Abstract
Background
The intestinal microbiota plays a critical role in the pathophysiology of pouchitis, a major complication after ileal pouch anal anastomosis in patients with ulcerative colitis. Recently, controlled trials have demonstrated that probiotics are effective in maintenance of remission in pouchitis patients. However, the mechanism by which therapy with probiotics works remains elusive. This study explores the role of the bacterial and fungal flora in a controlled trial for maintenance of remission in pouchitis patients with the probiotic VSL#3 compound.
Methods
The mucosa associated pouch microbiota was investigated before and after therapy with VSL#3 by analysis of endoscopic biopsies using ribosomal DNA/RNA based community fingerprint analysis, clone libraries, real time polymerase chain reaction (PCR), and fluorescence in situ hybridisation. Patients were recruited from a placebo controlled remission maintenance trial with VSL#3.
Results
Patients who developed pouchitis while treated with placebo had low bacterial and high fungal diversity. Bacterial diversity was increased and fungal diversity was reduced in patients in remission maintained with VSL#3 (p = 0.001). Real time PCR experiments demonstrated that VSL#3 increased the total number of bacterial cells (p = 0.002) and modified the spectrum of bacteria towards anaerobic species. Taxa specific clone libraries for Lactobacilli and Bifidobacteria showed that the richness and spectrum of these bacteria were altered under probiotic therapy.
Conclusions
Probiotic therapy with VSL#3 increases the total number of intestinal bacterial cells as well as the richness and diversity of the bacterial microbiota, especially the anaerobic flora. The diversity of the fungal flora is repressed. Restoration of the integrity of a “protective” intestinal mucosa related microbiota could therefore be a potential mechanism of probiotic bacteria in inflammatory barrier diseases of the lower gastrointestinal tract.
Keywords: bacteria, fungi, microflora, pouchitis, probiotics
Probiotics are living microorganisms that, on ingestion in sufficient numbers, exert health benefits beyond the metabolic effect of nutritional components. It has been suggested that probiotic bacteria are effective and promising agents for the treatment of inflammatory gastrointestinal barrier disorders, including infectious colitis, antibiotic associated diarrhoea, and inflammatory bowel diseases (IBD).1,2,3,4,5,6 In recent years a series of placebo controlled randomised clinical trials have been conducted demonstrating the clinical efficacy of probiotics.7,8,9,10,11 However, understanding of the mechanism of action of probiotic bacteria is still incomplete.12 In vitro and animal studies have shown that probiotics influence epithelial barrier function and gut permeability, restore the commensal bacterial microbiota, and reduce the production of inflammatory cytokines.13,14,15,16 Induction of anti‐inflammatory cytokine expression, probably through modification of bacterial signalling in epithelial cells, has also been suggested.11,17 This hypothesis is supported by the efficacy of local delivery of anti‐inflammatory cytokines in animal models.18
Functional experiments using animal models suggest that probiotics may have profound effects on the composition of the faecal flora well beyond insertion of new bacterial strains.19,20 However, the effects of probiotic bacteria on the resident mucosa related intestinal microbiota that is directly interacting with the host mucosal barrier organ is poorly understood. Culture based methods have identified a maximum of 10–30% of the bacterial spectrum (and probably less than 10% of fungal species) of the complex bacterial community in the human gut. Molecular techniques may overcome these limitations.21,22,23,24,25 The 16S/18S rRNA marker gene system has been employed previously to analyse the bacterial communities of the intestine and to investigate the dominant species using oligonucleotide hybridisation, real time polymerase chain reaction (PCR), and fluorescent in situ hybridisation (FISH).22,26,27,28,29,30,31,32,33,34,35 Community fingerprinting techniques have been shown to be a powerful tool for determining microbial diversity in complex samples.36,37,38,39
Ulcerative colitis (UC) is a subform of IBD, a group of chronic recurrent gastrointestinal inflammatory barrier diseases. Restorative proctocolectomy with ileal pouch anal anastomosis is a well established procedure in the surgical treatment of UC and familial adenomatous polyposis.40,41,42 Approximately 15–46% of patients with UC develop pouchitis, a major complication after ileal pouch anal anastomosis, within five years of operation.43,44,45,46 A chronic relapsing form of pouchitis can be distinguished from a chronically active form.47 The aetiology and pathophysiology of pouchitis are still unknown. Interaction between the mucosal immune system and the gut flora appears to play a crucial role in the initiation of pouchitis. Alterations of the bacterial microbiota may be an important factor triggering the disease process48,49 and may explain the efficacy of antibiotic treatment in pouchitis.50,51 However, remission periods are often short and the condition is complicated by frequent relapses. Recent studies have shown that probiotic treatment with VSL#3, a mixture of eight different probiotic bacterial strains, is effective in maintaining remission in pouchitis.52,53,54
A subset of patients (n = 15), who had participated in a double blind, randomised, placebo controlled clinical trial, were investigated.52 Patients with pouchitis in remission that had been induced by antibiotic therapy were recruited to receive either the VSL#3 probiotic compound or placebo for maintenance of remission. Biopsies were obtained before and two months after initiation of VSL#3 or placebo treatment. Probiotic therapy with VSL#3 increased the total number of intestinal bacterial cells as well as the richness and diversity of the bacterial microbiota, especially the anaerobic flora, whereas the fungal flora was repressed. In contrast, patients who relapsed while receiving placebo showed reduced diversity of the mucosal flora.
Material and methods
Patients
Patients with recurrent or chronic active pouchitis, who were enrolled in a placebo controlled, randomised, remission maintenance trial with the probiotic preparation VSL#352 were recruited for this substudy. The main inclusion criterion was prior successful induction of remission (defined as a pouchitis activity index of 0 or 1) by therapy with metronidazole and ciprofloxacin.52 Fifteen of 36 patients in the trial agreed to have additional biopsies for this study, taken for exploration of the mucosal microbiota. Pouchitis was defined by histological and endoscopic criteria using the pouchitis disease activity index (PDAI).55 Age ranged from 22 to 64 years (median 34) in the15 patients; there were six females and nine males. Total PDAI (expressed as medians) was 3 (1–7) at study entry in both groups.
Patients received VSL#3 (VSL pharmaceuticals Inc., Gaitherburg, Maryland, USA) 6 g once daily or identical placebo for 12 months. VSL#3 contains 300 billions viable lyophilised bacteria per gram, comprising four strains of lactobacilli (Lactobacillus plantarum, Lactobacillus casei, Lactobacillus acidophilus, and Lactobacillus delbrueckii subspecies bulgaricus), three strains of bifidobacteria (Bifidobacterium infantis, Bifidobacterium longum, and Bifidobacterium breve), and one strain of streptococcus (Streptococcus salivarius subspecies thermophilius). Placebo sachets contained maize without bacteria but did not differ in taste or physical appearance from VSL#3.
