Abstract
Background
Attenuated familial adenomatous polyposis (AFAP) is associated with germline mutations in the 5′, 3′, and exon 9 of the adenomatous polyposis coli (APC) gene. These mutations probably encode a limited amount of functional APC protein.
Methods and results
We found that colonic polyp number varied greatly among AFAP patients but members of the same family tended to have more similar disease severity. 5′ Mutants generally had more polyps than other patients. We analysed somatic APC mutations/loss of heterozygosity (LOH) in 235 tumours from 35 patients (16 families) with a variety of AFAP associated germline mutations. In common with two previous studies of individual kindreds, we found biallelic changes (“third hits”) in some polyps. We found that the “third hit” probably initiated tumorigenesis. Somatic mutation spectra were similar in 5′ and 3′ mutant patients, often resembling classical FAP. In exon 9 mutants, in contrast, “third hits” were more common. Most “third hits” left three 20 amino acid repeats (20AARs) on the germline mutant APC allele, with LOH (or proximal somatic mutation) of the wild‐type allele; but some polyps had loss of the germline mutant with mutation leaving one 20AAR on the wild‐type allele.
Conclusions
We propose that mutations, such as nt4661insA, that leave three 20AARs are preferentially selected in cis with some AFAP mutations because the residual protein function is near optimal for tumorigenesis. Not all AFAP polyps appear to need “three hits” however. AFAP is phenotypically and genetically heterogeneous. In addition to effects of different germline mutations, modifier genes may be acting on the AFAP phenotype, perhaps influencing the quantity of functional protein produced by the germline mutant allele.
Keywords: genetic pathways, familial adenomatous polyposis, germline mutation
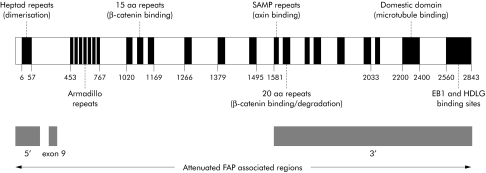
Classical familial adenomatous polyposis (FAP) is caused by germline mutations in the adenomatous polyposis coli (APC) gene between codons 178 and 1580. FAP patients typically develop hundreds to thousands of adenomatous polyps in the colon and rectum by the third decade of life. If left untreated, one or more adenomas progress to carcinoma by 45 years of age. Extracolonic features, such as polyps of the upper gastrointestinal tract, desmoid tumours, and osteomas, are also common. Attenuated FAP (AFAP or AAPC) patients generally present with a lower number (<100) of colorectal adenomas by their fourth decade and have a later age of onset of colorectal cancer (mean age 55 years).1,2,3 In some AFAP patients, extracolonic features have been reported to be infrequent4 although other AFAP patients—such as those with hereditary desmoid disease—have severe extracolonic disease.5,6 AFAP is associated with germline mutations in specific regions of the APC gene (fig 1): the 5′ end (codons 1–177, exons 1–4); the 3′ end (distal to codon 1580); and the alternatively spliced region of exon 9 (codons 311–408).3,7,8 The molecular mechanism(s) underlying these genotype‐phenotype associations for APC remains largely unknown.
Figure 1 Representation of the adenomatous polyposis coli protein comprising important functional domains and showing regions of the protein germline mutation which are associated with attenuated familial adenomatous polyposis (FAP).
APC is a tumour suppressor gene and almost all mutations truncate the protein or take the form of allelic loss (loss of heterozygosity (LOH)). Several genetic studies of colorectal adenomas from FAP patients have shown that somatic APC mutations are dependent on the position of the germline APC mutation (fig 1).9,10,11 The APC protein contains seven 20 amino acid repeats (20AARs) which are involved in degrading the transcriptional cofactor beta‐catenin and hence negatively regulate Wnt signalling. In colorectal polyps, germline mutations between codons 1285 and 1378 leave only one 20AAR intact and are strongly associated with somatic loss of the wild‐type APC allele. LOH usually occurs through mitotic recombination, thus leaving two identical alleles and a total of two 20AARs in the tumour cell.12 FAP patients who carry germline mutations before codon 1285 (no 20AARs) tend to have somatic mutations which leave one or, more commonly, two 20AARs in the protein. Finally, patients with germline mutations after codon 1398 (two or three 20AARs) tend to have somatic mutations before codon 1285. The same associations are also found in sporadic colorectal tumours.13 This interdependence of “first” and “second hits” shows that selective constraints on APC mutations are active and that an optimum level of beta‐catenin mediated signalling must be achieved for the tumour cell to grow.10 There is no reason to expect that AFAP polyps are not subject to the same selection for optimal Wnt signalling as other colorectal adenomas.
The “first hit‐second hit” associations can explain why FAP patients with germline APC mutations between codons 1285 and 1378 have particularly severe colorectal disease because the associated allelic loss occurs at a higher spontaneous frequency than the somatic truncating mutations selected in other FAP patients.9 Conversely, the milder disease in AFAP patients may be explained if the mutations required to give the polyp cell a strong selective advantage are difficult to acquire. Spirio and colleagues1 studied colorectal tumours from a single AFAP family with a germline APC mutation in the 5′ end of the gene (codon 142FS). Approximately 12% of their polyps showed loss of the germline mutant allele, implying that this was a “third hit” subsequent to a mutation on the germline wild‐type allele. Furthermore, a large proportion (36%) of the truncating somatic mutations detected were 1 bp insertions at an A6 tract between nucleotides 4661–4666 (codons 1554–1556). Spirio and colleagues1 concluded that germline mutations in the 5′ region of APC encode proteins that retain residual activity, owing to alternative splicing or initiation of translation. Somatic mutations would be required not only to inactivate the wild‐type allele but also to reduce the residual activity of the mutant germline allele. Su and colleagues14 studied nine adenomas from an AFAP family with a germline mutation (R332X) in exon 9. They found “third hits”, including loss of the germline mutant allele and 4661insA, and showed the latter to occur on the germline mutant chromosome. The APC isoprotein lacking exon 9 retained at least partial ability to downregulate beta‐catenin mediated transcription, providing a reason for the “three hits” and thus attenuation of the phenotype. Su and colleagues14 suggested that exon 9 mutant AFAP patients develop more tumours than the general population because the germline mutant APC allele could be inactivated by a broad spectrum of somatic mutations, including some, such as nt4661insA, that would not normally affect a wild‐type APC allele.
