Abstract
Background
Although prostaglandin E2 (PGE2), cyclooxygenase 2 (COX‐2), and microsomal prostaglandin E synthase 1 (mPGES‐1) are known to play a role in various inflammatory events, their roles in the pathogenesis of gastro‐oesophageal reflux disease are not known.
Aims
We examined the dynamics of COX‐1, COX‐2, mPGES‐1, mPGES‐2, cytosolic PGES (cPGES), and PGE2 synthetic activity in rat acid reflux oesophagitis and the effects of COX‐2 inhibitors on the severity of oesophagitis.
Methods
Acid reflux oesophagitis was induced by ligating the transitional region between the forestomach and the glandular portion and wrapping the duodenum near the pylorus. Rats were killed on day 3 (acute phase) or day 21 (chronic phase) after induction of oesophagitis.
Results
Expression of COX‐2 and mPGES‐1 was markedly increased in oesophagitis while modest changes in COX‐1, cPGES, and mPGES‐2 expression were observed. COX‐2 and mPGES‐1 were colocalised in epithelial cells of the basal layer, as well as inflammatory and mesenchymal cells in the lamina propria and submucosa. COX‐2 inhibitors significantly reduced the severity of chronic oesophagitis but did not affect acute oesophageal lesions. COX‐2 inhibitors significantly inhibited the increase in PGE2 synthesis in oesophageal lesions on both days 3 and 21. Epithelial proliferation was significantly increased in the basal layer on day 21. Inflammatory cells and epithelial cells of the basal layer exhibited reactions for EP4 in oesophagitis.
Conclusion
PGE2 derived from COX‐2 and mPGES‐1 plays a significant role in the pathogenesis of chronic acid reflux oesophagitis, and possibly in basal hyperplasia and persistent inflammatory cell infiltration.
Keywords: cyclooxygenase 2, microsomal prostaglandin E synthesis 1, prostaglandin E2, cyclooxygenase 2 inhibitors, reflux oesophagitis
Gastric acid is one of the most important pathogenic factors of gastro‐oesophageal reflux disease (GORD) as proton pump inhibitors are effective in treating most patients with GORD.1 Continuous acid reflux induces several degrees of oesophageal mucosal injury, as well as mucosal thickening, with elongation of lamina papillae and basal cell hyperplasia.2 In addition to the strong regenerative responses of the oesophageal epithelium, inflammation with marked leucocytic infiltration is observed in lamina propria and submucosa in oesophagitis.
Prostaglandins (PGs) have various biological bioactivities, and there is evidence of their involvement in numerous pathological events. Human studies have shown that oesophageal PG levels are significantly increased in GORD patients compared with healthy adults,3 and that oesophageal PGE2 levels are correlated with the severity of oesophageal mucosal injury.4 However, the detailed role of PGE2 in the pathogenesis of GORD remains unclear. PGE2 is produced through three sequential enzymatic reactions: release of arachidonic acid from membrane glycerophospholipids by phospholipase A2; conversion of arachidonic acid to the unstable intermediate prostanoid PGH2 by cyclooxygenase (COX); and isomerisation of PGH2 to PGE2 by PGE synthase (PGES).5,6 COX has two isoforms: COX‐1 is constitutively expressed in various cells and tissues and plays an important role in maintaining homeostasis while COX‐2 is inducible and plays a key role in the process of inflammation.5 Recently, three different forms of PGES have been identified: cytosolic PGES (cPGES), microsomal PGES (mPGES)‐1, and mPGES‐2.6 cPGES is constitutively expressed in a wide variety of cells and tissues and is functionally coupled with COX‐1 whereas mPGES‐1 is a glutathione dependent enzyme which is preferentially coupled with COX‐2.7 Induced expression of mPGES‐1 has been postulated to be associated with various inflammatory conditions, such as rheumatoid arthritis8 and inflammatory bowel disease.9 mPGES‐2 has been shown to be glutathione independent, in contrast with mPGES‐1, but its functions are still unclear.6 As acid reflux causes oesophageal inflammation in GORD patients, it is possible that COX‐2, mPGES‐1, and PGE2 are associated with the pathogenesis of GORD. The present study was designed to examine: (1) the expression and dynamics of COXs and PGES as well as PGE2 levels in rat acid reflux oesophagitis, and (2) the effects of COX‐2 inhibitors on the severity of oesophagitis and oesophageal PGE2 levels.
Methods
Animals and induction of oesophagitis
Specific pathogen free male Wistar rats (Japan SLC, Hamamatsu, Japan) weighing approximately 200 g at the start of the experiment were used. Acid reflux oesophagitis was induced by the methods of Omura and colleagues.10 In brief, the duodenum near the pyloric ring was covered with a 2 mm wide piece of 18Fr Nelaton catheter (Terumo Co, Tokyo, Japan), and the transitional region between the forestomach and the glandular portion was ligated to enhance reflux of gastric contents into the oesophagus. Solid food was withdrawn for two days after induction of oesophagitis but rats were allowed drinking water. Rats were killed three or 21 days after induction of oesophagitis. Sham operated rats were used as controls. Oesophageal tissues were excised and immediately frozen in liquid nitrogen and stored at −80°C until real time reverse transcription‐polymerase chain reaction (RT‐PCR) or western blotting. For histological studies, samples were gently rinsed with saline and fixed in 10% buffered formalin. Samples were embedded in paraffin and 4 μm thick sections were cut. To examine the effects of COX‐2 inhibitors on the severity of oesophagitis, rats were subcutaneously given vehicle, 10 mg/kg/day of celecoxib (kindly provided by Pfizer Inc., New York, USA), or 10 mg/kg/day of nimesulide (Cayman Chemical Co., Ann Arbor, Michigan, USA) after induction of oesophagitis. Lesion index was defined as the total area of oesophageal lesions. The experimental protocol was approved by the Animal Care and Use Committees of Osaka City University.