Mucosal biopsies were obtained during pouch endoscopy and immediately snap frozen in liquid nitrogen. Patients were studied before and after two months of therapy with study medication. Of the 15 patients recruited for the substudy, 10 received VSL#3 and five placebo. All VSL#3 patients in this substudy were still in remission after two months while all placebo patients showed signs of active pouchitis. All clinical and molecular examinations were carried out, and the results collated before the trial was unblinded.
Treatment of biopsy samples and DNA extraction
Biopsy and stool specimens were immediately snap frozen in liquid nitrogen after colonoscopy. DNA was extracted following a protocol adapted to the characteristics of the microorganisms56: biopsies were incubated with 200 μl TL‐buffer (200 mM HEPES, pH 7.5, 1 M KCl, 100 mM MgCl2, 1 mM EDTA, 0.2% NaN3) and 25 μl proteinase K (PeqLab, Erlangen, Germany) at 55°C for two hours. DNA was extracted using the FastDNA Spin Kit for soil after mechanical homogenisation (FastPrep FP 120 instrument) according to the manufacturer's instructions (both BIO 101, Carlsbad, California, USA). For sensitive and accurate quantification, DNA concentrations were determined using PicoGreen according to the manufacturer's guidelines (dsDNA quantification kit; Molecular Probes, Leiden, the Netherlands).
Real time PCR
The real time PCR assay used in this study has been described previously.57 Amplification and detection were carried out in 96 well optical plates on an ABI PRISM 7700 Sequence Detector with TaqMan Universal PCR 2× Master Mix, primer (0.4 μM), probe (0.2 μM), and 20 ng of sample DNA in a final volume of 50 μl per reaction. The universal probe UNI (5′‐ACT GAG ACA CGG TCC A‐3′) binds to position 321‐37 and is VIC labelled. In the present study, minor groove binder fluorescent probes with non‐fluorescent quencher dyes (also called “dark” quencher) were used (Applied Biosystems, Foster City, California, USA). SDS software v1.7 or later was used to support non‐fluorescent quencher probes. The universal primers used in this study hybridise to conserved regions on the 16S gene. The forward primer TPU1 (5′‐AGA GTT TGA TCM TGG CTC AG‐3′) binds to position 8‐27 and the reverse primer RTU8 (5′‐ AAG GAG GTG ATC CAN CCR CA‐3′) binds to position 1522‐41 (Escherichia coli reference numbering). The absolute number of cells was normalised to the total amount of DNA extracted from each biopsy. The total number of cells was interpolated from the averaged standard curve as described.57,58
Single strand conformational polymorphism (SSCP) analysis
For initial PCR, the two conserved primers COM‐1 (5′‐CAG CAG CCG CGG TAA TAC‐3′, position 519–536 on the reference E coli 16S gene) and Com2‐Ph (5′‐CCG TCA ATT CCT TTG AGT TT‐3′, position 907–926 on the reference E coli 16S gene) were used, as published by Schwieger and Tebbe.36 PCR products were controlled for size and products by agarose gel electrophoresis. Preparation of single stranded DNA and silver staining of SSCP gels were performed according to Schwieger and Tebbe.36 Image editing and normalisation were performed using GelCompare II‐software (Applied Maths, Kortrijk, Belgium). The general diversity of bacterial species was calculated according to Shannon and Weaver, as described previously.56
Denaturing gradient gel electrophoresis (DGGE) analysis
For PCR amplification of 18S rRNA fragments (1.650 bp), the two fungus specific primers NS1 (5′‐GTA GTC ATA TGC TTG TCT C‐3′) and FR1‐GC (5′‐CCC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GCC GAI CCA TTC AAT CGG TAI T‐3′) were used, as published by Vainio and Hantula.59 DGGE analysis and silver staining were performed according to Newton and colleagues.60 Image editing, normalisation, and calculation of diversity indices were carried out as described above.
Sequencing of bands from SSCP/DGGE community profiles
To identify the bacteria correlated with the bands, selected products from the polyacrylamide gel after silver staining were excised with sterile scalpels. Gel slices were then mixed with elution buffer. After incubation, 5–20 μl were subjected to a PCR with COM primers (see above). After control with 2% agarose gel electrophoresis, 8 μl of the PCR products were digested with 0.3 U SAP (shrimp alkaline phosphatase) and 1.5 U ExoI. The sequencing reaction was performed on an ABI PRISM 3700 DNA Analyser using 1 μl of ABI PRISM BigDye (Applied Biosystems), 30 μM concentration of each sequencing primer (Eurogentec Seraing, Belgium), and 2 μl of digested PCR product. Forward and reverse sequences were aligned with Sequencher software package (Gene Codes Corp., Ann Arbor, Michigan, USA). Fragments were identified by NCBI BLAST database search.
Group specific clone libraries
Group specific clone libraries for the following bacterial taxa were generated: Lactobacillus, Bifidobacterium, Bacteroides/Prevotella, and γ‐Proteobacteria/Enterobacteriaceae (for primer information see table 1). More than 600 clones were sequenced from 12 independent taxa specific libraries (Lactobacillus n = 135, Bifidobacterium n = 136, Bacteroides/Prevotella n = 124, and γ‐Proteobacteria/Enterobacteriaceae n = 236) that were generated from pooled disease related (before study therapy, placebo, VSL#3) PCR products. A two step reconditioning PCR approach (35/10 cycles) was used as described elsewhere.61,62 The specific fragment of the 16S rDNA was amplified and cloned into E coli cells using the pCR 2.1 TOPO TA Cloning Kit for sequencing (Invitrogen, Karlsruhe, Germany), as described previously.56
Table 1 Oligonucleotides used for the group specific clone libraries.
| Bacterial group | Primer | Direction | Position | Sequence (5′‐3′) | Reference |
|---|---|---|---|---|---|
| Lactobacillus | Lac1 | Forward | 352–370* | AGCAGTAGGGAATCTTCCA | Walter31 |
| Lac2 | Reverse | 679–680* | ATTYCACCGCTACACATG | Walter31 | |
| Bifidobacterium | Bif_164 | Forward | 164–181* | GGGTGGTAATGCCGGATG | Langendijk23 |
| Bif_662 | Reverse | 679–662* | TTCCACCGTTACACCGGGAA | Langendijk23 | |
| Bacteroides/Prevotella | Bac‐32 | Forward | 32–50* | AACGCTAGCTACAGGCTT | Bernhard77 |
| Bac_303 | Reverse | 303–321* | CCAATGTGGGGGACCTTC | Manz63 | |
| γ‐Proteobacteria/Enterobacteriaceae | TPU5_F | Forward | 906–26* | AAACTCAAATGAATTGACGG | von Wintzingerode78 |
| Enter1416 | Reverse | 1416–23* | CTTTTGCAACCCACTCC | Sghir79 | |
| Sequencing | M13 (21) | Forward | 389–404† | TGTAAAACGACGGCCAGT | Guttman80 |
| M13 (24) | Reverse | 208–25† | AACAGCTATGACCATG | Guttman80 |
*Position referring to the Esherichia coli reference sequence.