The existing studies only analysed single families but established the important principle that “third hits” can occur in AFAP. These “third hits” could be LOH or mutation at codon 4661. In this study, we analysed a larger number of AFAP families with the following aims:
to search for phenotypic differences among AFAP families, both between and within kindreds with mutations in each of the three AFAP associated regions of APC;
to determine whether the two families reported were typical of AFAP;
to find out the somatic APC mutation spectrum in AFAP patients with 3′ mutations and to compare this with the other AFAP associated regions of APC;
to find out why 4661insA is such a common “third hit”;
to delineate the pathways of somatic APC mutation in AFAP, with emphasis on whether polyps end up with the optimal genotype, as predicted by studies of classical FAP;
to determine whether “three hits” are always needed in AFAP.
Materials and methods
Study population and samples
We contacted the polyposis registries in the UK, Switzerland, Germany, and Denmark with a request to study colorectal tumours from AFAP patients with characterised germline APC mutations in the 5′ or 3′ regions of the gene (codons 1–177 and 1580–2843) or in the alternatively spliced region of exon 9 (codons 311–408). In total, 235 fresh frozen or formalin fixed, paraffin embedded colorectal tumours were obtained from 35 individuals in 16 families. All patients gave written informed consent. A total of 231 of the tumours were colorectal adenomas, almost all of tubular morphology and with a median diameter of 3 mm (range 1–17 mm); four tumours were colorectal carcinomas (median diameter 5 mm (range 2–20)). Thirty tumours were from six AFAP patients from five families with germline APC mutations in the 5′ region of the gene (G126X, 141FS, Q163X, 170FS, 173FS). Seventy nine tumours were from 10 AFAP patients from five families, each of which carried the relatively common R332X nonsense mutation in the alternatively spliced region of APC exon 9. A total of 126 tumours were from 19 AFAP patients from six families with germline APC mutations in the 3′ region of the gene (1597FS, 1738FS, 1919FS, 1943FS, 1982FS, 2078FS). Clinical details (APC germline mutation, sex, age at presentation, polyp count) were obtained and are being analysed as part of a larger study of phenotype in AFAP (AL Knudsen, in preparation); numbers of polyps analysed per patient are summarised in table 1. Paired normal tissue was available for all patients. Haematoxylin‐eosin stained sections were prepared from each tumour to confirm the presence of at least 60% neoplastic tissue. DNA was extracted from tumour and normal tissue using standard methods.
Table 1 Characteristics of the 35 patients with germline adenomatous polyposis coli (APC) mutations in the three attenuated familial adenomatous polyposis (AFAP) associated regions (5′, exon 9, and 3′; codons 1–177, 311–408, and >1580).
| Patient ID | Germline APC mutation | Sex | Age at presentation (y) | Polyp count | Polyps analysed |
|---|---|---|---|---|---|
| AFX MK | G126X | M | 36 | 834 | 6 |
| DFAP48 | 141FS | F | 56 | 2 | 1 |
| AVC.III.2 | Q163X | M | 51 | 2100 | 4 |
| 554.iv.2/1112 | 170FS | F | 32 | 1357 | 11 |
| 554.iii.2 | 170FS | M | 50 | 1077 | 5 |
| 1464/1 | 173FS | M | 39 | “Multiple” | 3 |
| 673.iii.3/1132 | R332X | F | 49 | 200–300 | 6 |
| 1571.ii.2 | R332X | F | 68 | 5 | 5 |
| 578.AA | R332X | F | n/a | n/a | 4 |
| 578.FPL | R332X | F | 27 | 20 | 6 |
| 578.iii.9 | R332X | M | 41 | n/a | 17 |
| 578.iv.1 | R332X | F | 43 | 130 | 14 |
| 578.iv.4 | R332X | F | 27 | n/a | 9 |
| 578.iv.7 | R332X | M | n/a | n/a | 15 |
| DFAP16 | R332X | M | 52 | “Multiple” | 1 |
| DFAP81 | R332X | M | 32 | <100 | 2 |
| 344–40 | 1597FS | M | 47 | 99 | 4 |
| 344–44 | 1597FS | M | 43 | 50 | 3 |
| 01/266 | 1738FS | F | 53 | 29 | 19 |
| 2233/3 | 1919FS | n/a | n/a | n/a | 15 |
| MD2976 | 1919FS | F | 43 | >100 | 21 |
| 77–11 | 1943FS | M | 66 | 500 | 24 |
| 77–12 | 1943FS | M | 56 | 500 | 2 |
| 77–40 | 1943FS | M | 39 | 500 | 2 |
| 1460/6 | 1982 FS | F | 65 | >100 | 5 |
| 1460/28 | 1982 FS | F | 33 | 8 | 1 |
| 1460/42 | 1982 FS | F | 33 | >100 | 4 |
| 1460/88 | 1982 FS | M | 48 | <100 | 1 |
| 1489/10 | 1982 FS | F | 29 | n/a | 1 |
| 1624/04 | 1982 FS | F | 40 | <100 | 3 |
| S73119 | 2078FS | F | 52 | <100 | 1 |
| DW20284 | 2078FS | F | n/a | n/a | 1 |
| J42424 | 2078FS | F | n/a | n/a | 3 |
| L12562 | 2078FS | F | 60 | “Numerous” | 3 |
| 110.2.vi | 2078FS | F | n/a | 33 | 14 |
FS, frameshift; n/a, not available.