Assessment of epithelial proliferation
Oesophageal epithelial proliferation was assessed by proliferating cell nuclear antigen (PCNA) staining. Sections were treated with antigen retrieval solution (target retrieval solution; Dako Japan, Kyoto, Japan) in a water bath for 40 minutes at 94°C. After immersion of the sections in a blocking agent solution (protein block serum‐free; Dako) for 20 minutes, endogenous peroxidase was inactivated by immersion of sections in 3% hydrogen peroxide for 15 minutes. Sections were then incubated with monoclonal mouse anti‐PCNA antibody (Dako) at a dilution of 1:200 overnight at 4°C. Immunohistochemical staining was performed with an LSAB‐2 kit (Dako) according to the manufacturer's instructions. Counterstaining was performed with methyl green. Oesophageal epithelial proliferation was quantified by PCNA labelling index, defined as the mean number of PCNA positive cells in five randomly selected high power fields.
RNA isolation and real time RT‐PCR
Total RNA was extracted and purified from frozen oesophageal tissues using Isogen (Nippon Gene, Toyama, Japan) according to the manufacturer's instruction. PCR primers and TaqMan probes for COX‐1, COX‐2, cPGES, mPGES‐1, and mPGES‐2 were used according to a previous report.11 Real time quantitative RT‐PCR analyses were performed using an ABI prism 7700 sequence detection system instrument and software (PE Applied Biosystems, Inc., Foster City, California, USA). The reaction mixture was prepared according to the manufacturer's protocol using the platinum quantitative RT‐PCR Thermoscript one‐step system (Invitrogen, Carlsbad, California, USA). Thermal cycling conditions included 50°C for 30 minutes and 95°C for five minutes, followed by 45 cycles of amplification at 95°C for 15 seconds and 60°C for one minute. Total RNA was subjected to real time quantitative RT‐PCR for measurement of target genes and glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) was used as an internal standard with TaqMan GAPDH control reagents (PE Applied Biosystems). Each resulting gene amount was divided by GAPDH gene amount to obtain a normalised value. mRNA levels are expressed as ratios to the mean value for normal oesophageal tissue.
Western blot analysis
Oesophageal tissues were homogenised and total protein separated in TNE buffer containing 10 μg/ml PMSF, 60 μg/ml aprotinin, and 1 mmol/l sodium orthovanadate. Target protein was separated with SuperSep 15% (Wako, Osaka, Japan) for PGES or SuperSep 5–20% (Wako) for COX‐1, COX‐2, and EP4, and transferred to PVDF membrane (Nippon Genetics, Tokyo, Japan). The membrane was blocked with blocking reagent and incubated with specific polyclonal rabbit antibodies to COX‐1, COX‐2, mPGES‐1, mPGES‐2, cPGES (Cayman), and EP4 (Santa Cruz Biotechnology Inc., California, USA) at dilutions of 1:500, 1:500, 1:250, 1:200, 1:1000, and 1:500, respectively, overnight. After washing with Tris buffered saline containing 0.1% Tween 20, the membrane was incubated with antirabbit IgG conjugated with horseradish peroxidase (Amersham Pharmacia Biotech, UK) at 1:1000 dilution for one hour at room temperature. Detection of target protein was achieved with an ECL western blotting detection system (Amersham) and visualised with an LAS‐1000 plus image reader (Fuji Photo‐Film, Tokyo, Japan).
Immunohistochemical study
After antigen retrieval, endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 15 minutes at 37°C. After immersion of the sections in a blocking agent solution for 20 minutes, they were incubated with primary antibody overnight at 4°C. The primary antibodies used were polyclonal goat anti‐COX‐1 (Santa Cruz) at a dilution of 1:200, goat anti‐COX‐2 antibody (Santa Cruz) at a dilution of 1:200, rabbit anti‐mPGES‐1 antibody (Cayman) at a dilution of 1:200, rabbit anti‐mPGES‐2 antibody (Cayman) at a dilution of 1:50, rabbit anti‐cPGES antibody (Cayman) at a dilution of 1:200, and rabbit anti‐EP4 antibody (Santa Cruz) at a dilution of 1:100. Immunohistochemical staining was performed by a streptavidin‐biotin peroxidase method using the ImmunoCruz staining system (Santa Cruz) or Dako LSAB 2 system HRP (Dako Japan). Sections were counterstained with haematoxylin.
To evaluate colocalisation of COX‐2 and mPGES‐1, double labelling by immunofluorescence methods was performed using confocal laser scanning microscopy. Each primary antibody pair was applied to separate rat oesophagitis tissue samples and incubated overnight at 4°C. The primary antibody to COX‐2 was allowed to react with a secondary antibody (donkey antigoat IgG at a dilution of 1:100) labelled with Alexa Fluor 568 (Invitrogen). The primary antibody to mPGES‐1 was allowed to react with a secondary antibody (donkey antirabbit IgG at a dilution of 1:100) labelled with Alexa Fluor 488 (Invitrogen). Samples were examined with a confocal microscope (model TCS4D/DMIRBE; Leica, Heidelberg, Germany) equipped with argon and argon‐krypton laser sources.
