Abstract
Background and aims
Perturbation of differentiation of the crypt‐villus axis of the human small intestine is associated with several intestinal disorders of clinical importance. At present, differentiation of small intestinal enterocytes in the crypt‐villus axis is not well characterised.
Subjects and methods
Expression profiling of microdissected enterocytes lining the upper part of crypts or the middle of villi was performed using the Affymetrix X3P arrays and several methods for confirmation.
Results
A total of 978 differentially expressed sequences representing 778 unique UniGene IDs were found and categorised into four functional groups. In enterocytes lining the upper part of crypts, cell cycle promoting genes and transcription/translation related genes were predominantly expressed, whereas in enterocytes lining the middle of villi, high expression of cell cycle inhibiting genes, metabolism related genes, and vesicle/transport related genes was found.
Conclusion
Two types of enterocytes were dissected at the molecular level, the non‐absorptive enterocyte located in the upper part of crypts and the absorptive enterocyte found in the middle of villi. These data improve our knowledge about the physiology of the crypt‐villus architecture in human small intestine and provide new insights into pathophysiological phenomena, such as villus atrophy, which is clinically important.
Keywords: enterocyte, small intestine, molecular characterisation
In human small intestine, differentiation can be found in two distinct axes, the cephalocaudal axis and the crypt‐villus axis (CVA).1 The CVA is a vertical axis with epithelial cells extending bidirectionally upwards and downwards from the stem cells anchored adjacent to the crypt basis. Enterocytes, mucous producing goblet cells, and enteroendocrine cells migrate out of the crypt to the villus, while Paneth cells migrate towards the crypt‐base. Structural differentiation and functional specialisation of each of the four cell lineages occurs along the CVA in a few days, a phenomenon that is described as the third axis, the developmental time axis (DTA).2
To elucidate key mechanisms in the differentiation of enterocytes, the CVA of the human small intestine is a very important model because two dimensional separation of regulatory pathways is found. The first dimension consists of simple cellular differentiation at a defined location in the CVA and the second dimension consists of the DTA resulting in a spatial distribution of different mRNA species along the CVA.3,4,5,6
The CVA is apparently one important example of the strong link between structure and function in biological systems. Disorders of the CVA, like villus atrophy, are associated with several clinical symptoms. Villus atrophy with simultaneous hyperplasia of crypts preferentially found in coeliac disease is associated with maldigestion and malabsorption, which generally indicates the existence of different types of enterocytes between villi and crypts in the human small intestine. Clinical evidence from patients with severe coeliac disease suggests that crypt lining enterocytes do not compensate for the function of the villus epithelium.
The aim of the present study was to perform molecular dissection of enterocytes in the CVA in human small intestine with regard to the phenomenon of maldigestion and malabsorption, which are both of highly important clinically. Our experiments distinguished two types of enterocytes in the CVA, the non‐absorptive (located in the upper part of crypts) and the absorptive (located in the middle of villi) type of enterocytes. The complete data set is available as supplementary material online with this article (http://www.gutjnl.com/supplemental).
Materials and methods
Tissues
In the study, surgical resections of small intestinal tissues from 11 patients (six men, mean age 65 years (range 39–79); five women, mean age 72 years (range 48–80)) with sporadic adenocarcinoma of the colon were included. Normal unaffected small intestinal mucosa was mechanically dissected from the adjacent tissue layers, immediately cooled in liquid nitrogen, and stored at −80°C until use. All surgical resections were indicated by principles and practice of oncological therapy. All diagnoses were established by conventional clinicopathological criteria. The use of human tissues for study purposes was approved by each patient and permitted by Heidelberg University. From all tissues (n = 11), total RNA was extracted using the guanidinium‐isothiocyanate acid‐phenol technique. RNA quality and integrity of whole tissue samples (mucosa) was assessed by 2100‐Bioanalyzer runs using the RNA‐6000‐Nano‐LabChip (Agilent, Böblingen, Germany), and samples showing intact 18S and 28S bands were further processed. RNA from six surgical resections (four men aged 39, 66, 77, and 79 years; two women aged 74 and 80 years) was used for hybridisation of X3P arrays (see below).