Mutation screening
All samples were screened for somatic APC mutations using fluorescence single strand conformational polymorphism (SSCP) analysis on the ABI3100 sequencer (details available from the authors on request). Fresh frozen samples were screened between codons 1 and 1779. Owing to the limiting quantity of DNA, formalin fixed, paraffin embedded samples were screened between codons 1220 and 1603, an area encompassing the somatic mutation cluster region15 and extending beyond the first SAMP repeat involved in axin binding. Samples with bandshifts on SSCP analysis were sequenced in both forward and reverse orientations from a new polymerase chain reaction (PCR) product.
Cloning
We wished to determine the phase of somatic APC mutations with respect to the germline wild‐type or mutant allele but the quality of DNA available from archival tumours was insufficient to allow long range PCR amplification. We therefore identified a single nucleotide polymorphism (SNP) (nt4479 A>G) within APC which was close enough to most somatic mutations of interest to be PCR amplified and which was informative and linked to the disease causing mutation. After amplification of a region encompassing the somatic APC mutation and the SNP, the PCR product was cloned and multiple clones were sequenced using the pGEM‐T Easy Vector System II (Promega, Madison, Wisconsin, USA).
Loss of heterozygosity analysis
In the case of germline nonsense mutations in APC, LOH (allelic loss) analysis was performed using three microsatellite markers, D5S346, D5S421, and D5S656, which map close to APC. Where linkage information was available for the microsatellites studied, the allele targeted by the allelic loss was assigned as germline mutant or wild‐type. Where no linkage information was available, the allele targeted was determined by inspection of the sequencing electropherogram in constitutional and tumour DNA for the region containing the mutation. In the case of germline (and somatic) frameshift mutations, LOH analysis was performed using oligonucleotide primers which encompassed the germline insertion/deletion, which was then used as a pseudopolymorphism for assessing loss. Standard methods of fluorescence based genotyping on the ABI3100 sequencer were used. Allelic loss was scored at any informative marker if the area under one allelic peak in the tumour was reduced by more than 50% relative to the other allele, after correction for the relative peak areas of the alleles found in constitutional DNA of the same patient.
Multiplex ligation dependent probe amplification (MLPA) analysis and real time quantitative multiplex (RQM)‐PCR
MLPA analysis to determine the copy number of the APC promoter and individual exons was performed on polyps with allelic loss at APC using the Salsa MLPA kit P043 APC (MRC‐Holland) according to manufacturer's instructions. RQM‐PCR to determine the copy number of APC exon 14 (normalised against human serum albumin (Alb) exon 12) was performed as previously described.16 The assay has previously been shown to be sensitive for tumour samples containing less than 30% contaminating normal tissue.11
Results
Overall phenotypic assessment
We have previously shown that disease severity (number of colorectal adenomas) in classical FAP patients varies considerably, independent of the germline mutation, but that family members tend to have similar severities of disease.17 In order to test for the same tendency in AFAP, we searched the published literature (details available from authors) for all patients who had germline mutations in the AFAP associated regions of APC and with precisely reported colorectal polyp counts at presentation. We then combined these data with our own. Patient age had no significant effect on polyp number. We then tested for familial aggregation of disease severity and found good evidence for this, both when all families were considered together (p<0.00001, Kruskal‐Wallis test) and when families with germline mutations in the three AFAP associated regions of APC were analysed separately (p = 0.0002, p = 0.045, p = 0.0005, respectively, for 5′, exon 9, and 3′ mutants, Kruskal‐Wallis test). While some effects of local clinical practice are possible, such strong associations are unlikely to result from systematic errors in polyp counting. We then calculated median polyp count for each family irrespective of size, and tested whether this varied among the three groups with mutations in different regions of the APC gene. The 33 families with 5′ mutations had significantly more severe disease (median of medians 69 polyps (interquartile range (IQR) 45–475); p = 0.047, Kruskal‐Wallis test) than the 16 exon 9 mutant and 26 3′ mutant families, who had similar disease severities (median of medians 16 (IQR 8–130) for exon 9; median of medians 15 (IQR 4–150) for 3′).
Somatic mutations in tumours of patients with AFAP associated germline APC mutations
Given that our data showed aggregation of disease severity within families, it became more likely that individual kindreds analysed by previous studies1,14 had provided only a partial description of the genetic pathways of tumorigenesis in AFAP. We first screened colorectal tumours from 5′ mutant patients for somatic APC changes (see supplementary table 1 which can be viewed on the Gut website at http://www.gutjnl.com/supplemental). We found truncating somatic mutations in nine of 30 (30%) adenomas. Similar to adenomas from classical FAP patients with germline mutations before codon 1285,9,10,11 all of the truncating mutations left either one or two 20AARs in the protein. Just two of the adenomas (7%) harboured a detected “third hit”, each in the form of loss of the germline mutant allele (see supplementary table 1 which can be viewed on the Gut website at http://www.gutjnl.com/supplemental). Our results were consistent with those reported by Albuquerque and colleagues10 on the polyps of a single 5′ mutant patient but differed from those of Spirio and colleagues1 in that we found no mutations at nucleotides 4661–6 or at any other site after the third 20AAR. It was notable that while most of the patients of Spirio and colleagues1 had presented with attenuated polyposis, Albuquerque's patient had been reported to have approximately 100 adenomas and most of our 5′ mutant patients had presented with a classical FAP phenotype (table 1). The family of Spirio and colleagues1 cannot therefore be considered representative of all patients with mutations in the AFAP associated 5′ region of APC.