†M13 priming sites on pCR 2.1 plasmid vector.
Cloning and sequencing of inserts was performed using 3.2 pmol of M13F and M13R sequencing primers (table 1), as described previously.56 Alignment, assembly, and trimming of vector sequences were performed using the Sequencher software package (Gene Codes Corp.). Sequences were checked for vector contamination using the NCBI VecScreen tool. OTUs were identified by NCBI BLAST analysis using search results of at least 97% similarity. Sequences were examined for chimera using the Chimera Check tool of the Ribosomal Data Projects (RDP) of the Center for Microbial Ecology, Michigan State University, Michigan, USA.
Fluorescent in situ hybridisation (FISH) and oligonucleotide probes
Biopsies were fixed in 500 μl of freshly prepared 4% buffered formalin and embedded in Histoplast (Sigma, St Louis, Missouri, USA) according to routine procedures. Cross sections (2 μm) were cut and placed on coated microscope slides (Superfrost*/Plus). Sections were hybridised as described previously35 using the following 16S/23S rRNA targeted oligonucleotide probes: (1) an equimolar mixture of five bacteria directed probes EUB 338, EUB 785, EUB 927, EUB 1055, and EUB 1088 (58, 59), referred to as EUB mix, to detect all bacteria; (2) Bac 303 to detect the Bacteroides/Prevotella cluster63; (3) Erec 482 specific for most of the clostridia and eubacteria belonging to the Clostridium coccoides‐Eubacterium rectale group64; (4) bifidobacterial probe Bif 16423; (5) Lab 158 for nearly all species of the genera Lactobacillus and Enterococcus in the gut65; and Ec 1531 specific for a number of Enterobacteriaceae (for example, Escherichia coli, Klebsiella pneumonia). The oligonucleotides were 5′ labelled with the indocarbocyanine dye Cy3 (Thermo Hybaid GmbH, Waltham, Massachusetts, USA).
Expression of data
Normally distributed data are expressed as mean (SD), if not indicated otherwise. Statistical significance of the differences was examined using the Student's t test for normally distributed data and the Mann Whitney U test or the Wilcoxon matched pairs test for non‐normally distributed data. Distribution of data was evaluated by calculating Lilliefors probabilities based on the Komolgorov‐Smirnov test.
Results
Patients
Fifteen patients in remission induced by antibiotic therapy participated in this substudy (out of 36 participating in a double blind, randomised, placebo controlled trial of VSL#3 in pouchitis maintenance). No signs of morphological or histological inflammation were seen, as required by the inclusion criteria. The 10 patients who received VSL#3 were verified as being in remission at the time of the second biopsy (two months) while all five patients who were receiving placebo showed clinical and endoscopic signs of recurrent inflammation.
SSCP analysis and real time PCR of the bacterial microbiota
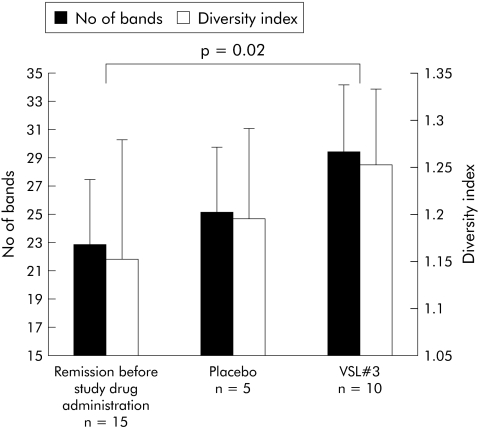
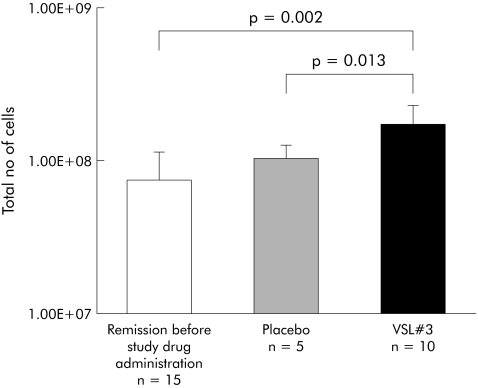
SSCP fingerprinting analysis of 16S rDNA fragments showed low bacterial richness and diversity in mucosal biopsies from patients at study entry after antibiotic induction of remission in comparison with normal colon. Probiotic therapy with VSL#3 increased bacterial richness and diversity of the pouch mucosal flora compared with both patients in remission before therapy and patients developing pouchitis while receiving placebo. The difference in bacterial diversity between patients before and after therapy with VSL#3 was statistically significant (p = 0.02) (fig 1). A universal real time PCR that amplified 16S rDNA was then used to compare the total number of bacterial cells. Treatment with VSL#3 caused a statistically significant increase in the number of 16S rDNA copies compared with pretreatment remission (n = 10, p = 0.002) and placebo treatment (n = 5, p = 0.013), as shown in fig 2. Separate statistical analysis showed no correlation between results of the SSCP analysis and real time PCR and PDAI (clinical, endoscopic, and histological subscores).
Figure 1 Bacterial diversity indicated as richness (No of bands) and diversity indices, obtained from single strand conformational polymorphism profiles of pouchitis patients in remission before randomisation (after treatment with antibiotics), after placebo (no therapy), and after VSL#3. For each group, the mean (SD) number of bands are shown. Bacterial diversity of the probiotic group was significantly higher compared with the starting value (p = 0.02). Weighted diversity scores (SD) showed the same tendency.
Figure 2 Treatment with VSL#3 significantly increased the number of 16Sr DNA copies compared with pretreatment samples and placebo (p = 0.002, p = 0.013, respectively).
To investigate the nature of the species contributing to the increase in bacterial richness and diversity, which were consistently different between pouches in remission before and during VSL#3 treatment, bands were excised from the SCCP gel, reamplified, and sequenced. A BLAST suggested the presence of enterobacillus, Enterococcus sp, Enterobacter sp, and Clostridium sp in mucosal biopsies. Bacterial strains contained in VSL#3 were not detected as part of the mucosa adherent flora.