Laser microdissection (LMD)
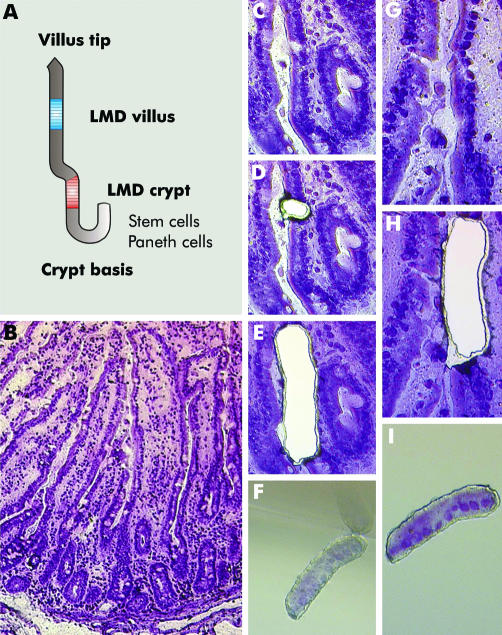
Serial cryosections of 8 µm were mounted on 1.2 µm polyethylene terephthalate (PET) foil slides designed for laser microdissection, haematoxylin stained, air dried, and then laser microdissected using a Leica AS LMD System. Two main areas of interest were defined for microdissection: the first located in the upper part of crypts (above proliferative cells with mitotic figures); the second in the middle of villi (above the crypt, but below the villus tip) (fig 1). Mitotic enterocytes of crypts, goblet cells, and intraepithelial lymphocytes were morphologically identified and then excluded from microdissection by laser ablation. Contiguous epithelial cell groups from 200 crypts and 200 villi per sample were laser microdissected from a 40× field of view and collected in one tube cap each, which was filled with 30 µl of guanidine isothiocyanate containing lysis buffer (Buffer RLT, RNeasy Mini Kit; Qiagen, Hilden, Germany). Laser parameters for microdissection were set to aperture 2, intensity 30, and speed 5. Total RNA from microdissected enterocytes was extracted using the RNeasy Micro Kit following the manufacturer's instructions (Qiagen). All steps were carried out under RNase free conditions.
Figure 1 Principles of laser microdissection (LMD). (A) Scheme for LMD of enterocytes in two distinct locations in the crypt‐villus axis. (B) Overview of haematoxylin stained cryosection of small intestinal mucosa. (C‐F) LMD of enterocytes from the upper part of crypts. In (D), ablation of a goblet cell by laser prior to LMD of enterocytes (E) and sampling of the probe (F) is shown. (G–J) LMD of enterocytes from the middle of villi without prior laser ablation.
Preparation of hybridisation samples and hybridisation
RNA of LMD samples was isolated using the RNeasy kit and applied to two consecutive rounds of linear amplification following essentially the Eberwine protocol7 whereas reverse transcription of the second round cDNA was started by random oligonucleotide primer. Antisense RNA (aRNA) was synthesised taking advantage of the T7‐MegaScript kit (Ambion, Huntingdon, UK) whereas biotinylated nucleotides were incorporated during the second round of amplification. Purified and biotinylated aRNA (15 µg) was hybridised to X3P arrays (61 359 probe sequences printed representing approximately 47 000 transcripts (Affymetrix, Wooburn Green, UK)) according to standard procedures (Hybridization Oven 640; Fluidics Station 450; GeneChip Scanner 3000).
Data analysis
Twelve microarrays (six patients, one crypt, and one villus sample each patient) were utilised. The Affymetrix raw data (12× CEL files representing the six villus samples and the corresponding crypt samples) were analysed using the BioConductor software package (release 1.4)8 with the R environment (version 1.9.1; www.r‐project.org). The procedure comprised the RMA algorithm (package affy),9 linear modelling with moderate t test based on empirical Bayesian statistics (package limma),10 and a multiple testing correction (false discovery rate by Benjamini and Hochberg (p<0.05 was considered as significant)). Further analyses using pathway and gene ontology information were performed with the GenMapp software11 and the GOTree Machine.12
Confirmation of differential gene expression
Using small intestinal tissues from the other patients in the study (n = 5; two men and three women), confirmation of array data was performed using several techniques such as western blot, quantitative reverse transcription‐polymerase chain reaction (RT‐PCR), immunohistochemistry, and mRNA in situ hybridisation, as previously described.13,14 In addition, the array extracted villus and crypt specific genes were compared with a public database of in situ hybridisation results (CVD)15 on the internet (http://pc113.imbg.ku.dk/ps/).
