Although slow transit constipation (STC) may not be a congenital disease, the frequent onset in adolescence and strong female predominance suggest that STC could be a result of a sex modified multifactorial disorder of the gastrointestinal tract with a genetic basis. Several genes such as RET proto‐oncogene and the neurturin gene have been analysed in STC. Unfortunately, few mutations were found to be associated with STC.1,2 Our previous studies described a decrease in volume in interstitial cells of Cajal (ICC) in patients with STC, and downregulation of c‐kit mRNA and c‐kit protein expression in the colonic tissues of STC patients.3,4 At present, we do not know why ICCs are lost from colonic tissues of patients with STC. Evidence suggests that the c‐kit/SCF signal pathway plays a crucial role in ICC development and maintenance of its phenotype. An example of loss of function mutations of the c‐kit gene is mice that lack the network of ICCs and show abnormal intestinal pacemaker activity.
To date, no study has explored whether the c‐kit gene is a candidate for STC. Therefore, we screened a series of patients with chronic idiopathic STC for germline mutations of c‐kit.
The STC group included 23 patients who had a history of longstanding intractable constipation, with bowel movements ranging between once per five and 15 days. Colon transit time, determined by radio‐opaque marker tests, was markedly increased by more than 120 hours, and conventional medical therapy had failed in all cases. The control group included eight patients undergoing partial colectomy for non‐obstructive carcinoma (T1‐T2) or adenoma. Genomic DNA extracted from the resected colon of patients and controls was screened by direct DNA sequencing using the fluorescent dideoxy terminator method. The coding region between exon 9 and exon 21 of the c‐kit gene was fully sequenced in both directions, including some intron and intron–exon boundaries. Results are summarised in table 1.
Table 1 Polymorphisms in the c‐kit gene (exon 9 to exon 21).
| Position | Nucleotide | Amino acid | Allele frequency | χ2 | p Value | |
|---|---|---|---|---|---|---|
| Controls | STC | |||||
| Exon 10 | 75515T→C | 531Ile→Thr | 0 | 0.022 | 0.748 | 0.387 |
| Exon 10 | 75561A→G | 546Lys→Lys | 0 | 0.022 | 0.748 | 0.387 |
| Intron 11 | 75794T→A | — | 0 | 0.174 | 6.570 | 0.018* |
| Intron 16 | 81240G→A | — | 0 | 0.063 | 2.304 | 0.129 |
| Intron 17 | 81517C→T | — | 0.118 | 0.261 | 2.506 | 0.159 |
| Intron 19 | 85240A→G | — | 0 | 0.109 | 3.942 | 0.069 |
| Intron 20 | 86548T→A | — | 0.029 | 0.196 | 4.940 | 0.038* |
—, No amino acid changes.
*p<0.05 versus controls.
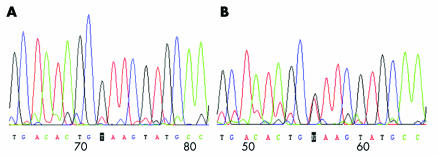
The results were compared with published sequence data and eight control DNAs. Seven genovariation sites were detected. Only one mutation was found in one case at the c‐kit gene 75515T→C, which resulted in codon 531‐Ile to Thr in exon 10 but allele frequency was comparable between patients and controls. Two polymorphism sites in the intron region (base substitution at 75794T→A and the heterozygous mutation at 86548T→A) exhibited a significantly higher frequency compared with controls (fig 1).
Figure 1 DNA sequencing of the c‐kit gene at intron 20. (A) Controls. (B) STC patients with the mutation at base 86548T→A.
This study evaluated for the first time whether c‐kit gene variations were related to the pathogenesis of STC. Seven genovariation sites were detected; only two (81240G→A, 85240A→G) had been published previously. The only missense mutation was found in one case at c‐kit gene 75515T→C, which result in codon 531‐Ile to Thr in exon 10. This point mutation may be pathological for individuals but its significance in the aetiology of STC needs further evaluation in large samples. Moreover, two polymorphism sites in the intron region were remarkable. The base substitution at 75794T→A (just six bases downstream of intron 11) and the heterozygous mutation at 86548T→A (located at 48 bases upstream of intron 21) are expected to affect exon–intron splicing, and are thus worthy of further exploration. The unique published missense single nucleotide polymorphism site of the c‐kit gene at base 75544 A→G, the codon 541‐Met to Val, were not detectable in all subjects.5 This implies that the codon 541 polymorphism is not the cause of STC. Moreover, exons 1–8 were not detected in this study. It is also possible that the extracellular region of the c‐kit gene and other candidate genes, especially the gene encoding SCF, may be involved. Future studies should address these hypotheses.
In conclusion, mutation of the c‐kit gene is not a frequent cause of STC but some polymorphisms in the intron region are worthy of further study.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (No 30300156) and the Science Committee of Chongqing (No 7438).
Footnotes
Conflict of interest: None declared.
References
- 1.De Miguel M P, Cheng L, Holland E C.et al Dissection of the c‐Kit signaling pathway in mouse primordial germ cells by retroviral‐mediated gene transfer. Proc Natl Acad Sci U S A 20029910458–10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knowles C H, Gayther S A, Scott M.et al Idiopathic slow‐transit constipation is not associated with mutations of the RET proto‐oncogene or GDNF. Dis Colon Rectum 200043851–857. [DOI] [PubMed] [Google Scholar]
- 3.Tong W D, Liu B H, Zhang L Y.et al Expression of c‐kit messenger ribonucleic acid and c‐kit protein in sigmoid colon of patients with slow transit constipation. Int J Colorectal Dis 200520363–367. [DOI] [PubMed] [Google Scholar]
- 4.Tong W D, Liu B H, Zhang L Y.et al Decreased interstitial cells of Cajal in the sigmoid colon of patients with slow transit constipation. Int J Colorectal Dis 200419467–473. [DOI] [PubMed] [Google Scholar]
- 5.Nagata H W A, Metcalfe D D. Identification of a polymorphism in the transmembrane domain of the protooncogene c‐kit in healthy subjects. Exp Clin Immunogenet 199613210–214. [PubMed] [Google Scholar]