Taxa specific clone libraries
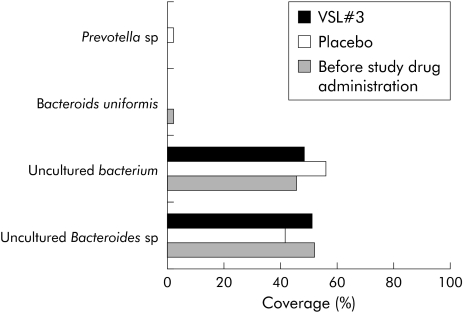
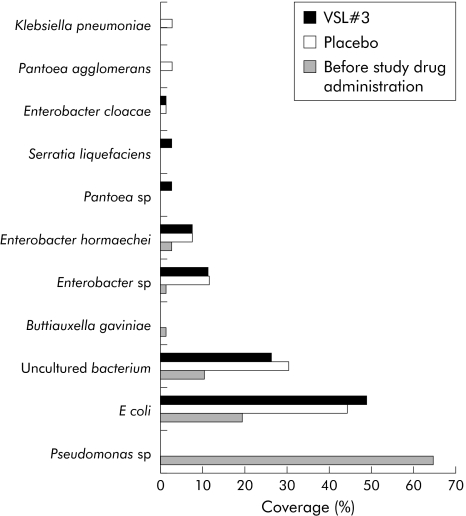
Community fingerprinting techniques are used for formal assessment of the richness and diversity of complex bacterial/fungal habitats, but for technical reasons often fail to provide sufficient taxonomic data. Clone libraries yield a realistic taxonomic representation of a complex microbial habitat that can be further narrowed and optimised by taxa specific amplification. To obtain data on the qualitative composition of the intestinal mucosa related microbiota after VSL#3 therapy, clone libraries with more than 600 sequenced clones were performed for the following taxa: Lactobacillus, Bifidobacterium, Bacteroides/Prevotella, and γ‐Proteobacteria/Enterobacteriaceae. For each of the three experimental groups (before study treatment, but after antibiotic induction therapy, placebo treatment, and probiotic VSL#3 treatment) separate libraries were generated to assess differences in bacterial composition. As shown in figs 3 and 4, the spectrum of Lactobacillus and Bifidobacterium species was more diverse after VSL#3 treatment in comparison with placebo and samples before randomisation. With regard to lactobacilli and bacteroides species, patients at inclusion had only a narrow bacterial spectrum, with Lactobacillus mucosae and Bifidobacterium longum as the predominant species before treatment. The Bacteroides/Prevotella and γ‐Proteobacteria/Enterobacteriaceae libraries showed only gradual differences between patients treated with placebo and VSL#3 (figs 5, 6). Most importantly, little differences were seen during treatment in general with regard to bacterial composition in the Bacteroides/Prevotella group, with the exception of the γ‐Proteobacteria/Enterobacteriaceae flora. High levels of Pseudomonas species were observed after induction of remission by antibiotic therapy (probably as a result of overgrowth due to antibiotic selection pressure through preinclusion induction of remission).
Figure 3 Lactobacillus flora in remission (after antibiotic therapy), after placebo, and after VSL#3 therapy, as assessed by analysis of clone libraries. A total of 135 clones were randomly picked and sequenced. The graph shows the percentage of single species identified by BLAST analysis. Lactobacillus flora was more diverse under VSL#3 therapy in comparison with placebo. At inclusion (that is, before randomisation, after induction therapy with antibiotics), only two Lactobacillus species were detected, most of them Lactobacillus mucosae (>97%).
Figure 4 Bifidobacterium flora in remission (after antibiotic therapy), after placebo, and after VSL#3 therapy, as assessed by analysis of clone libraries. A total of 136 clones were randomly picked and sequenced. The graph shows the percentage of single species identified by BLAST analysis. As seen for the Lactobacillus species (fig 3), the Bifidobacterium flora was more diverse in the VSL#3 group compared with both the placebo and samples before therapy. Before therapy (that is, before randomisation, after induction with antibiotics) Bifidobacterium longum bv infantis were seen almost exclusively.
Figure 5 Bacteroides/Prevotella flora in remission (after antibiotic therapy), after placebo, and after VSL#3 therapy, as assessed by analysis of clone libraries. A total of 135 clones were randomly picked and sequenced. The graph shows the percentage of single species identified by BLAST analysis. The diversity of Bacteroides/Prevotella bacteria was low (n = 4), with the majority of sequences being assigned as uncultured Bacteroides species. There were no significant differences between the groups.
Figure 6 γ‐Proteobacteria/Enterobacteriaceae flora in remission (after antibiotic therapy), after placebo, and after VSL#3 therapy, as assessed by clone libraries. A total of 236 clones were randomly picked and sequenced because of the expected higher diversity. The graph shows the percentage of single species identified by BLAST analysis. The diversity of Enterobacteriaceae, especially Enterobacter species and Escherichia coli, was higher in the placebo and VSL#3 groups but there were only slight differences between these two groups. Remarkably, there was a high proportion of Pseudomonas species in the group before study therapy, which could be due to overgrowth induced by antibiotic selection pressure.
Detection of mucosal bacteria by FISH
To complement the PCR based analysis, a second independent technique was used to identify and localise bacterial species. Colonisation was investigated in tissue sections of six patients in each group (before and during treatment) using the EUB mix of different fluorescence labelled 16S rRNA probes. With the different sets of probes described in the methods section it was found that nearly all bacteria detected by this technique represented Enterobacteriaceae. Bacteria were mainly detected within the epithelium and only rarely within the lamina propria. In uninflamed pouch (before treatment), intraepithelial Enterobacteriaceae (mainly Ecoli) were seen (fig 7). During treatment with VSL#3, a high mucosal count of these organisms was found. In contrast, recurrent inflammation during placebo treatment was associated with a reduction in the presence of Enterobacteriaceae (fig 4). No signals were detected with the Bacteroides/Prevotella, Clostridium coccoides‐Eubacterium rectale, or bifidobacterial probes in any of the tissue sections. Lactobacillus/Enterococcus signals were seen only occasionally in tissue sections of VSL#3 treated patients.
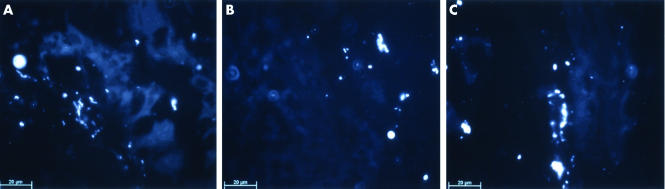
Figure 7 The 16S rRNA targeted oligonucleotide mixture EUB that globally detects bacterial sequences was used in tissue sections from pouch mucosal biopsies. Subdifferentiation was carried out using a set of specific oligonucleotide probes. Almost all bacteria detected were classified as Enterobacteriaceae whereas the Lactobacilli/Enterococcus probes resulted only in occasional detection of signals in VSL#3 treated patients. The figure is a representation of six experiments per group (VSL#3 and placebo, before and after treatment). Mucosal biopsies in patients in remission prior to study drug therapy showed high numbers of intraepithelial bacteria (A). Colonisation was reduced after placebo treatment, which was accompanied by relapse of inflammation (B). Bacterial colonisation of the epithelium was maintained during VSL#3 treatment (C).