Results
We used a t test based statistical analysis in order to identify genes with differential expression along the CVA in crypt‐villus pairs of six independent tissue samples, which would allow for inclusion of genes with significant differences smaller than a twofold change. Details of each result obtained are given in supplementary table 1 (supplementary material can be viewed on the Gut website at http://www.gutjnl.com/supplemental).
Table 1 Validation of microarray experiments using 10 arbitrarily selected genes.
| Gene | X3P ID | p‐fdr | M (fold) sample pairwise | Localisation | Confirmation | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |||||
| ACSL5 | Hs.11638.0.S1_3p_a_at | 0.045 | 3.3 | 1.7 | 4.4 | 2.3 | 1.9 | 1.2 | Villus | P, ISH, IHC, W |
| ALPI | g13787191_3p_at | 0.001 | 6.4 | 3.6 | 9.7 | 3.5 | 3.9 | 5.1 | Villus | P, IHC |
| CKAP4 | g5803112_3p_a_at | 0.021 | −5.6 | −5.7 | −4.2 | 1.4 | −4.9 | −4.6 | Crypt | P, IHC |
| DFNA5 | g4758153_3p_a_at | 0.012 | 11.1 | 6.7 | 37.7 | 3.9 | 45.1 | 30.9 | Villus | P, W |
| DMBT1 | g4758169_3p_a_at | 0.045 | −1.6 | −3.9 | −8.7 | −3.2 | −8.1 | −8.4 | Crypt | P, ISH, IHC, W |
| DPP4 | g4503366_3p_at | 0.048 | 2.7 | 1.6 | 6.7 | 2.9 | 2.2 | 2.0 | Villus | P |
| ISG20 | g6857799_3p_at | 0.005 | 7.3 | 3.2 | 11.4 | 3.8 | 4.0 | 9.4 | Villus | P |
| MKi67 | Hs.80976.1.S1_3p_a_at | 0.022 | −9.6 | −3.3 | −6.0 | 1.0 | −10.1 | −4.7 | Crypt | P, IHC |
| SLC5A1 | Hs.149429.0.A1_3p_at | 0.047 | 3.9 | 1.8 | 5.8 | 1.6 | 1.8 | 1.4 | Villus | P |
| TP53 | g8400737_3p_at | 0.024 | −3.6 | −3.5 | −4.0 | −2.5 | −6.4 | −1.8 | Crypt | P, IHC |
Fold‐change (M) of expression levels along the crypt‐villus axis in the six sample pairs used for the array experiments are shown; positive values indicate villus specific genes, negative values crypt specific genes. The tissues used for validation are from independent samples (n = 11 laser microdissection samples for quantitative reverse transcription‐polymerase chain reaction (RT‐PCR), and n = 5 for other analyses).
p‐fdr, p value including multiple testing correction according to Benjamini‐Hochberg; P, qRT‐PCR; ISH, mRNA in situ hybridisation; IHC, immunohistochemistry; W, western blotting.
Following the rma‐limma algorithm, 978 sequences were found that satisfied the criteria for significant differential expression between enterocytes lining the upper part of crypts and enterocytes lining villi in the tissues investigated (the whole data set is available in supplementary tables 2 and 3 which can be viewed on the Gut website at http://www.gutjnl.com/supplemental). Of these, 382 genes displayed a greater than twofold change in five of six tissue samples, and 124 genes were more than twofold differentially expressed along the CVA in six of six tissue samples. The 978 sequences represent 778 unique UniGene IDs (62 sequences had no respective UniGene ID). Of these, 363 genes were predominantly expressed in crypts and 415 genes were predominantly expressed in villi.
Table 2 Comparison of X3P data with data from the crypt/villus mRNA in situ hybridisation database (CVD) provided by Olsen and colleagues15.
| Gene | X3P | CVD | Concordance |
|---|---|---|---|
| ALPI | Villus: 5.0‐fold | Villus | + |
| APOA1 | Villus: 10.1‐fold | Villus | + |
| APOA4 | Villus: 4.1‐fold | Villus | + |
| APOBEC1 | Villus: 3.8‐fold | Villus | + |
| MME | Villus: 1.9‐fold | Villus | + |
| MYB | Crypt: 6.7‐fold | Crypt | + |
| RPL3 | Crypt: 2.4‐fold | Crypt | + |
| RPS4X | Crypt: 2.1‐fold | Villus | − |
| SLC15A1 | Villus: 3.7‐fold | Villus | + |
| SLC25A5 | Villus: 1.5‐fold | Villus | + |
| SLC5A1 | Villus: 2.7‐fold | Villus | + |
| SLC6A8 | Villus: 3.3‐fold | Villus | + |
| TOP2A | Crypt: 14.9‐fold | Crypt | + |
| VIL1 | Villus: 2.0‐fold | Villus | + |
X3P ratio of crypt versus villus with p‐fdr<0.05 are given.