Measurement of PGE2 production in oesophageal tissue
PGE2 production by oesophageal tissue was assessed using a vortex method previously described.12 The amount of PGE2 was determined by enzyme immunoassay (PGE2 EIA kit; Cayman Chemical). PGE2 production was expressed as picograms of PGE2 per gram of tissue per minute.
Statistical analysis
Statistical analysis was performed with Statview software version 5.0 J (SAS Institute Japan, Tokyo, Japan). Differences between groups were examined for significance by one way ANOVA followed by the Fisher PLSD test. A p value of less than 0.05 was considered significant.
Results
Macroscopic and histological assessment
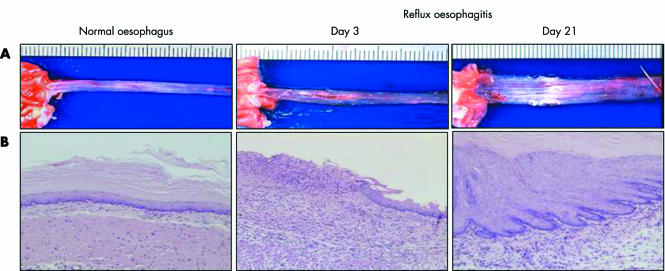
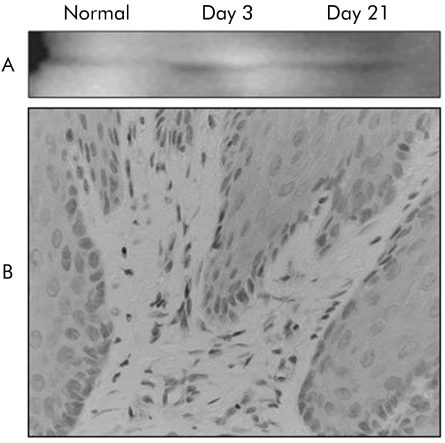
As there were no oesophageal lesions on day 2 after induction of oesophagitis, possibly due to lack of food administration, we examined oesophageal lesions on day 3 to determine acute phase changes. Figure 1 shows the macroscopic and histological appearances of the normal oesophagus and reflux oesophagitis in the rat model. Normal oesophagus exhibited a thin epithelial layer with squamous cells, and few inflammatory cells were found in the submucosal layer. In contrast, there were several erosions and oedematous mucosa in the middle and lower parts of the oesophagus on day 3 with histological defects of the epithelial layer and marked infiltration of inflammatory cells in the lamina propria, submucosal layer, and bases of ulcers. On day 21, several oesophageal lesions associated with marked thickening of the oesophageal wall exhibited mucosal thickening with elongation of the lamina propria papillae and basal cell hyperplasia, as well as persistent inflammatory cell infiltration.
Figure 1 Macroscopic (A) and histological appearance (B) of normal oesophagus and reflux oesophagitis. Normal oesophagus has a thin epithelial layer with few inflammatory cells. However, several erosions and ulcers, and histological defects of the epithelium and marked inflammatory cell infiltration are observed on day 3 (acute phase). Oesophageal lesions with mucosal thickening with basal cell hyperplasia, elongation of lamina propria papillae, and inflammatory cell infiltration were found in the middle and lower parts of the oesophagus on day 21 (chronic phase).
Oesophageal epithelial proliferation in rat oesophagitis
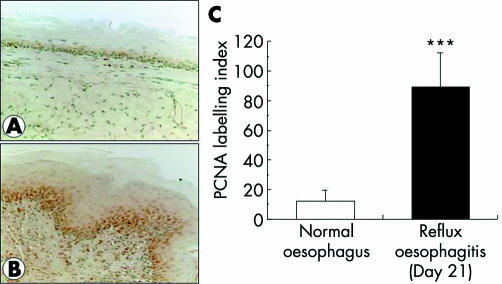
A few PCNA positive cells were found in normal oesophageal epithelium (fig 2A) while most epithelial cells in the basal layer and some inflammatory cells exhibited a reaction for PCNA on day 21 of reflux oesophagitis (fig 2B). The PCNA labelling index was significantly increased in chronic oesophagitis (on day 21) compared with normal oesophagus (fig 2C).
Figure 2 Oesophageal epithelial proliferation assessed by proliferating cell nuclear antigen (PCNA) staining. There were few PCNA positive cells in the normal oesophagus (A) while most epithelial cells of the basal layer and some inflammatory cells exhibited a reaction for PCNA on day 21 after induction of oesophagitis (B). The PCNA labelling index was significantly higher in oesophagitis than in normal oesophagus (C). Values are mean (SD) of six experiments.***?p <0.0001 versus normal oesophagus.