For patients with exon 9 germline mutations, we found truncating somatic mutations in 47/79 (59%) adenomas (table 2 and supplementary table 2; supplementary table 2 can be viewed on the Gut website at http://www.gutjnl.com/supplemental). Of the total of 50 truncating mutations, 33 (66%) were nt4661insA at codon 1554, and this change was always present on the germline mutant allele where assignment was possible. (An uncharacterised defect in DNA mismatch repair as a cause for this observation was excluded by analysing the microsatellite marker BAT26.) Three other mutations leaving three 20AARs (at codons 1518, 1530, and 1537) were found. LOH was found in 13/79 (16%) adenomas; this affected the wild‐type allele in nine cases and the mutant allele in four cases. Thirty one (39%) adenomas had evidence of “thirds hits”, either two detected somatic changes or a single identified somatic change on the germline mutant allele. The data allowed three main genetic pathways to be identified in exon 9 mutant patients' polyps with evidence of “third hits” (table 2):
Table 2 Numbers of tumours with evidence of “third hits” (somatic mutation of germline mutant allele) at the adenomatous polyposis coli (APC) gene in exon 9 and 3′ mutant patients' polyps.
| Somatic mutation on germline wild‐type allele (“second hit”) | Somatic mutation on germline mutant allele (“third hit”) | No of 5′ mutant patients | No of exon 9 mutant patients | No of 3′ mutant patients |
|---|---|---|---|---|
| LOH | 20AAR3 | 0 | 6 | 0 |
| LOH | 20AAR2 | 0 | 0 | 2 |
| 20AAR0 | 20AAR3 | 0 | 2 | 1 |
| 20AAR1 | 20AAR3 | 0 | 2 | 0 |
| 20AAR0 | LOH | 0 | 0 | 1 |
| 20AAR1 | LOH | 2 | 4 | 3 |
| 20AAR2 | LOH | 0 | 0 | 2 |
| Not found | 20AAR3 | 0 | 20 | 0 |
| Not found | 20AAR2 | 0 | 2 | 2 |
| Not found | LOH | 0 | 0 | 9 |
20AAR1, truncating mutation before first 20 amino acid beta‐catenin binding and degradation repeats (20AAR), etc. Note that these are minimum estimates of the true frequency, not only because we could not screen the entire gene for mutations in small archival polyps but also because it was not possible to assign all mutations to the germline mutant or wild‐type allele.
LOH, loss of heterozygosity.
mutation leaving three 20AARs on germline mutant allele, plus loss of the wild‐type allele;
mutation leaving three 20AARs on germline mutant allele, with undetectable mutation of the wild‐type allele (most likely towards the 5′ end of the gene, which could not be screened in all polyps, and leaving zero 20AARs);
mutation leaving one 20AAR on the wild‐type allele plus loss of the germline mutant allele.
For patients with 3′ germline mutations, we found truncating somatic mutations in 35/126 (28%) adenomas (table 2 and supplementary table 3; supplementary table 3 can be viewed on the Gut website at http://www.gutjnl.com/supplemental). Of the total of 36 truncating mutations, only two (6%) were nt4661insA. Three other mutations leaving three 20AARs (at codons 1537, 1576, and 1570) were found. LOH was found in 30/126 (23%) adenomas, equally affecting the wild‐type and mutant alleles. Twenty (16%) adenomas had either two detected somatic changes or an identified somatic change of the germline mutant allele. There was no clear tendency for different families to acquire different somatic mutations (supplementary table 3; supplementary table 3 can be viewed on the Gut website at http://www.gutjnl.com/supplemental). The data only allowed one consistent genetic pathway to be identified in the 3′ mutant patients' polyps with evidence of “third hits”—namely a mutation leaving one (or two) 20AARs on the germline wild‐type allele plus loss of the mutant allele (table 2).
Table 3 Observed and expected frequencies of somatic mutant APC alleles in attenuated familial adenomatous polyposis (AFAP) polyps with “three hits” for “kick‐start” and “step wise” models of tumorigenesis. Polyp with one somatic insertion/deletion and loss of the germline wild‐type allele.
| Model | Allele frequencies | Polyp 1 | Polyp 2 | Polyp 3 | Polyp 4 | Polyp 5 | Polyp 6 | Polyp 7 | χ2 |
|---|---|---|---|---|---|---|---|---|---|
| Observed αgl | 0.21 | 0.27 | 0.21 | 0.29 | 0.30 | 0.26 | 0.28 | ||
| Observed βsom | 0.64 | 0.52 | 0.49 | 0.52 | 0.33 | 0.40 | 0.27 | ||
| “Kick‐start”, LOH first | Expected βsom = [(1−2αgl)/2] | 0.29 | 0.23 | 0.29 | 0.21 | 0.20 | 0.24 | 0.22 | 1.59 |
| “Kick‐start”, LOH second | Expected βsom = (1−2αgl) | 0.58 | 0.46 | 0.58 | 0.42 | 0.40 | 0.48 | 0.44 | 0.14 |
| “Step wise” | Expected minimum βsom* | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 2.72 |
| Expected maximum βsom = [(1−2αgl)/2] | 0.29 | 0.23 | 0.29 | 0.21 | 0.20 | 0.24 | 0.22 | 1.59 |
αgl = proportion of germline wild‐type allele in polyp; βsom = proportion of somatic mutant allele in polyp. Observed αgl frequency was determined from the loss of heterozygosity (LOH) ratios. Observed βsom frequencies were similarly determined from LOH ratios generated by polymerase chain reaction amplification of a region encompassing the somatic insertion/deletion and subsequent Genescan analysis (using constitutional DNA from patients with germline mutations identical to the somatic change for normalisation). The observed “third” to “second hit” ratio for polyp 3 was determined by sequencing 58 clones of a polymerase chain reaction product encompassing both somatic changes.
*Assuming that the somatic insertion/deletion can be detected if it comprises >20% of all alleles in the polyp.