+, concordance.
Table 3 Functional group analysis based on cellular or biological gene ontology terms (GO‐term)12.
| Observed | Expected | Enrichment ratio | p Value | p‐fdr | p‐bonf | |
|---|---|---|---|---|---|---|
| Cellular GO‐term | ||||||
| Cytosolic ribosome (sensu Eukaryota) | 15 | 2.15 | 6.98 | <0.00001 | 0.00135 | 0.00674 |
| Intracellular non‐membrane bound organelle | 113 | 56.23 | 2.01 | <0.00001 | 0.00135 | 0.00674 |
| Ribosome | 33 | 8.22 | 4.01 | <0.00001 | 0.00135 | 0.00674 |
| Vacuole | 20 | 5.55 | 3.6 | <0.00001 | 0.00135 | 0.00674 |
| Large ribosomal subunit | 11 | 2.04 | 5.39 | <0.00001 | 0.00135 | 0.00674 |
| Chromosome | 28 | 11.22 | 2.5 | 0.00001 | 0.00225 | 0.01348 |
| Chromosome, pericentric region | 9 | 1.7 | 5.29 | 0.00003 | 0.00578 | 0.04044 |
| Spindle | 11 | 2.72 | 4.04 | 0.00006 | 0.01011 | 0.08088 |
| Nucleolus | 12 | 4.08 | 2.94 | 0.00066 | 0.09885 | 0.88968 |
| Biological GO‐term | ||||||
| Cell proliferation | 105 | 55.28 | 1.9 | <0.00001 | 0.00733 | 0.01466 |
| DNA metabolism | 57 | 26.68 | 2.14 | <0.00001 | 0.00733 | 0.01466 |
| Nucleotide metabolism | 23 | 9.48 | 2.43 | 0.00007 | 0.05131 | 0.20524 |
| Macromolecule biosynthesis | 63 | 38.75 | 1.63 | 0.00007 | 0.05131 | 0.20524 |
| Nucleotide biosynthesis | 18 | 6.77 | 2.66 | 0.00013 | 0.07623 | 0.38116 |
Significance value (p) is corrected for multiple testing by the method of Benjamini‐Hochberg (p‐fdr) and the method of Bonferroni (p‐bonf). Only significantly enriched gene ontology terms with p‐fdr <0.1 are shown.
The following results verified the absence of contaminating cells (for example, goblet cells, Paneth cells, apoptotic enterocytes) from the enterocytes microdissected from crypts and villi:
only a few apoptosis related mRNA species (TNFRSF1A, TNFRSF1B, and TNFSF5) were found;
TTF3, a molecular marker for goblet cell differentiation,16 was very weakly but homogeneously expressed;
the same was found for expression of marker molecules of enteroendocrine cells; and
expression of lysozyme, a Paneth cell marker, was not found.
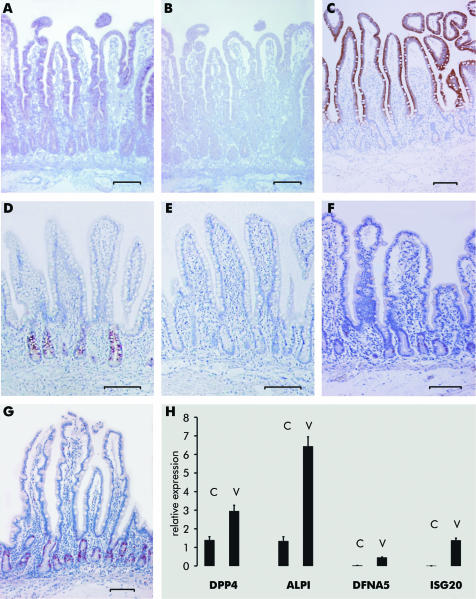
In order to validate the results obtained from chip analyses, we studied 10 arbitrarily selected genes by quantitative RT‐PCR, mRNA in situ hybridisation, western blotting, and/or immunohistochemistry using independent samples. Differential expression of all 10 genes along the CVA was confirmed (data summarised in table 1; examples in fig 2).