DGGE analysis of the fungal microbiota
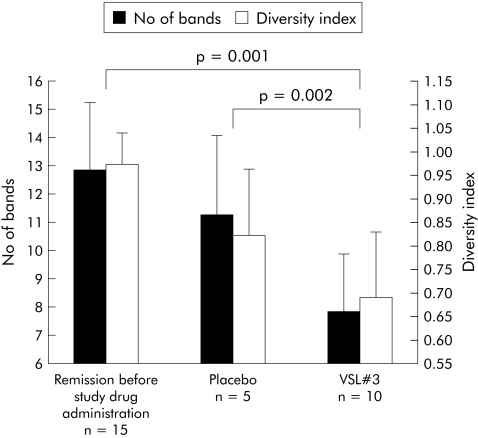
Because fungi are a potent, yet little explored, element of the intestinal microbiota, DGGE analysis was used to investigate fungal diversity. Due to the specific characteristics of the 18S rRNA gene, separation of 18S rDNA fragments by genetic fingerprinting requires longer amplicons. Therefore, DGGE was used because the length of the amplicons for SSCP analysis is restricted to 400–500 bp. Fungal diversity, indicated as number of bands and as weighted diversity index, was reciprocal compared with bacterial diversity. In remission before therapy (that is, following induction by antibiotics), fungal diversity was high (n = 15). During both placebo (n = 5) and VSL#3 (n = 10) treatment, fungal diversity decreased. In the VSL#3 group, a marked reduction in fungal diversity was seen both in comparison with pretreatment levels (p = 0.001) and with the placebo group (p = 0.002) (fig 8). Individual DGGE profiles showed wide interindividual variation in banding patterns. No consistent bands were identified using band matching analysis (fig 9). Separate statistical analysis of clinical, endoscopic, and histological PDAI scores revealed no significant correlation with fungal diversity.
Figure 8 Fungal diversity, indicated as richness (No of bands) and diversity indices, obtained from denaturing gradient gel electrophoresis profiles of pouchitis patients in remission (before study therapy, after induction with antibiotics), after placebo, and two months after treatment with VSL#3. For each group, the mean (SD) number of bands are shown. Fungal diversity in the VSL#3 group was significantly higher compared with both the starting value in remission (p = 0.001) and placebo (p = 0.002). Weighted diversity scores (SD) showed the same tendency.
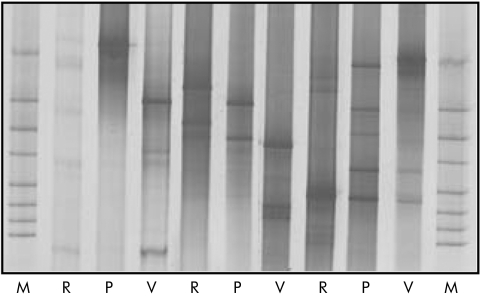
Figure 9 Subset of denaturing gradient gel electrophoresis (DGGE) profiles showing wide interindividual variation in the composition of the fungal microflora The gel demonstrates patients in remission at study start (R), and after two months of treatment with VSL#3 (V) or placebo (P). The fingerprints of fungal communities were generated by separation of 18S rDNA fragments. Defined fungal species were used as standards (lanes M), as described at Newton and colleagues60 DGGE analysis was carried out for all samples and numeric calculations are shown in fig 5.
Discussion
The aetiology and pathogenesis of pouchitis is still unclear although ulcerative colitis and pouchitis share many similarities.66,67,68,69 This study investigated the influence of the probiotic agent VSL#3 on the bacterial microbiota in a placebo controlled clinical trial52 which was designed to assess the value of VSL#3 in maintaining antibiotic induced remission in patients with pouchitis.
In pouchitis, bacterial overgrowth and a decrease in beneficial bacteria, in particular lactobacilli and bifidobacteria, in comparison with conventional ileostomy flora, has been described.69,70,71 It appears likely that faecal stasis and immune stimulation by stool bacteria play an important role in the development of pouch inflammation. The mucosa associated flora is probably of immediate relevance to the disease process28,35 and differs significantly from the composition of the ileal and midstream/faecal flora.29,72
Morphological data from FISH analysis indicate that normal Enterobacteriaceae species (mainly E coli) are found in uninflamed epithelium in ileoanal pouch biopsies (that is, from pouchitis patients who are in remission) in contrast with the normal ileum and colon of healthy volunteers in which no E coli was found within the epithelium.28 When recurrent pouch inflammation occurred during placebo treatment, low bacterial diversity and a marked reduction in intramucosal Enterobacteriaceae species were found. The increase in Enterobacteriaceae within the mucosa during VSL#3 therapy indicates that remission maintenance under probiotics is associated with restoration of parts of the normal pouch flora.
For other relevant groups of intestinal bacteria, such as Bacteroides/Prevotella or the Lactobacilli/Bifidobacteria group, only sporadic signals were detected by FISH. The limited amount of mucosal biopsy material as well as the confined sensitivity of FISH analysis for single bacteria prompted us to complement the FISH results with PCR based molecular techniques.
The FISH results were supported by diversity analysis with SSCP and, at least in part, by taxa specific clone libraries. Successful maintenance of remission after VSL#3 treatment appeared to result in higher diversity of bacterial species that included members of the normal anaerobic enteric flora and not merely colonisation with strains contained only in VSL#3. With the exception of the Bacteroides/Prevotella group, the richness of bacteria was increased in the VSL#3 group compared with pretreatment levels. As diversity was increased during VSL#3 maintained remission, we suggest that this effect may be a component of the therapeutic mechanism of probiotics. The suggestion that low bacterial diversity could be an important mechanism for mucosal inflammation is supported by earlier investigations56 in which mucosal inflammation in IBD was associated with loss of normal anaerobic bacteria such as Bacteroides species, Eubacterium species, and Lactobacillus species.