Comparison between somatic mutations in the three groups of patients
The somatic mutation spectra of the 5′ and 3′ mutant patients' tumours did not differ significantly from each other as regards: (i) proportion of mutations leaving one, two, or three 20AARs (p = 0.074, χ2 test); (ii) overall LOH frequency (2/9 v 30/126; p = 0.64); and (iii) proportion of tumours with detected “third hits” or an identified somatic change on the germline mutant allele (2/9 v 20/126; p = 0.45). However, while exon 9 mutant patients had a similar frequency of LOH (13/79, 22%; p = 0.14) to other patients, germline exon 9 mutants had a higher frequency of mutations that left three 20AARs (36/50 v 5/45; p<0.001, χ2 test) and a higher frequency of tumours with detected “third hits” (31/79 v 22/156; p<0.001, χ2 test). In large part, these associations reflected the fact that nt4661insA was particularly common in exon 9 mutant patients' tumours and exclusively targeted the mutant germline allele. Overall, our data were consistent with a large proportion of polyps in the 5′ and 3′ mutant patients developing along the “classical” FAP pathway, their polyps showing similar somatic mutations to individuals with germline mutations which leave zero 20AARs.9 Exon 9 mutant patients were, however, significantly different from the other two groups of patients. Although not all nt4661insA mutations could be assigned to a germline allele, if we made the reasonable assumption that all of these mutations were on the germline mutant allele, over half of all tumours from exon 9 mutant patients had “third hits” (supplementary table 2; supplementary table 2 can be viewed on the Gut website at http://www.gutjnl.com/supplemental). These differences could not readily be explained by features such as the size or dysplasia of the tumours analysed, which did not differ significantly among the three patient groups (details not shown).
Mechanism of LOH
We tested the possibility that different LOH events (for example, those involving the germline wild‐type and mutant alleles) were caused by different mechanisms, such as mitotic recombination and deletion, which resulted in different gene dosages and functional consequences. However, none of 17 tumours with allelic loss (10 with mutant LOH and seven with wild‐type LOH) showed copy number changes in the APC promoter region or exons using MLPA analysis. We selected for RQM‐PCR analysis 10 further tumours (two with mutant LOH and eight with wild‐type LOH) with mean LOH ratios below 0.3 (indicating that contamination with normal tissue was low enough not to confound the detection of deletion11) but all adenomas showed copy number values between 0.79 and 0.97, consistent with diploid APC copy number and LOH by mitotic recombination.
Early pathways of tumorigenesis in AFAP polyps with “three hits”
Our data, combined with previous findings,1,14 showed that a substantial proportion of AFAP adenomas have acquired two somatic APC changes, one targeting the germline wild‐type and one the germline mutant allele. Consideration of the order in which these somatic changes occur and their respective effects on tumour growth has important implications for determining the molecular genetic mechanism underlying AFAP. In AFAP adenomas, initiation of tumour growth might require all “three hits” to be present in the tumour cell of origin (“kick‐start” model). In this case, the two somatic mutations could occur in either order without functional consequence. The “kick‐start” model implies that a mechanism exists which results in an increase in the intrinsic or effective mutation rate in order to explain the relatively high frequency of such tumours compared with the general population. Alternatively, the “second hit”—necessarily involving the germline wild‐type allele—might be sufficient for early adenoma growth, with the “third hit” (involving the germline mutant allele) being required for subsequent tumorigenesis prior to clinical presentation. This “step wise” model postulates that mutation of the germline wild‐type allele induces limited clonal expansion (thereby increasing the effective mutation rate), and is followed by mutation of the germline mutant allele to give an optimal APC genotype.
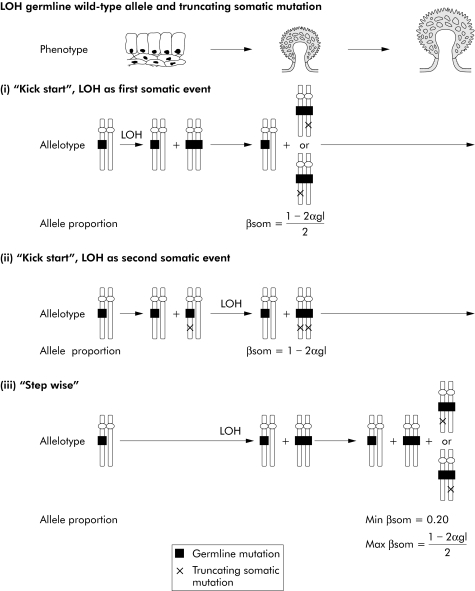
APC mutation data from individual adenomas can be used to distinguish between these possibilities, because the “kick‐start” and “step wise” models are expected to leave distinct footprints as regards the proportion(s) of somatic mutant allele(s), as these proportions depend on the order in which the somatic changes have occurred and some residual adenoma with “two hits” is expected in the “step wise” case (figs 2–4).
Figure 2 Pathways of tumorigenesis in attenuated familial adenomatous polyposis (AFAP) polyps with “three hits”, illustrating the possible sequences in which somatic mutations/allelic loss may occur in AFAP polyps with “three hits” as well as the possible functional effects of these changes. Loss of the germline wild‐type allele and truncating somatic mutation are shown. In a “kick‐start” model, these changes can occur in either order (i) or (ii) and tumour growth ensues once both somatic changes have occurred; in a “step wise” model (iii), loss of the germline wild‐type allele leads to limited clonal expansion and is followed by the truncating somatic mutation which promotes further tumour growth. The expected proportions of βsom and αgl are shown. βsom = proportion of somatic mutant allele in polyp; αgl = proportion of germline wild‐type allele in polyp. LOH, loss of heterozygosity by mitotic recombination. For each model, the expected proportion of the somatic mutant allele (βsom) in the polyp can be determined from the proportions of the germline wild‐type (αgl) allele as shown. αgl can be estimated from the LOH ratio.