Figure 2 Expression of different genes in enterocytes along the crypt‐villus axis (CVA) visualised with mRNA in situ hybridisation on paraffin embedded tissues (A, B, D, E; A, D antisense probe; B, E sense probe), immunohistochemistry (C, F, G), or quantitative reverse transcription‐polymerase chain reaction (H). (A‐C) ACSL5. (D, E) DMBT1 (tissues were counterstained with haematoxylin). (F) TP53. (G) MKi67. (H) DPP4, ALPI, DFNA5, and ISG20; C, expression in crypt; V, expression in villus. Tissues investigated are from other patients than the probes analysed with the X3P arrays. Bars indicate 100 µm.
In addition, a public intestinal crypt/villus mRNA in situ hybridisation database (CVD)15 was screened to confirm the crypt‐villus gene expression pattern out of the X3P arrays. Of the genes contained in CVD, 53 were available for comparison because they showed unambiguous differential expression between the upper crypt (segment 2 in CVD) and the villus (segment 3 in CVD) of the small intestine. Fourteen of the 53 genes were also present in the gene set identified by our chip analyses. Of these, 13 (93%) showed a concordant pattern (table 2) while the ribosomal gene RPS4X showed a different expression (crypt top according to our data).
X3P detected genes were categorised into two functional groups (cell proliferation and metabolism) based on analyses of gene ontology terms12 (table 3).
Subcategorisation into four functional groups was suggested by advanced analysis of constantly enriched genes by functional terms: (i) cell cycle; (ii) transcription/ translation; (iii) metabolism; and (iv) vesicle/ transport.
Cell cycle inhibiting genes are preferentially expressed in enterocytes lining villi
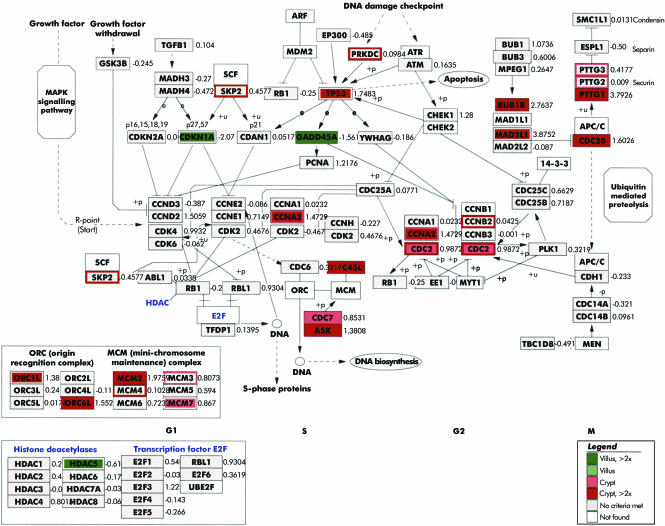
Despite the fact that mitotic enterocytes were excluded from analysis, genes preferentially involved in cell cycle promotion were expressed in enterocytes isolated from crypts. In enterocytes of villi, the cell cycle was negatively regulated because strong expression of CDKN1A (p21), a cyclin dependent kinase inhibitor, GADD45A/B, an important player in cell cycle G2‐M arrest, Q69LE1, a putative cyclin dependent kinase inhibitor, and HDAC5, a suppressor of the transcriptional activity was found (data are summarised in fig 3).
Figure 3 Expression profiling of cell cycle related genes in non‐absorptive (crypt, shown in red) and absorptive (villus, shown in green) enterocytes superimposed onto the cell cycle pathway map.11
Transcription/translation related genes are predominantly expressed in crypts
Genes encoding molecules important for transcription/ translation, such as RPLs, MRPLs (for example, MRPL12),17 IARS, Q96A35, and RPLP0, were predominantly expressed in crypts (see supplementary fig 1; supplementary material can be viewed on the Gut website at http://www.gutjnl.com/supplemental). Spatial distribution of expression of the genes MYC, MAD, and MAX along the CVA was identical to that found in mice.6 Strong expression of MYC was found in the crypt lining enterocytes, but MAD and MAX were preferentially expressed in villi.