Dynamics of COX and PGES in rat oesophagitis
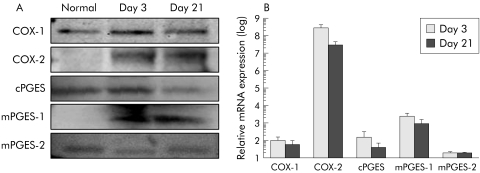
COX‐1, COX‐2, cPGES, mPGES‐1, and mPGES‐2 proteins was observed as bands of 70 kDa, 72 kDa, 23 kDa, 16 kDa, and 32 kDa, respectively (fig 3A). COX‐2 and mPGES‐1 protein expression was hardly detected in normal oesophageal tissues but was markedly increased in reflux oesophagitis on days 3 and 21. Expression of COX‐1, cPGES, and mPGES‐2 proteins was detected in tissues of both normal oesophagus and oesophagitis, with modest changes in levels of these proteins observed in oesophagitis (fig 3A). Real time RT‐PCR revealed that mRNA expression of COX‐1, COX‐2, cPGES, and mPGES‐1, but not of mPGES‐2, was significantly increased in oesophagitis compared with normal oesophagus (fig 3B). In particular, COX‐2 and mPGES‐1 mRNA expression in oesophagitis exhibited approximately 2×108‐fold and 250‐fold increases on day 3 and approximately 2×107‐fold and 100‐fold increases on day 21, respectively, compared with normal oesophageal tissue. These marked increases in COX‐2 and mPGES‐1 mRNA expression in oesophageal lesions were correlated with increased expression of these proteins, as detected on western blotting.
Figure 3 Expression and dynamics of cyclooxygenases (COXs) and prostaglandin E synthase (PGES) in rat oesophagitis. Western blotting (A) showed that COX‐2 and microsomal PGES (mPGES)‐1 expression was significantly increased in rat oesophagitis while modest changes in COX‐1, cytosolic PGES (cPGES), and mPGES‐2 expression were observed. Real time reverse transcription‐polymerase chain reaction (RT‐PCR) (B) revealed that expression of COX‐2, cPGES, and mPGES‐1 mRNA but not mPGES‐2 mRNA was significantly increased compared with normal oesophagus, with a particularly marked increase in COX‐2 mRNA expression observed. Relative mRNA levels are ratios to the mean value for normal oesophagus. Values are mean (SD) of 6–8 experiments.
To determine whether COX‐2 mRNA induction in rat oesophagitis was due to acid exposure, rats were given 30 mg/kg of rabeprazole intraperitoneally once daily. COX‐2 mRNA expression in oesophageal lesions was significantly decreased compared with that in oesophageal tissue of vehicle treated rats with oesophagitis, by 5.7% and 1.4% on days 3 and 21, respectively.
Localisation of COX and PGES in rat chronic oesophagitis
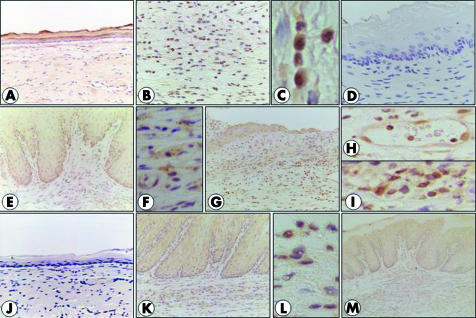
COX‐1 expression was observed in normal oesophageal epithelium (fig 4A) but stronger expression was found in some inflammatory cells in regions with oesophagitis (fig 4B, C). There was little expression of COX‐2 in normal oesophagus (fig 4D) while inflammatory cells or mesenchymal cells and epithelial cells in regions with basal hyperplasia exhibited reactions for COX‐2 in oesophagitis (fig 4E, F). A similar pattern of localisation was found for mPGES‐1 (fig 4J–L). cPGES expression was found in the epithelium of both normal oesophagus and oesophagitis (fig 4G), as well as in endothelial cells (fig 4H) and inflammatory cells (fig 4I) in oesophagitis. mPGES‐2 expression was found in oesophageal epithelial cells of normal oesophagus and oesophagitis, as well as in inflammatory cells in regions with oesophagitis (fig 4M).
Figure 4 Localisation of cyclooxygenases (COXs) and prostaglandin E synthase (PGES) in normal oesophagus and oesophagitis. COX‐1 expression was found in normal oesophageal epithelium (A), and inflammatory cells exhibited a reaction for COX‐1 in oesophagitis (B, C). There was no expression of COX‐2 in normal oesophagus (D) while epithelial cells of the basal layer and inflammatory cells in the lamina propria and submucosa exhibited a reaction for COX‐2 in oesophagitis (E). COX‐2 expression was found in mesenchymal cells (fibroblasts) in lamina propria (F). Cytosolic PGES expression in oesophagitis (G) was found in epithelial cells, endothelial cells (H), and some inflammatory cells (I). Microsomal PGES (mPGES)‐1 was not expressed in normal oesophagus (J) while epithelial cells and inflammatory and mesenchymal cells exhibited a reaction for it (K, L). mPGES‐2 expression was found in epithelial cells and inflammatory cells (M).
Colocalisation of COX‐2 and mPGES‐1 in rat oesophagitis
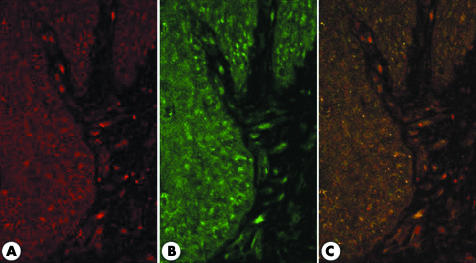
On immunofluorescence study using confocal laser microscopy, COX‐2 and mPGES‐1 expression was observed in epithelial cells of the basal layer, as well as in inflammatory cells and mesenchymal cells in the lamina propria and submucosa in a cytoplasmic pattern (fig 5A, B). Double immunostaining of COX‐2 and mPGES‐1 with immunofluorescence conjugated antibodies revealed that COX‐2 and mPGES‐1 were colocalised in these cells (fig 5C).