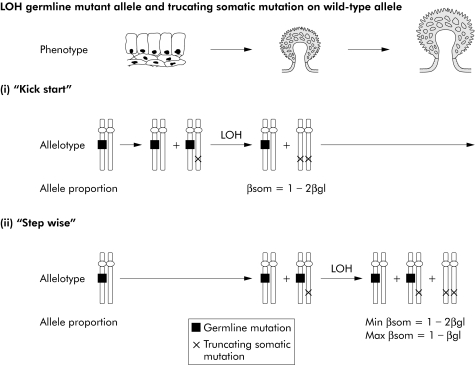
Figure 3 Pathways of tumorigenesis in attenuated familial adenomatous polyposis (AFAP) polyps with “three hits”, illustrating the possible sequences in which somatic mutations/allelic loss may occur in AFAP polyps with “three hits” as well as the possible functional effects of these changes. Loss of the germline mutant allele and truncating somatic mutation are shown. In both the “kick‐start” (i) and “step wise” (ii) models, the truncating somatic mutation precedes loss of the germline mutant allele but in the “kick‐start” model, both changes are required for tumour growth. The expected proportions of βgl and βsom are shown. βsom = proportion of somatic mutant allele in polyp; βgl = proportion of germline mutant allele in polyp. LOH, loss of heterozygosity by mitotic recombination. For each model, the expected proportion of the somatic mutant allele (βsom) in the polyp can be determined from the proportions of the germline mutant (βgl) allele as shown. βgl can be estimated from the LOH ratio.
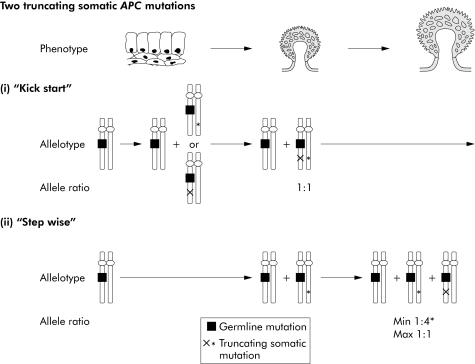
Figure 4 Pathways of tumorigenesis in attenuated familial adenomatous polyposis (AFAP) polyps with “three hits”, illustrating the possible sequences in which somatic mutations/allelic loss may occur in AFAP polyps with “three hits” as well as the possible functional effects of these changes. Two truncating somatic mutations are shown. In a “kick‐start” model (i), these changes can occur in either order and tumour growth ensues once both somatic changes have occurred; in a “step wise” model (ii), somatic mutation of the germline wild‐type allele causes limited clonal expansion and is followed by somatic mutation of the germline mutant allele which promotes further tumour growth. The expected ratio of the two somatic alleles is 1:1 for the “kick‐start model” but lies between 1:4 and 1:1 for the “step wise” model with the minimum estimate (*) assuming a mutation detection sensitivity of 20%.
Consider, for example, polyps with loss of the germline wild‐type allele and a somatic insertion/deletion mutation on the germline mutant allele. We can measure two ratios of relative allelic dosage, one for the germline mutation and the other for the truncating somatic mutation, and use these to estimate the proportion of each allelotype in the tumour. Furthermore, we can calculate the expected values of these ratios by predicting the proportion of somatic mutant allele expected in the tumour under different models of tumorigenesis (figs 2–4). By comparing the observed proportion of the somatic mutant allele with that expected, we can determine whether the “step wise” or “kick‐start” model fits better (see figs 2–4 for details). Similarly, observed and expected allele proportions can be determined for adenomas with one somatic insertion/deletion mutation on the germline wild‐type allele and loss of the germline mutant allele (figs 2–4). For tumours with two truncating somatic mutations, the expected ratio of the two mutant alleles under each model can be compared with the ratio measured directly by cloning a polymerase chain reaction product encompassing both changes, sequencing multiple clones, and ounting how many times each allele is represented (figs 2–4).
For seven tumours with loss of the germline wild‐type allele and a somatic insertion/deletion mutation, the observed and expected proportions of the somatic mutant allele were very similar to those expected under the “kick‐start” model, assuming that the “second hit” was the insertion/deletion (on the somatic mutant allele) and the “third hit” was allelic loss (tables 3–5). Similar results in favour of a “kick‐start” model were obtained for three adenomas with one somatic insertion/deletion and loss of the germline mutant allele, and for one adenoma with two truncating somatic mutations (tables 3–5).
Table 4 Observed and expected frequencies of somatic mutant APC alleles in attenuated familial adenomatous polyposis (AFAP) polyps with “three hits” for “kick‐start” and “step wise” models of tumorigenesis. Polyp with one somatic insertion/deletion and loss of the germline mutant allele.
| Model | Allele frequencies | Polyp 8 | Polyp 9 | Polyp 10 | χ2 |
|---|---|---|---|---|---|
| Observed βgl | 0.33 | 0.10 | 0.24 | ||
| Observed βsom | 0.41 | 0.83 | 0.61 | ||
| “Kick‐start” (LOH must be second) | Expected βsom = (1−2βgl) | 0.34 | 0.80 | 0.52 | 0.03 |
| “Step wise” | Expected minimum βsom = (1−2βgl) | 0.34 | 0.80 | 0.52 | 0.03 |
| Expected maximum βsom = (1−βgl) | 0.67 | 0.90 | 0.76 | 0.14 |
βgl = proportion of germline mutant allele in polyp; βsom = proportion of somatic mutant allele in polyp. Observed βgl frequency was determined from the loss of heterozygosity (LOH) ratios. Observed βsom frequencies were similarly determined from LOH ratios generated by polymerase chain reaction amplification of a region encompassing the somatic insertion/deletion and subsequent Genescan analysis (using constitutional DNA from patients with germline mutations identical to the somatic change for normalisation).
Table 5 Observed and expected frequencies of somatic mutant APC alleles in attenuated familial adenomatous polyposis (AFAP) polyps with “three hits” for “kick‐start” and “step wise” models of tumorigenesis. Two truncating somatic mutations.
| Model | “Third” to “second hit” ratios* | Polyp 11 | χ2 |
|---|---|---|---|
| Observed | 27:31 | ||
| “Kick‐start” | Expected | 1:1 | 0.03 |
| “Step wise” | Expected minimum | 1:4 | 14.4 |
| Expected maximum | 1:1 | 0.03 |
*Assuming a detection sensitivity of 20% for the somatic “third hit”.