Metabolism related genes are predominantly expressed in enterocytes lining villi
In enterocytes lining villi, expression of a high number of genes related to carbohydrate metabolism (for example, FUCA1, GLO1, HUM2DD, ALDOB, PFKP, ACLY), fatty acid metabolism (for example, APOA1, APOA4, APOBEC1, ADIPOR2, FACL5), or protein metabolism (for example, DPP4, RAB17) was preferentially found, which was indicative of the absorptive enterocyte (see supplementary fig 2; supplementary material can be viewed on the Gut website at http://www.gutjnl.com/supplemental).
Strong expression of vesicle/transport related genes in enterocytes lining villi
Genes encoding for proteins with putative functions in vesicles and/or transport processes were predominantly expressed in enterocytes lining villi (see supplementary fig 3; supplementary material can be viewed on the Gut website at http://www.gutjnl.com/supplemental). Genes belonging to the cytochrome P450 “superfamily”, which encodes many proteins with enzymatic activities involved in the detoxification metabolism,18 together with CST3, PGRMC2, and HMOX1, were additionally found in enterocytes lining villi. HMOX1 is highly inducible by its substrate haeme and various non‐haeme substances such as glutamine, heavy metals, bromobenzene, and endotoxin.19 Strong expression of lysosome related genes such as CTSB, LGMN, and FUCA1 was additionally found in villi together with membrane transport/endocytosis related genes, such as several members of the SLC family, the SNX4 gene, RAB17, CORO1A, and HFE, and genes involved in cellular organisation and biogenesis (for example, VILL, Q9H6Z0, GGH, SLC12A2, KPNA2).
Discussion
In order to investigate differentiation of enterocytes along the time dependent CVA in human small intestine, expression profiling was performed. Our analysis revealed 778 genes that were differentially expressed between the crypt top and villus epithelium. Compared with the study of Mariadason and colleagues,6 the number of UniGene IDs found was lower, probably due to several facts, including the use of human tissues with a heterogeneous genetic background, individual composition of ingests of the patients, and differences in sex and age, and experimental parameters. Furthermore, the spatial distribution of cells under investigation was important because in the mouse studies by Mariadason et al the gene profile found in the most basal intestinal cells was compared with the profile in the most apical cells6 whereas the aim of our study was to distinguish between different types of enterocytes in the crypt top versus the mid‐villus epithelium with regard to maldigestion and malabsorption, which are both clinically relevant. The strong concordance (93% for overlapping genes) of the X3P array data with the in situ hybridisation data provided by Olsen and colleagues15 further supports the integrity of our approach. In the present study four functional groups were identified which characterise non‐absorptive enterocytes found in the upper part of crypts and absorptive enterocytes found in the middle of villi. The gene profile of each functional group and the results of the gene ontology analysis indicated an active mechanism in cell cycle arrest, a prerequisite for the transition from the non‐absorptive to the absorptive state. In addition to the Wnt control of differentiation in the intestinal epithelium,20 one further regulating transcriptional mechanism probably underlying this complex differentiation programme was identified, the MYC‐MAD‐MAX network. This finding is in agreement with results obtained for the mouse small intestine.6
In our experimental setting, the spatial distribution of the non‐absorptive type was restricted to enterocytes above the proliferative pool of crypts whereas the absorptive type was exclusively found lining villi, a phenomenon which is of potential importance for the pathophysiology of diseases associated with disturbances in the crypt‐villus architecture, such as coeliac disease. There is some experimental evidence21 that in Marsh IIIc coeliac disease, hyperplasia of the non‐absorptive but not of the absorptive type is found. After reconstitution of villi under a gluten free diet, differentiation to the absorptive type is seen, again in the villus lining enterocytes.21 The main clinical symptoms associated with villus atrophy are probably due to this molecular setting. Gene profiling of enterocytes microdissected from small intestinal tissues with injured villi architecture could be helpful to further elucidate the underlying pathways for cellular transition.
In conclusion, our study provides evidence for the existence of different types of enterocytes characterised by their gene profile and their spatial distribution within the CVA in human small intestine. The molecular discrimination of non‐absorptive and absorptive enterocytes is an important step towards improving our knowledge of the pathophysiology of villi injury which is found in several intestinal diseases.
The three supplementary tables and three supplementary figs can be viewed on the Gut website at http://www.gutjnl.com/supplemental.
Supplementary Material
Acknowledgements
The authors are grateful to S Herberger, M Haak, M Keith, and S Thys for excellent technical work. The help of J Moyers is acknowledged. The study was supported in part by the Tumorzentrum Heidelberg/ Mannheim (I/1.2.).