Figure 5 Confocal double immunostaining for cyclooxygenase 2 (COX‐2) and microsomal prostaglandin E synthase 1 (mPGES‐1). (A) COX‐2 (red) was stained in epithelial cells of the basal layer, and inflammatory and mesenchymal cells in the lamina propria. (B) The pattern of expression of mPGES‐1 (green) was similar to that of COX‐2. (C) Merging panels (A) and (B) revealed that COX‐2 and mPGES‐1 were colocalised.
Effects of COX‐2 inhibitors on severity of oesophagitis
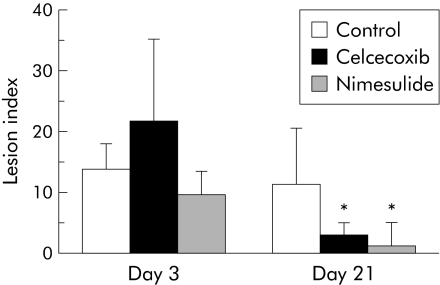
Figure 6 shows the effects of celecoxib and nimesulide on the lesion index of reflux oesophagitis. Neither celecoxib nor nimesulide affected acute oesophageal lesions on day 3. Histologically, marked infiltration of inflammatory cells was observed in both control and COX‐2 inhibitor treated groups, and there were no significant differences in histological findings between the control and COX‐2 inhibitor treated groups. Treatment with COX‐2 inhibitors prevented the development of chronic oesophagitis and significantly reduced lesion index on day 21.
Figure 6 Effects of cyclooxygenase 2 (COX‐2) inhibitors on severity of rat oesophagitis. The COX‐2 inhibitors celecoxib (10 mg/kg/day) and nimesulide (10 mg/kg/day) did not affect acute oesophageal lesions (day 3) but significantly reduced the severity of chronic oesophagitis (day 21). Values are mean (SD) of 6–8 experiments. *p<0.001 versus control on day 21. Control = vehicle treated group.
PGE2 synthetic activity in rat oesophagitis and effect of celecoxib on PGE2 levels
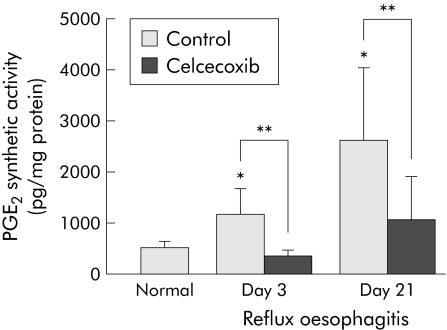
PGE2 synthetic activity was significantly increased in oesophagitis on both days 3 and 21 compared with normal oesophagus (fig 7). In particular, the increase in PGE2 synthetic activity on day 21 was higher than that on day 3. Treatment with celecoxib almost completely inhibited the increase in PGE2 synthetic activity in regions with oesophagitis on days 3 and 21, suggesting that the increase in PGE2 in rat oesophagitis was mainly due to COX‐2.
Figure 7 Effect of celecoxib on oesophageal prostaglandin E2 (PGE2) synthetic activity. PGE2 levels were significantly increased in oesophagitis on both days 3 and 21. Celcecoxib (10 mg/kg/day) significantly inhibited the increase in PGE2 levels in oesophagitis. Values are mean (SD) of six experiments. *p<0.001 versus normal oesophagus; **p<0.001. Control = vehicle treated group.
Expression and localisation of EP4
EP4 protein was detected as a band of 53 kDa by western blotting, and EP4 expression was significantly increased in oesophagitis compared with normal oesophagus (fig 8A). Immunohistochemical staining revealed that oesophageal epithelial cells in the basal layer and inflammatory and mesenchymal cells exhibited reactions for EP4 (fig 8B).
Figure 8 Expression of EP4 protein in rat oesophagitis. Western blotting (A) showed that EP4 protein expression was increased in oesophagitis. Immunohistochemical staining (B) revealed that epithelial cells and inflammatory and mesenchymal cells exhibited a reaction for EP4 in chronic oesophagitis.
Discussion
The present study demonstrated that mRNA and protein expression of COX‐2 and mPGES‐1 was markedly increased in rat acid reflux oesophagitis compared with normal oesophagus, while modest changes in expression of COX‐1, cPGES, and mPGES‐2 were observed. PGE2 levels were significantly elevated in oesophagitis, with increased expression of COX‐2 and mPGES‐1 on both days 3 and 21. Immunohistochemical study revealed that COX‐2 and mPGES‐1 were expressed in inflammatory and mesenchymal cells in the lamina propria and submucosa, as well as in the epithelial cells of the basal layer. Confocal double immunostaining confirmed colocalisation of COX‐2 and mPGES‐1 in these cells. As PGE2 is efficiently produced in cells cotransfected with COX‐2 and mPGES‐1 cDNAs,7 the increase in PGE2 levels in oesophagitis was due to COX‐2 and mPGES‐1. This conclusion is further supported by the finding that acid induced expression of the COX‐2 gene and stimulated PGE2 synthesis in human cultured normal oesophageal epithelial cells.13 Interestingly, COX‐2 inhibitors such as celecoxib and nimesulide significantly reduced the severity of chronic oesophagitis (on day 21) but did not affect acute oesophageal lesions (on day 3). As celecoxib significantly reduced the increase in PGE2 synthetic activity on both days 3 and 21, PGE2 derived from COX‐2 and mPGES‐1 plays a significant role in the development of chronic oesophagitis but not in acute oesophageal lesions.