Discussion
Our analysis of a relatively large set of AFAP families has shown complexity in the phenotype and early genetic pathways of tumorigenesis. The two previous analyses of somatic APC mutations in AFAP each focused on single families, one with a germline mutation in the 5′ region of the gene1 and the other with a mutation in exon 9.14 These two studies unequivocally provided the important and original finding that “three hits” (that is, two somatic mutations, including loss or mutation of the germline mutant allele) can occur in AFAP tumours. The restricted size of the two studies meant, however, that they were unable to provide further conclusions.
We have found that patients with germline APC mutations in the 5′ and 3′ regions of the gene or the alternatively spliced region of exon 9 have a highly variable large bowel phenotype, in that the number of colorectal adenomas varies from almost none to the hundreds or thousands of lesions found in classical FAP.3 Although assessment methods necessarily differ among clinical centres, our analysis shows that patients with 5′ APC mutations (codons 1–177) are likely to have a more severe phenotype than those with mutations in exon 9 or the 3′ end of the gene (>codon 1580). Phenotypic severity also tends to be similar within families, suggesting that restricting analyses to single kindreds may not provide accurate assessment of AFAP patients.
Our study has confirmed that “three hits” at APC often occur in AFAP adenomas. In such polyps, the “third hit” appears to be required for initiation of tumorigenesis. Although “third hits” might occur at loci other than APC, we have previously found no mutations at beta‐catenin in AFAP polyps (unpublished data). In polyps with “three hits” from exon 9 mutant and 3′ mutant patients, we have been able to identify specific combinations of APC mutations which tend to occur. Exon 9 is alternatively spliced in all normal and neoplastic tissues which we have examined (not shown). The combinations of APC mutations almost certainly produce a near optimal level of Wnt signalling, comparable with those found in classical FAP.9 Some of the combinations (such as R332X‐nt4661insA/LOH ) strongly suggest that the tumour has developed as a result of the functional effects of the germline mutant allele, but other combinations of mutations (such as truncating mutation leaving one 20AAR on the wild‐type with LOH of the germline mutant) might simply be indicative of a “sporadic” tumour occurring on the background of AFAP.
In our families, “third hits” were much rarer in 5′ and 3′ mutant patients than in the exon 9 mutants. These former families' somatic mutations usually, but not always, resembled those of classical FAP patients who have germline mutations before the first 20AAR of the APC protein. In many ways, this is the result which would be predicted were the 5′ or 3′ mutations simply to cause absent or non‐functional protein. 5′ APC mutations probably produce a small amount of partially functional APC through use of an internal ribosome entry site (IRES) at codon 18418. 3′ Mutant proteins have been reported as being unstable19 although the reasons for this are unknown. It is entirely plausible that levels of functional APC protein vary among individuals with both 5′ and 3′ mutations, for example as a result of modifier alleles. Thus for an adenoma to form, some patients would tend to require “third hits” and others would not. The family of Spirio and colleagues,1 for example, may have been relatively efficient at use of the IRES. Formal testing of this hypothesis in vivo would require an exceptionally large unselected series of tumours and patients.
Our analysis of exon 9 mutant cases provides further evidence to show that not all AFAP patients are the same. “Third hits” were common in these patients' tumours. There was a markedly increased frequency of mutations which left three 20AARs on the germline mutant allele, particularly—but not exclusively—at nt4661, which appears to be a relatively hypermutable site. Our view differs somewhat from that of Su and colleagues14 who proposed that insAnt4661 mutations were overrepresented in AFAP polyps because both “strong” and “weak” mutations were sufficient to severely reduce the function of the exon 9 mutant allele. We suggest that mutations leaving three 20AARs on the germline mutant allele are common because the resulting allelotype R332X‐4661insA gives a near optimal genotype, taking into account loss of the germline wild‐type allele and alternative splicing of exon 9. Variation in splicing efficiency, again through modifier allele action, could explain phenotypic variability in exon 9 mutant AFAP but it appears that many of these patients produce sufficient functional protein by splicing out exon 9 that “third hits” are necessary in most polyps.
The reason why AFAP patients develop fewer polyps than classical FAP patients is evident, in that “three hits” are often needed to produce the near optimal genotype. We do not however claim that all polyps from patients with AFAP associated APC mutations require “three hits”. Even allowing for the imperfections of mutation screening and LOH analysis in archival specimens, we were able to analyse the fresh frozen adenomas comprehensively and found many without “three hits”. Moreover, several polyps from our patients had somatic mutations which would have been predicted from a “two hit” model of optimal Wnt signalling. Currently, we cannot explain why in a single patient some polyps seem to require “three hits” and others do not, but it is possible that “third hits” at other loci can substitute for APC mutation. Another possibility is that selective constraints on the diminished APC function needed for tumorigenesis are “just right”1,10 at some times, but weaker at others, for example during development or when tissue is undergoing repair.
Genetic analysis of colorectal tumours from patients with germline mutations in AFAP associated regions of APC, in this study and others, has revealed a novel mechanism underlying the genotype‐phenotype association in this tumour syndrome—namely, a requirement for “three hits” in at least some AFAP adenomas. This finding must be viewed in the framework of the model of optimal combinations of APC mutations, rather than simple loss of protein function. More than one different combination of APC mutations can provide near optimal Wnt signalling in AFAP. However, not all AFAP patients are the same. Given that assembling a very large series of AFAP patients is extremely difficult, it is not easy to decide on what is the “typical” AFAP phenotype or somatic genotype. In the seven families with 5′ APC mutations studied to date (Spirio and colleagues,1 Albuquerque and colleagues,10 and this study), approximately 15–20% of polyps seem to acquire “three hits”, but only Spirio and colleagues1 found a high frequency of nt4661insA. In the six 3′ mutant families studied (all from this study), the frequency of “third hits” seems similar to that of the 5′ mutants. Six exon 9 mutant families have been studied (Su and colleagues14 and this study) and almost all of these show evidence of a high frequency of “third hits”—we estimate a minimum of 50% in our study. In addition, there appear to be genetic factors apart from the germline APC mutation that influence disease severity, as evidenced by the tendency for polyp numbers to be similar within families. The phenotypic and somatic molecular heterogeneity in AFAP means that clinical management of patients with AFAP associated mutations must be empirical. Accurate prediction of phenotype may only be possible when factors, such as modifier genes, that influence genetic pathways and disease severity are identified.