Abbreviations
CVA - crypt‐villus axis
CVD - intestinal crypt/villus mRNA in situ hybridisation database
DTA - developmental time axis
LMD - laser microdissection
RT‐PCR - reverse transcription‐polymerase chain reaction
Footnotes
Conflict of interest: None declared.
The three supplementary tables and three supplementary figs can be viewed on the Gut website at http://www.gutjnl.com/supplemental.
References
- 1.Parnis S, Nicoletti C, Ollendorff V.et al Enterocytin: a new specific enterocyte marker bearing a B30.2‐like domain. J Cell Physiol 2004198441–451. [DOI] [PubMed] [Google Scholar]
- 2.Dusing M R, Brickner A G, Lowe S Y.et al A duodenum‐specific enhancer regulates expression along three axes in the small intestine. Am J Physiol Gastrointest Liver Physiol 2000279G1080–G1093. [DOI] [PubMed] [Google Scholar]
- 3.Traber P, Silberg D. Intestine‐specific gene transcription. Annu Rev Physiol 199658275–297. [DOI] [PubMed] [Google Scholar]
- 4.Madison B B, Dunbar L, Qiao X T.et al Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem 200227733275–33283. [DOI] [PubMed] [Google Scholar]
- 5.Tadjali M, Seidelin J B, Olsen J.et al Transcriptome changes during intestinal cell differentiation. Biochim Biophys Acta 20021589160–167. [DOI] [PubMed] [Google Scholar]
- 6.Mariadason J M, Nicholas C, L'Italien K E.et al Gene expression profiling of intestinal epithelial cell maturation along the crypt‐villus axis. Gastroenterology 20051281081–1088. [DOI] [PubMed] [Google Scholar]
- 7.Van Gelder R N, von Zastrow M E, Yool A.et al Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A 1990871663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gentleman R C, Carey V J, Bates D M.et al Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 20045R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irizarry R A, Hobbs B, Collin F.et al Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 20034249–264. [DOI] [PubMed] [Google Scholar]
- 10.Smyth G K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Appl Genetics Mol Biol. 2004;3: No1, article 3, [DOI] [PubMed]
- 11.Dahlquist K D, Salomonis N, Vranizan K.et al GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet 20023119–20. [DOI] [PubMed] [Google Scholar]
- 12.Zhangl B, Schmoyer D, Kirovl S.et al GOTree Machine (GOTM): a web‐based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics 2004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gassler N, Kopitz J, Tehrani A.et al Expression of acyl‐CoA synthetase 5 reflects the state of villus architecture in human small intestine. J Pathol 2004202188–196. [DOI] [PubMed] [Google Scholar]
- 14.Mollenhauer J, Herbertz S, Holmskov U.et al DMBT1 encodes a protein involved in the immune defense and in epithelial differentiation and is highly unstable in cancer. Cancer Res 2000601704–1710. [PubMed] [Google Scholar]
- 15.Olsen L, Hansen M, Ekstrom C T.et al CVD: the intestinal crypt/villus in situ hybridization database. Bioinformatics 2004201327–1328. [DOI] [PubMed] [Google Scholar]
- 16.Iwakiri D, Podolsky D K. Keratinocyte growth factor promotes goblet cell differentiation through regulation of goblet cell silencer inhibitor. Gastroenterology 20011201372–1380. [DOI] [PubMed] [Google Scholar]
- 17.Sylverster J E, Fischel‐Ghodisan N, Mougey E B.et al Mitochondrial ribosomal proteins: candidate genes for mitochondrial disease. Genet Med 2004673–80. [DOI] [PubMed] [Google Scholar]
- 18.Patel H R, Hewer A, Hayes J D.et al Age‐dependent change of metabolic capacity and genotoxic injury in rat intestine. Chem Biol Interact 199811327–37. [DOI] [PubMed] [Google Scholar]
- 19.Coeffier M, Le Pessot F, Leplingard A.et al Acute entereal glutamine infusion enhances heme oxygenase‐1 expression in human duodenal mucosa. J Nutr 20021322570–2573. [DOI] [PubMed] [Google Scholar]
- 20.Pinto D, Clevers H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp Cell Res 2005306357–363. [DOI] [PubMed] [Google Scholar]
- 21.Obermüller N, Keith M, Kopitz J.et al Coeliac disease is associated with impaired expression of acyl‐CoA synthetase 5. Int J Colorectal Dis 200621130–134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.