Restoration of Enterobacteriaceae species in the mucosa and the increase in bacterial diversity are observed in the setting of the clinical trial, which demonstrated successful remission maintenance by VSL#3.52 The effects were observed on the specific background of remission induction with antibiotics. Antibiotics lead to significant alteration of the enteric bacterial balance. In neonates, antibiotic treatment induces complete eradication of Lactobacillus species together with a marked reduction in colonic total aerobic and anaerobic bacteria, in particular Enterobacteriaceae and Enterococcus.73 The net effect of antimicrobial therapy therefore results in (i) a decrease in the total number of bacterial cells and (ii) a reduction in specific bacterial populations. Colonisation of the mucosa by Enterobacteriaceae species and diversification of Lactobacillus species and Bifidobacterium species in the VSL#3 group might be facilitated by eradication of complex populations by the preceding antibiotic therapy and the diminished competition by other dominant bacterial consortia, such as Enterobacteriaceae or Enterococcus. Therefore, expansion and diversification of the bacterial microbiota observed in the VSL#3 group could be specific to pretreatment with antibiotics on the background of the dysbiotic flora in IBD.56,74 In fact, this may also be the case for the clinical efficacy of the treatment in pouchitis.52
It is well documented that pouchitis is associated with loss of beneficial intestinal bacteria, especially the subgroups lactobacilli and bifidobacteria.69,70,71 We observed, as a net effect of VSL#3 associated remission, recolonisation and diversification of the potentially beneficial Lactobacillus and Bifidobacterium flora. As most patients in the placebo group relapsed and almost all patients in the VSL#3 group remained in remission, the specific effects of VSL#3 and the effect of an inflammation free mucosa interact and therefore cannot be separated in this study. Additional therapeutic mechanisms induced by VSL#3 treatment are possible that may be either primary or secondary to alteration of the constitution of the pouch microbiota. Specific induction of anti‐inflammatory cytokines by probiotics could decrease the inflammatory activity in the mucosa and thereby provide better growth conditions for diversification of the flora.11 Rachmilewitz et al demonstrated in Toll‐like receptor and MyD88 deficient mice that the immune stimulatory effect of probiotic bacteria isolated from the VSL#3 compound was dependent on Toll‐like receptor 9 signalling.75 Therefore, direct interaction of probiotic bacteria with mucosal immunoregulation could either induce anti‐inflammatory pathways11 or directly interact with innate immune mechanisms75 as the primary mechanisms altering the growth conditions of the mucosal microbiota, or could be secondary to changes in microbial constitution and diversity. The significant difference between the placebo and the VLS#3 groups indicates that VSL#3 induced remission is associated with reconstruction of the intestinal flora, but investigation of the hierarchy of mechanistic events has to await further exploration in model systems.
Fungi are important elements of the human flora. However, little is known about the composition of comensal fungal intestinal species. Fungal microbiotic diversity was inversely related to bacterial diversity. However, it has to be pointed out that fungal species represent only a small fraction of the total microbiota. Immunocompetent hosts have an efficient phagocytic capacity to prevent fungal invasion.76 It can be hypothesised that alteration in the balance of bacterial and fungal species in the mucosal flora reflects a metabolic dysbalance of the complex microbial ecosystem with pathophysiological consequences for the mucosal barrier. Most importantly, restored balance between bacterial and fungal diversity was seen in VSL#3 treated patients. The question of whether the increase in fungal diversity is a direct result of higher bacterial diversity or an independent parallel effect remains unclear. Although fungi are normal members of the intestinal microbiota to a certain extent, fungal overgrowth is a typical complication of any bacterial imbalance following antibiotic therapy or a specific dietary regimen, as for example in intensive care units. Most likely the fungal overgrowth seen here was a direct consequence of bacterial dysbalance supported by diminished control mechanisms and reduced competition through the total bacterial microbiota. Studies on the mutual dependencies of the different parts of the intestinal microbiota are urgently needed to clarify this point.
Our study demonstrated that VSL#3 maintained remission was accompanied by a higher bacterial and a reduced fungal diversity in comparison with placebo treatment. The increase in bacterial diversity was not caused by colonisation with bacterial strains contained in VSL#3 but represents an independent effect. It is not unclear at present whether the anti‐inflammatory and immunoregulatory effects of probiotics are primary or secondary to induction of changes in the diversity of the mucosal microbiota. Increase in bacterial diversity may be a therapeutic mechanism for the probiotic mixture VSL#3 in maintenance of antibiotic induced remission in pouchitis. Methods for manipulating the complex microecology of the mucosal flora may be an important field for future therapeutic developments, especially addressing the secondary or primary prevention of pouchitis.
Electronic database and website information
URLs used in this article are as follows:
ARB software, ribosomal sequence database, and handling: http://www.arb‐home.de
NCBI BLAST homepage: http://www.ncbi.nlm.nih.gov/BLAST/
Ribosomal Database Project II: http://rdp.cme.msu.edu/
VecScreen database: http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html
Acknowledgements
This paper describes a substudy to a randomised, controlled, clinical trial of VSL#3 versus placebo as maintenance therapy for pouchitis in remission, which has been reported separately.52 The work was supported by grants from the Deutsche Crohn und Colitis Vereinigung (DCCV), the Crohn's and Colitis Foundation of America (CCFA), the Deutsche Forschungsgemeinschaft (SFB 415), by Training and Mobility of Researchers (TMR‐II) grant of the European commission, by the German Competence network IBD, and by the National Genome Research Network (both supported by the German Ministery of Education and Science (BMBF)). The authors thank the clinical investigators and the patients who provided research material for this study. The authors also thank Kornelia Smalla (Biologische Bundesanstalt für Land‐ und Forstwirtschaft, Institut für Pflanzenvirologie, Mikrobiologie und biologische Sicherheit, Braunschweig, Germany) for technical assistance with the DGGE analysis.
Abbreviations
IBD - inflammatory bowel disease
UC - ulcerative colitis
PDAI - pouchitis disease activity index
PCR - polymerase chain reaction
FISH - fluorescent in situ hybridisation
SSCP - single strand conformational polymorphism
DGGE - denaturing gradient gel electrophoresis
Footnotes
Conflict of interest: None declared.
