite instability
LOD - logarithm of odds
HLOD - heterogeneity LOD
NPL - non‐parametric LOD
PCR - polymerase chain reaction
Footnotes
Conflict of interest: None declared.
References
- 1.Wei E, Giovannucci E, Wu K.et al Comparison of risk factors for colon and rectal cancer. Int J Cancer 2004108433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannon Albright L, Skolnick M, Bishop D.et al Common inheritance of susceptibility to colonic adenomatous polyps and associated colorectal cancers. N Engl J Med 1988319533–537. [DOI] [PubMed] [Google Scholar]
- 3.Houlston R, Collins A, Slack J.et al Dominant genes for colorectal cancer are not rare. Ann Hum Genet 19925699–103. [DOI] [PubMed] [Google Scholar]
- 4.Lichtenstein P, Holm N, Verkasalo P.et al Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark and Finland. N Engl J Med 200034378–85. [DOI] [PubMed] [Google Scholar]
- 5.Lynch H, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med 2003348919–932. [DOI] [PubMed] [Google Scholar]
- 6.Lewis C, Neuhausen S, Daley D.et al Genetic heterogeneity and unmapped genes for colorectal cancer. Cancer Res 1996561382–1388. [PubMed] [Google Scholar]
- 7.Liu T, Wahlberg S, Burek E.et al Microsatellite instability as a predictor of mutation in a DNA mismatch repair gene in familial colorectal cancer. Genes Chromosomes Cancer 20002717–25. [DOI] [PubMed] [Google Scholar]
- 8.Wahlberg S, Liu T, Lindblom P.et al Various mutation screening techniques in the DNA mismatch repair genes hMSH2 and hMLH1. Genet Test 19993259–264. [DOI] [PubMed] [Google Scholar]
- 9.Olsson L, Lindblom A. Family history of colorectal cancer in a Sweden country. Fam Cancer 2003287–93. [DOI] [PubMed] [Google Scholar]
- 10.Bodmer W, Bailey C, Bodmer J.et al Localisation of the gene for familial adenomatous polyposis on chromosome 5. Nature 1987328614–616. [DOI] [PubMed] [Google Scholar]
- 11.Groden J, Thliveris A, Samovitz W.et al Identification and characterisation of the familial adenomatous polyposis coli gene. Cell 199166589–600. [DOI] [PubMed] [Google Scholar]
- 12.Peltomaki P, Aaltonen L, Sistonen P.et al Genetic mapping of a locus predisposing to human colorectal cancer. Science 1993260810–812. [DOI] [PubMed] [Google Scholar]
- 13.Lindblom A, Tannergård P, Werelius B.et al Genetic mapping of a second locus predisposing to hereditary non‐polyposis colon cancer. Nat Genet 19935279–282. [DOI] [PubMed] [Google Scholar]
- 14.Lindgren G, Liljegren A, Jaramillo E.et al Adenoma prevalenc and cancer risk in familial non‐polyposis colorectal cancer. Gut 200250228–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liljegren A, Lindgren G, Brandberg Y.et al Individuals with an increased risk for colorectal cancer‐perceived benefit and psychological aspect on surveillance by means of regular colonoscopies. J Clin Oncol 2004221736–1742. [DOI] [PubMed] [Google Scholar]
- 16.Williams A, Balasooriya B, Day D. Polyps and cancer of the large bowel: a neropsy study in Liverpool. Gut 198223835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jass J, Young P, Robinson E. Predictors of presence, multiplicity, size and dysplasia of colorectal adenomas. A necropsy study in New Zeland. Gut 1992331508–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syrigos K, Charalampopoulos A, Ho J.et al Colonoscopy in asymptomatic individuals with a family history of colorectal cancer. Ann Surg Oncol 20029439–443. [DOI] [PubMed] [Google Scholar]
- 19.ÒConell J, Weeks D. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 199863259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores and marker‐sharing statistics. Am J Hum Genet 1996581323–1337. [PMC free article] [PubMed] [Google Scholar]
- 21.Cottingham R J, Iduru R, Schaffer A. Faster sequential genetic linkage computations. Am J Hum Genet 199353252–263. [PMC free article] [PubMed] [Google Scholar]
- 22.Lindblom A, Zhou X, Liu T.et al Colorectal cancer as a complex disease: defining at‐risk subjects in the general population. A preventive strategy. Expert Rev Anticancer Ther 2004489–97. [DOI] [PubMed] [Google Scholar]
- 23.Jaeger E, Woodford‐Richens K, Locket M.et al An ancestral Ashkenazi haplotype at the HMPS/CRAC1 locus on 15q13–q14 is associated with hereditary mixed polyposis syndrome. Am J Hum Genet 2003721261–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiesner G, Daley D, Lewis S.et al A subset of familial colorectal neoplasia kindreds linked to chromosome 9q22.2–31.2. Proct Natl Acad Sci USA 200310012961–12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fearnhead N S, Winney B, Bodmer W F. Rare variant hypothesis for multifactorial inheritance: susceptibility to colorectal adenomas as a model. Cell Cycle 20054521–526. [DOI] [PubMed] [Google Scholar]