Our previous study using the same model of oesophagitis showed that mRNA expression of cytokines such as interleukin 1β (IL‐1β) and tumour necrosis factor α (TNF‐α) was significantly increased in oesophagitis on both days 3 and 21.14 Immunohistochemically, these cytokines were found in inflammatory cells in the lamina propria and submucosa.14 As rabeprazole almost completely inhibited the development of oesophagitis from day 3 and significantly inhibited inflammatory cytokines14 as well as COX‐2 gene expression, acid must have triggered acute oesophageal mucosal injury with marked infiltration of inflammatory cells that produced cytokines and COX‐2. Taken together, these findings and those of the present study indicate that the increase in expression of COX‐2 and PGE2 on day 3 is associated with an initial response to acid induced oesophageal inflammation, followed by oesophageal mucosal injury, and is not directly related to the development of acute oesophageal lesions.
In the chronic phase, continuous acid exposure induced persistent infiltration of inflammatory cells and an increase in epithelial proliferation in the basal layer, which are histological findings similar to those in patients with GORD.2 As EP4, a PGE2 receptor subtype, was expressed in epithelial cells in regions with basal hyperplasia and inflammatory cells, PGE2 may be associated with epithelial proliferation of the basal layer and persistent inflammatory cell infiltration via binding to EP4 in chronic oesophagitis. This type of association between PGE2 derived from COX‐2 and epithelial proliferation has been found in human Barrett's oesophagus.15,16 The question arises as to why acute oesophageal lesions were healed in COX‐2 inhibitor treated rats even though production of PGE2 by COX‐2 was completely blocked. Several studies have shown that growth factors such as epidermal growth factor,17 transforming growth factor α,18 hepatocyte growth factor,17 and keratinocyte growth factor19 are associated with oesophageal mucosal healing through stimulation of epithelial proliferation and restitution. These factors may thus be responsible for oesophageal mucosal healing, and excess PGE2 derived from COX‐2 might be related to the development of basal hyperplasia and persistent infiltration of inflammatory cells in chronic oesophagitis.
Several animal studies have shown that indomethacin reduces oesophageal epithelial injury caused by acid or radiation.20,21 A recent study by Lanas and colleagues22 showed that DFU (a selective COX‐2 inhibitor), but not indomethacin or piroxicam, inhibited acidified pepsin induced rabbit oesophagitis. They found that COX‐2 activity was significantly increased in oesophagitis, especially high grade oesophagitis characterised by diffuse erosion/ulceration, inflammation, bleeding, and reactive epithelial changes. These results are consistent with the present findings for the chronic phase of acid reflux oesophagitis. However, Baatar and colleagues23 demonstrated that COX‐2 expression was increased in epithelial cells at the margin of acetic acid induced oesophageal ulcers and that celecoxib significantly delayed oesophageal ulcer healing by reducing epithelial proliferation, possibly through downregulation of HGF‐cMet‐ERK2 pathway activity. Although the reasons for the differences in effects of COX‐2 between our study and others are unclear, they may be related to the type of experimental model used as the oesophagus was continuously exposed to acid in the present study. As gastric acid regulated inflammatory responses, including cytokine gene expression, in scarred gastric mucosa during rat gastric ulcer recurrence,24 continuous acid exposure might affect oesophageal responses to COX‐2.
Although recent studies have clarified the roles of mPGES‐1 in various pathophysiological events such as fever, pain, disorders of bone metabolism, and Alzheimer's disease,6 only one such study has been reported for the oesophagus. Jang and colleagues25 found that mPGES‐1 expression was significantly increased in stromal cells with COX‐2 expression in rat oesophageal squamous dysplasia and Barrett's oesophagus using a model of duodenal content reflux. They suggested that mPGES‐1 plays a role in carcinogenesis in Barrett's oesophagus. Involvement of mPGES‐1 in tumorigenesis has been found in the colon26 and non‐small cell lung cancers.27 The finding that mPGES‐1 deficient mice displayed a significant reduction in inflammatory responses in an experimental model of arthritis28 indicates that mPGES‐1 also has important effects on inflammation. Several studies have shown that expression of mPGES‐1 is regulated by inflammatory cytokines such as IL‐1β and TNF‐α.29 As expression of these cytokines was increased in rat oesophagitis, they may be associated with induction of mPGES‐1 in acid oesophagitis. Further study using specific mPGES‐1 antagonists is needed to clarify whether PGE2 derived from mPGES‐1 is associated with the development of chronic oesophagitis.