References
- 1.Madsen K L. The use of probiotics in gastrointestinal disease. Can J Gastroenterol 200115817–822. [DOI] [PubMed] [Google Scholar]
- 2.Montaldo M, Arancio F, Izzi D.et al Probiotics: history, definition, requirements and possible therapeutic applications. Ann Ital Med Int 200217157–165. [PubMed] [Google Scholar]
- 3.Shanahan F. Probiotics in inflamatory bowel disease. Gut 200148609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hart A L, Stagg A J, Frame M.et al The role of the gut flora in health and disease, and its modification as therapy. Aliment Pharmacol Ther 2002161383–1393. [DOI] [PubMed] [Google Scholar]
- 5.Cremonimi E, Caro S, Santarelli L.et al Probiotics in antibiotic associated diarrhoea. Dig Liver Dis 200234S78–S80. [DOI] [PubMed] [Google Scholar]
- 6.Isolauri E, Kirjavainen P V, Salminen S. Probiotic: a role in the treatment of intestinal infection and inflammation. Gut 20005054–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gionchetti P, Rizzello F, Venturi A.et al Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double‐blind, placebo‐controlled trial. Gastroenterology 2000119305–309. [DOI] [PubMed] [Google Scholar]
- 8.Sen S, Mullan M M, Parker T J.et al Effect of Lactobacillus plantarum 299v on colonic fermentation and symptoms of irritable bowel syndrome. Dig Dis Sci 2002472615–2620. [DOI] [PubMed] [Google Scholar]
- 9.Pochapin M. The effect of probiotics on Clostridium difficile diarrhoea. Am J Gastroenterol 200095S11–S13. [DOI] [PubMed] [Google Scholar]
- 10.Kruis W, Schutz E, Fric P.et al Double‐blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther 199711853–858. [DOI] [PubMed] [Google Scholar]
- 11.Furrie E, Macfarlane S, Kennedy A.et al Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut 200554242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linskens R K, Huijsdens X W, Savelkoul P H.et al The bacterial flora in inflammatory bowel disease: current insights in pathogenesis and the influence of antibiotics and probiotics. Scand J Gastroenterol Suppl 200123429–40. [DOI] [PubMed] [Google Scholar]
- 13.Shibolet O, Karmeli F, Eliakim R.et al Variable response to probiotics in two models of experimental colitis in rats. Inflamm Bowel Dis 20028399–406. [DOI] [PubMed] [Google Scholar]
- 14.O'Mahony L, Freeney M, O'Halloran S.et al Probiotic impact on microbial flora, inflammation and tumour development in IL‐10 knock‐out mice. Aliment Pharmacol Ther 2001151219–1225. [DOI] [PubMed] [Google Scholar]
- 15.Madsen K, Cornish A, Soper P.et al Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology 2001121580–591. [DOI] [PubMed] [Google Scholar]
- 16.Ulisse S, Gionchetti P, DÁlo S.et al Expression of cytokines, inducible nitric oxide synthase, and matrix metalloproteinases in pouchitis: effects of probiotic treatment. Am J Gastroenterol 2001962691–2699. [DOI] [PubMed] [Google Scholar]
- 17.Borruel N, Carol M, Casellas F.et al Increased mucosal tumour necrosis factor {alpha} production in Crohn's disease can be downregulated ex vivo by probiotic bacteria. Gut 200251659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steidler L, Hans W, Schotte L.et al Treatment of murine colitis by Lactococcus lactis secreting interleukin‐10. Science 20002891352–1355. [DOI] [PubMed] [Google Scholar]
- 19.Tannock G W, Munro K, Harmsen H J.et al Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol 2000662578–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartosch S, Woodmansey E J, Paterson J C.et al Microbiological effects of consuming a synbiotic containing Bifidobacterium bifidum, Bifidobacterium lactis, and oligofructose in elderly persons, determined by real‐time polymerase chain reaction and counting of viable bacteria. Clin Infect Dis 20054028–37. [DOI] [PubMed] [Google Scholar]
- 21.Simon G L, Gorbach S L. Intestinal flora in health and disease. Gastroenterology 198486174–193. [PubMed] [Google Scholar]
- 22.Tannock G W. Molecular methods for exploring the intestinal ecosystem. Br J Nutr 200287S199–S201. [DOI] [PubMed] [Google Scholar]
- 23.Langendijk P, Schut F, Jansen G.et al Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus‐specific 16S rRNA‐targeted probes and its application in fecal samples. Appl Environ Microbiol 1995613069–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson K, Blitchington R. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol 1996622273–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McFarlene G, Gibson G. Metabolic activities of the normal colonic microflora. In: Gibson S, ed. Human health: contribution of microorganisms. Frankfurt: Springer, Frankfurt, Germany, 199417–38.
- 26.Zheng D, Alm E, Stahl D.et al Characterization of universal small‐subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol 1996624504–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zoetendal E G, Akkermans A D L, De Vos W M. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host‐specific communities of active bacteria. Appl Environ Microbiol 1998643854–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swidsinski A, Ladhoff A, Pernthaler A.et al Mucosal flora in inflammatory bowel disease. Gastroenterology 200212244–54. [DOI] [PubMed] [Google Scholar]
- 29.Zoetendal E G, von Wright A, Vilpponen‐Salmela T.et al Mucosa‐associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 2002683401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seksik P, Rigottier‐Gois L, Gramet G.et al Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 200352237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter J, Hertel C, Tannock G W.et al Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group‐specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol 2001672578–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tannock G W. The bifidobacterial and Lactobacillus microflora of humans. Clin Rev Allergy Immunol 200222231–253. [DOI] [PubMed] [Google Scholar]
- 33.Dore J, Sghir A, Hannequart‐Gramet G.et al Design and evaluation of a 16S rRNA‐targeted oligonucleotide probe for specific detection and quantitation of human faecal Bacteroides populations. Syst Appl Microbiol 19982165–71. [DOI] [PubMed] [Google Scholar]
- 34.Zoetendal E G, Ben‐Amor K, Harmsen H J.et al Quantification of uncultured Ruminococcus obeum‐like bacteria in human fecal samples by fluorescent in situ hybridization and flow cytometry using 16S rRNA‐targeted probes. Appl Environ Microbiol 2002684225–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleessen B, Kroesen A J, Buhr H J.et al Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol 2002371034–1041. [DOI] [PubMed] [Google Scholar]
- 36.Schwieger F, Tebbe C C. A new approach to utilize PCR‐single‐strand‐conformation polymorphism for 16S rRNA gene‐based microbial community analysis. Appl Environ Microbiol 1998644870–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tebbe C C, Schmalenberger A, Peters S.et al Single‐strand conformation polymorphism (SSCP) for microbial community analysis. In: Rochelle PA, ed. Environmental molecular microbiology: protocols and applications. Wymondham, UK: Horizon Scientific Press, 2001161–174.