Supplementary tables 1–3 can be viewed on the Gut website at http://www.gutjnl.com/supplemental.
Supplementary Material
Acknowledgements
We are grateful to the Equipment Park, Cancer Research UK, to the patients taking part in the study, and to several collaborating geneticists and histopathologists without whose help the study could not have been performed.
Abbreviations
FAP - familial adenomatous polyposis
AFAP - attenuated familial adenomatous polyposis
20AAR - 20 amino acid beta‐catenin binding and degradation repeats
MLPA - multiplex ligation dependent probe amplification analysis
RQM‐PCR - real time quantitative multiplex polymerase chain reaction
LOH - loss of heterozygosity
APC - adenomatous polyposis coli
SSCP - single strand conformational polymorphism
SNP - single nucleotide polymorphism
IQR - interquartile range
IRES - internal ribosome entry site
Footnotes
Conflict of interest: None declared.
Supplementary tables 1–3 can be viewed on the Gut website at http://www.gutjnl.com/supplemental.
References
- 1.Spirio L N, Samowitz W, Robertson J.et al Alleles of APC modulate the frequency and classes of mutations that lead to colon polyps. Nat Genet 199820385–388. [DOI] [PubMed] [Google Scholar]
- 2.Hernegger G S, Moore H G, Guillem J G. Attenuated familial adenomatous polyposis: an evolving and poorly understood entity. Dis Colon Rectum 200245127–134. [DOI] [PubMed] [Google Scholar]
- 3.Knudsen A L, Bisgaard M L, Bulow S. Attenuated familial adenomatous polyposis (AFAP): a review of the literature. Fam Cancer 2003243–55. [DOI] [PubMed] [Google Scholar]
- 4.Rozen P, Samuel Z, Shomrat R.et al Notable intrafamilial phenotypic variability in a kindred with familial adenomatous polyposis and an APC mutation in exon 9. Gut 199945829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eccles D M, van der Luijt R, Breukel C.et al Hereditary desmoid disease due to a frameshift mutation at codon 1924 of the APC gene. Am J Hum Genet 1996591193–1201. [PMC free article] [PubMed] [Google Scholar]
- 6.Scott R J, Froggatt N J, Trembath R C.et al Familial infiltrative fibromatosis (desmoid tumours) (MIM135290) caused by a recurrent 3′ APC gene mutation. Hum Mol Genet 199651921–1924. [DOI] [PubMed] [Google Scholar]
- 7.Young J, Simms L A, Tarish J.et al A family with attenuated familial adenomatous polyposis due to a mutation in the alternatively spliced region of APC exon 9. Hum Mutat 199811450–455. [DOI] [PubMed] [Google Scholar]
- 8.Soravia C, Berk T, Madlensky L.et al Genotype‐phenotype correlations in attenuated adenomatous polyposis coli. Am J Hum Genet 1998621290–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamlum H, Ilyas M, Rowan A.et al The type of somatic mutation at APC in familial adenomatous polyposis is determined by the site of the germline mutation: a new facet to Knudson's ‘two‐hit' hypothesis. Nat Med 199951071–1075. [DOI] [PubMed] [Google Scholar]
- 10.Albuquerque C, Breukel C, van der Luijt R.et al The ‘just‐right' signaling model: APC somatic mutations are selected based on a specific level of activation of the beta‐catenin signaling cascade. Hum Mol Genet 2002111549–1560. [DOI] [PubMed] [Google Scholar]
- 11.Crabtree M, Sieber O M, Lipton L.et al Refining the relation between ‘first hits' and ‘second hits' at the APC locus: the ‘loose fit' model and evidence for differences in somatic mutation spectra among patients. Oncogene 2003224257–4265. [DOI] [PubMed] [Google Scholar]
- 12.Sieber O M, Heinimann K, Gorman P.et al Analysis of chromosomal instability in human colorectal adenomas with two mutational hits at APC. Proc Natl Acad Sci U S A 20029916910–16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowan A J, Lamlum H, Ilyas M.et al APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci U S A 2000973352–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su L K, Barnes C J, Yao W.et al Inactivation of germline mutant APC alleles by attenuated somatic mutations: a molecular genetic mechanism for attenuated familial adenomatous polyposis. Am J Hum Genet 200067582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fearnhead N S, Britton M P, Bodmer W F. The ABC of APC. Hum Mol Genet 20017721–733. [DOI] [PubMed] [Google Scholar]
- 16.Sieber O M, Lamlum H, Crabtree M D.et al Whole‐gene APC deletions cause classical familial adenomatous polyposis, but not attenuated polyposis or “multiple” colorectal adenomas. Proc Natl Acad Sci U S A 2002992954–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crabtree M D, Tomlinson I P, Hodgson S V.et al Explaining variation in familial adenomatous polyposis: relationship between genotype and phenotype and evidence for modifier genes. Gut 200251420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heppner Goss K, Trzepacz C, Tuohy T M.et al Attenuated APC alleles produce functional protein from internal translation initiation. Proc Natl Acad Sci U S A 2002998161–8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Luijt R B, Meera Khan P, Vasen H F.et al Germline mutations in the 3′ part of APC exon 15 do not result in truncated proteins and are associated with attenuated adenomatous polyposis coli. Hum Genet 199698727–734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.