In this study, COX‐1 and cPGES mRNA expression in oesophageal lesions was significantly increased compared with that in normal oesophagus, but corresponding protein levels were not dramatically changed, in contrast with those of COX‐2 and mPGES‐1. As COX‐1 and cPGES were localised in epithelial and inflammatory cells, the increase in the number of these cells in oesophagitis might be associated with a slight increase in COX‐1 and cPGES gene expression. However, COX‐1 and cPGES may play a role in the pathogenesis of rat oesophagitis.
It is unknown whether COX‐2 inhibitors affect the development of GORD, although aspirin and other non‐steroidal anti‐inflammatory drugs have been found to be associated with oesophagitis or oesophageal stricture.30,31 A recent study by Hamoui and colleagues32 to determine the association between genetic changes and acid exposure in patients with GORD showed that the level of COX‐2 expression was positively correlated with oesophageal acid exposure, as assessed by 24 hour pH score, but found no significant increase in COX‐2 expression in patients with GORD or those who used non‐steroidal anti‐inflammatory drugs. Further clinical evidence is needed to determine whether COX‐2 inhibitors affect the development of GORD.
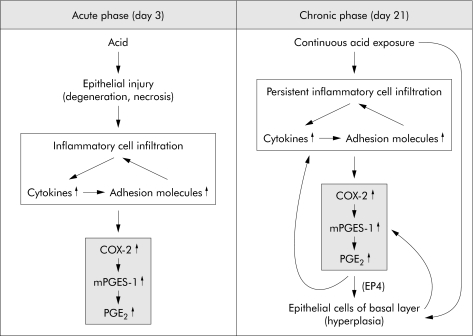
In conclusion, expression of COX‐2, mPGES‐1, and PGE2 was significantly increased in rat acid reflux oesophagitis. The effects of PGE2 derived from COX‐2 and mPGES‐1 might differ depending on the phase of rat oesophagitis, and include inflammatory responses to acid in the acute phase and an increase in epithelial proliferation of the basal layer and persistent inflammatory cell infiltration in the chronic phase (fig 9).
Figure 9 Roles of cyclooxygenase 2 (COX‐2), microsomal prostaglandin E synthase 1 (mPGES‐1), and prostaglandin E2 (PGE2) in rat oesophagitis during the acute and chronic phases.
Acknowledgments
This study was supported, in part, by a Grant‐in‐Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology in Japan.
Abbreviations
PG - prostaglandin
COX - cyclooxygenase
mPGES - microsomal prostaglandin E synthase
cPGES - cytosolic prostaglandin E synthase
GORD - gastro‐oesophageal reflux disease
PCNA - proliferating cell nuclear antigen
RT‐PCR - reverse transcription‐polymerase chain reaction
GAPDH - glyceraldehyde‐3‐phosphate dehydrogenase
IL‐1β - interleukin 1β
TNF‐α - tumour necrosis factor α
Footnotes
Conflict of interest: None declared.
References
- 1.Chiba N, De Gara C J, Wilkinson J M.et al Speed of healing and symptom relief in grade II to IV gastroesophageal reflux disease: a meta‐analysis. Gastroenterology 19971121798–1810. [DOI] [PubMed] [Google Scholar]
- 2.Ismail‐Beigi F, Horton P F, Pope C E I I.et al Histological consequences of gastroesophageal reflux in men. Gastroenterology 197058163–174. [PubMed] [Google Scholar]
- 3.Alber D, Moussard C, Toubin M.et al Gas chromatographic/mass spectrometric quantitative analysis of eicosanoids in human oesophageal mucosa. Biomed Environ Mass Spectrom 198816299–304. [DOI] [PubMed] [Google Scholar]
- 4.Marcinkiewicz M, Sarosiek J, Edmunds M.et al Monophasic luminal release of prostaglandin E2 in patients with reflux esophagitis under the impact of acid and acid/pepsin solutions. Its potential pathogenetic significance. J Clin Gastroenterol 199521268–274. [DOI] [PubMed] [Google Scholar]
- 5.Hla T, Neilson K. Human cyclooxygenase‐2 cDNA. Proc Natl Acad Sci USA 19922667384–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murakami M, Nakatani Y, Tanioka T.et al Prostaglandin E synthase. Prostaglandins Other Lipid Mediat 200268–69383–399. [DOI] [PubMed] [Google Scholar]
- 7.Murakami M, Naraba H, Tanioka T.et al Regulation of prostaglandin E2 biosynthesis by inducible membrane‐associated prostaglandin E2 synthase that acts in concert with cyclooxygenase‐2. J Biol Chem 200027532783. [DOI] [PubMed] [Google Scholar]
- 8.Trebino C E, Stock J L, Gibbons C P.et al Impaired inflammatory and pain responses in mice lacking an inducible prostaglandin E synthase. Proc Natl Acad Sci U S A. 2003: 22, 1009044–9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subbaramaiah K, Yoshimatsu K, Scherl E.et al Microsomal prostaglandin E synthase‐1 is overexpressed in inflammatory bowel disease. Evidence for involvement of the transcription factor Egr‐1. J Biol Chem 200427912647–12658. [DOI] [PubMed] [Google Scholar]
- 10.Omura N, Kashiwagi H, Chen G.et al Establishment of surgically induced chronic acid reflux esophagitis in rats. Scand J Gastroenterol 199934948–953. [DOI] [PubMed] [Google Scholar]
- 11.Claveau D, Sirinyan M, Guay J.et al Microsomal prostaglandin E synthase‐1 is a major terminal synthase that is selectively up‐regulated during cyclooxygenase‐2‐dependent prostaglandin E2 production in the rat adjuvant‐induced arthritis model. J Immunol 20031704738–4744. [DOI] [PubMed] [Google Scholar]