- 38.Valinsky L, Della Vedova G, Jiang T.et al Oligonucleotide fingerprinting of rRNA genes for analysis of fungal community composition. Appl Environ Microbiol 2002685999–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valinsky L, Della Vedova G, Scupham A J.et al Analysis of bacterial community composition by oligonucleotide fingerprinting of rRNA genes. Appl Environ Microbiol 2002683243–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stahlberg D, Gullberg K, Liljeqvist L.et al Pouchitis following pelvic pouch operation for ulcerative colitis. Incidence, cumulative risk, and risk factors. Dis Colon Rectum 1996391012–1018. [DOI] [PubMed] [Google Scholar]
- 41.Kock N G, Darle N, Hultén L.et al Ileostomy. Curr Probl Surg 1977141–52. [DOI] [PubMed] [Google Scholar]
- 42.Parks A G, Nicholls R J. Proctocolectomy without ileostomy. BMJ 1978285–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandborn W J. Pouchitis following ileal pouch‐anal anastomosis: definition, pathogenesis, and treatment. Gastroenterology 19941071856–1860. [DOI] [PubMed] [Google Scholar]
- 44.Svaninger G, Nordgren S, Oresland T.et al Incidence and characteristics of pouchitis in the Kock continent ileostomy and the pelvic pouch. Scand J Gastroenterol 199328695–700. [DOI] [PubMed] [Google Scholar]
- 45.Hurst R D, Molinari M, Chung T P.et al Prospective study of the incidence, timing and treatment of pouchitis in 104 consecutive patients after restorative proctocolectomy. Arch Surg 1996131497–500. [DOI] [PubMed] [Google Scholar]
- 46.Madden M V, Farthing M J, Nicholls R J. Inflammation in the ileal reservoir: pouchitis. Gut 199031247–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meagher A P, Farouk R, Dozois R R.et al J ileal pouch‐anal anastomosis for chronic ulcerative colitis: complication and long‐term outcome in 1310 patients. Br J Surg 199885800–803. [DOI] [PubMed] [Google Scholar]
- 48.Ruseler‐van Embden J G, Schouten W R, van Lieshout L M. Pouchitis: result of microbial imbalance? Gut 199435659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de‐Silva H J, Millard P R, Soper N.et al Effects of the fecal stream and stasis on the ileal pouch mucosa. Gut 1991321166–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madden M, McIntyre A, Nicholls R J. Double‐blind cross‐over trial of metronidazole versus placebo in chronic unremitting pouchitis. Dig Dis Sci 199439509–515. [DOI] [PubMed] [Google Scholar]
- 51.Mimura T, Rizzello F, Helwig U.et al Randomised, placebo‐controlled, double‐blind trial of once daily, high dose probiotic therapy (VSL#3) for maintaining remission in patients with recurrent or refractory pouchitis. Gut 200453108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gionchetti P, Rizzello F, Venturi A.et al Antibiotic combination therapy in patients with chronic, treatment‐resistant pouchitis. Aliment Pharmacol Ther 199913713–718. [DOI] [PubMed] [Google Scholar]
- 53.Campieri M, Gionchetti P. Probiotics in inflammatory bowel disease: new insight to pathogenesis or a possible therapeutic alternative? Gastroenterology 19991161246–1260. [DOI] [PubMed] [Google Scholar]
- 54.Gionchetti P, Rizello F, Helwig U.et al Prophylaxis of pouchitis onset with probiotic therapy: a double‐blind, placebo‐controlled trial. Gastroenterology 20031241202–1209. [DOI] [PubMed] [Google Scholar]
- 55.Sandborn W J, Tremaine W J, Batts K P.et al Pouchitis after ileal pouch‐anal anastomosis: a pouchitis disease activity index. Mayo Clin Proc 199469409–415. [DOI] [PubMed] [Google Scholar]
- 56.Ott S J, Musfeldt M, Wenderoth D F.et al Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 200453685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ott S J, Musfeldt M, Ullmann U.et al Quantification of intestinal bacterial populations by real‐time PCR with a universal primer set and minor groove binder probes: a global approach to the enteric flora. J Clin Microbiol 2004422566–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lyons S R, Griffen A L, Leys E J. Quantitative real‐time PCR for porphyromonas gingivalis and total bacteria. J Clin Microbiol 2000382362–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vainio E J, Hantula J. Direct analysis of wood‐inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol Res 2000104927–936. [Google Scholar]
- 60.Newton C, Gomes M, Fagbola O.et al Dynamics of fungal communities in bulk and Maize rhizosphere soil in the tropics. Appl Environ Microbiol 2003693758–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson J R, Marcelino L A, Polz M F. Heteroduplexes in mixed‐template amplifications: formation, consequence and elimination by ‘reconditioning PCR'. Nucl Acids Res 2002302083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Acinas S G, Klepac‐Ceraj V, Hunt D E.et al Fine‐scale phylogenetic architecture of a complex bacterial community. Nature 2004430551–554. [DOI] [PubMed] [Google Scholar]
- 63.Manz W, Amann R, Ludwig W.et al Application of a suite of 16S rRNA‐specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga‐flavobacter‐bacteroides in the natural environment. Microbiology 19961421097–1106. [DOI] [PubMed] [Google Scholar]
- 64.Franks A H, Harmsen H J M, Raangs G C.et al Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group‐specific 16S rRNA‐targeted oligonucleotide probes. Appl Environ Microbiol 1998643336–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harmsen H J, Raangs G C, He T.et al Extensive set of 16S rRNA‐based probes for detection of bacteria in human feces. Appl Environ Microbiol 2002682982–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luukkonen P, Jarvinen H, Tanskanen M.et al Pouchitis‐recurrence of the inflammatory bowel disease? Gut 199435243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Silva H J, Jones M, Prince C.et al Lymphocyte and macrophage subpopulations in pelvic ileal pouches. Gut 1991321160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mahida J R, Patel S, Gionchetti P.et al Macrophage subpopulations in the lamina propria of normal and inflamed colon and terminal leum. Gut 198930826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Onderdonk A B, Dvorak A M, Cisneros R L.et al Microbiologic assessment of tissue biopsy samples from ileal pouch patients. J Clin Microbiol 199230312–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kmiot W A, Youngs D, Tudor R.et al Mucosal morphology, cell proliferation, and fecal bacteriology in acute pouchitis. Br J Surg 1993801445–1449. [DOI] [PubMed] [Google Scholar]
- 71.Sandborn W J, McLeod R, Jewell D P. Medical therapy for induction and maintenance of remission in pouchitis: a systemic review. Inflamm Bowel Dis 1999533–39. [DOI] [PubMed] [Google Scholar]
- 72.Ott S J, Musfeldt M, Timmis K N.et al In vitro alterations of intestinal bacterial microbiota in fecal samples during storage. Diagn Microbiol Infect Dis 200450237–245. [DOI] [PubMed] [Google Scholar]
- 73.Schumann A, Nutten S, Donnicola D.et al Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. PhysiolGenomics200523235–245. [DOI] [PubMed] [Google Scholar]
- 74.Tamboli C P, Neut C, Desreumaux P.et al Dysbiosis in inflammatory bowel disease. Gut 2004531–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rachmilewitz D, Katakura K, Karmeli F.et al Toll‐like receptor 9 signaling mediates the anti‐inflammatory effects of probiotics in murine experimental colitis. Gastroenterology 2004126520–528. [DOI] [PubMed] [Google Scholar]
- 76.Lionakis M S, Kontoyiannis D P. Glucocorticoids and invasive fungal infections. Lancet 20033621828–1838. [DOI] [PubMed] [Google Scholar]
- 77.Bernhard A E, Field K G. Identification of nonpoint sources of fecal pollution in coastal waters by using host‐specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl Environ Microbiol 2000661587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.von Wintzingerode F, Landt O, Ehrlich A.et al Peptide nucleic acid‐mediated PCR clamping as a useful supplement in the determination of microbial diversity. Appl Environ Microbiol 200066549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sghir A, Gramet G, Suau A.et al Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl Environ Microbiol 2000662263–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guttman D S, Charlesworth D. An X‐linked gene with a degenerate Y‐linked homologue in a dioecious plant. Nature 1998393263–266. [DOI] [PubMed] [Google Scholar]