- 12.Tanigawa T, Watanabe T, Hamaguchi M.et al Anti‐inflammatory effect of two isoforms of COX in H. pylori‐induced gastritis in mice: possible involvement of PGE2, Am J Physiol Gastrointest Liver Physiol 2004286G148–G156. [DOI] [PubMed] [Google Scholar]
- 13.Kawabe A, Shimada Y, Soma T.et al Production of prostaglandinE2 via bile acid is enhanced by trypsin and acid in normal human esophageal epithelial cells. Life Sci 20047521–34. [DOI] [PubMed] [Google Scholar]
- 14.Hamaguchi M, Fujiwara Y, Takashima T.et al Increased expression of cytokines and adhesion molecules in rat chronic esophagitis. Digestion 200368189–197. [DOI] [PubMed] [Google Scholar]
- 15.Kaur B S, Khamnehei N, Iravani M.et al Rofecoxib inhibits cyclooxygenase 2 expression and activity and reduces cell proliferation in Barrett's esophagus. Gastroenterology 200212360–67. [DOI] [PubMed] [Google Scholar]
- 16.Shirvani V N, Ouatu‐Lascar R, Kaur B S.et al Cyclooxygenase 2 expression in Barrett's esophagus and adenocarcinoma: Ex vivo induction by bile salts and acid exposure. Gastroenterology 2000118487–496. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi M, Ota S, Ogura K.et al Hepatocyte growth factor stimulates wound repair of the rabbit esophageal epithelial cells in primary culture. Biochem Biophys Res Commun 1995216298–305. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara Y, Higuchi K, Hamaguchi M.et al Increased expression of transforming growth factor‐alpha and epidermal growth factor receptors in rat chronic reflux esophagitis. J Gastroenterol Hepatol 200419521–527. [DOI] [PubMed] [Google Scholar]
- 19.Baatar D, Kawanaka H, Szabo I L.et al Esophageal ulceration activates keratinocyte growth factor and its receptor in rats: implications for ulcer healing. Gastroenterology 2002122458–468. [DOI] [PubMed] [Google Scholar]
- 20.Eastwood G L, Beck B D, Castell D O.et al Beneficial effect of indomethacin on acid‐induced esophagitis in cats. Dig Dis Sci 198126601–608. [DOI] [PubMed] [Google Scholar]
- 21.Northway M G, Libshitz H I, Osborne B M.et al Radiation esophagitis in the opossum: radioprotection with indomethacin. Gastroenterology 198078883–892. [PubMed] [Google Scholar]
- 22.Lanas A, Jimenez P, Ferrandez A.et al Selective COX‐2 inhibition is associated with decreased mucosal damage induced by acid and pepsin in rabbit esophagitis. Inflammation 20032721–29. [DOI] [PubMed] [Google Scholar]
- 23.Baatar D, Jones M K, Pai R.et al Selective cyclooxygenase‐2 blocker delays healing of esophageal ulcers in rats and inhibits ulceration‐triggered c‐Met/hepatocyte growth factor receptor induction and extracellular signal‐regulated kinase 2 activation. Am J Pathol 2002160963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe T, Higuchi K, Tominaga K.et al Acid regulates inflammatory response in a rat model of induction of gastric ulcer recurrence by interleukin 1beta. Gut 200148774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jang T J, Min S K, Bae J D.et al Expression of cyclooxygenase 2, microsomal prostaglandin E synthase 1, and EP receptors is increased in rat oesophageal squamous cell dysplasia and Barrett's metaplasia induced by duodenal contents reflux. Gut 20045327–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamei D, Murakami M, Nakatani Y.et al Potential role of microsomal prostaglandin E synthase‐1 in tumorigenesis. J Biol Chem 200327819396–19405. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimatsu K, Altorki N K, Golijanin D.et al Inducible prostaglandin E synthase is overexpressed in non‐small cell lung cancer. Clin Cancer Res 200172669–2674. [PubMed] [Google Scholar]
- 28.Trebino C E, Stock J L, Gibbons C P.et al Impaired inflammatory and pain responses in mice lacking an inducible prostaglandin E synthase. Proc Natl Acad Sci U S A 20031009044–9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stichtenoth D O, Thoren S, Bian H.et al Microsomal prostaglandin E synthase is regulated by proinflammatory cytokines and glucocorticoids in primary rheumatoid synovial cells. J Immunol 2001167469–474. [DOI] [PubMed] [Google Scholar]
- 30.Ryan P, Hetzel D J, Shearman D J.et al Risk factors for ulcerative reflux oesophagitis: a case‐control study. J Gastroenterol Hepatol 199510306–312. [DOI] [PubMed] [Google Scholar]
- 31.Spechler S J. Epidemiology and natural history of gastro‐oesophageal reflux disease. Digestion 199251(Suppl 1)24–29. [DOI] [PubMed] [Google Scholar]
- 32.Hamoui N, Peters J H, Schneider S.et al Increased acid exposure in patients with gastroesophageal reflux disease influences cyclooxygenase‐2 gene expression in the squamous epithelium of the lower esophagus. Arch Surg 2004139712–717. [DOI] [PubMed] [Google Scholar]