Abstract
Background
High mobility group box 1 (HMGB1) is a non‐histone chromosomal protein implicated in a variety of biologically important processes, including transcription, DNA repair, V(D)J recombination, differentiation, and development. Overexpression of HMGB1 inhibits apoptosis, arguing that the molecule may act as an antiapoptotic oncoprotein. Indeed, increased expression of HMGB1 has been reported for several different tumour types. In this study, we analysed human colon carcinoma for HMGB1 as well as for c‐IAP2 expression levels. c‐IAP2 is an antiapoptotic protein which may be upregulated as a consequence of nuclear factor κB (NFκB) activation via HMGB1.
Methods
A comparative genomic hybridisation (CGH) database comprising 1645 cases from different human tumour types was screened to detect cytogenetic changes at the HMGB1 locus. Immunohistochemical staining of human colon tissue microarrays and tumour biopsies, as well as western blot analysis of tumour lysates, were performed to detect elevated HMGB1 and c‐IAP2 expression in colon carcinomas. The antiapoptotic potential of HMGB1 was analysed by measuring caspase activities, and luciferase reporter assays and quantitative polymerase chain reaction analysis were employed to confirm NFκB activation and c‐IAP2 mRNA upregulation on HMGB1 overexpression.
Results
According to CGH analysis, the genomic locus containing the HMGB1 gene was overrepresented in one third (35/96) of colon cancers. Correspondingly, HMGB1 protein levels were significantly elevated in 90% of the 60 colon carcinomas tested compared with corresponding normal tissues evaluable from the same patients. HMGB1 increased NFκB activity and led to co‐overexpression of the antiapoptotic NFκB target gene product c‐IAP2 in vitro. Furthermore, increased HMGB1 levels correlated with enhanced amounts of c‐IAP2 in colon tumours analysed by us. Finally, we demonstrated that HMGB1 overexpression suppressed caspase‐9 and caspase‐3 activity, suggesting that HMGB1 interferes with the apoptotic machinery at the level of apoptosomal caspase‐9 activation.
Conclusions
We identified in vitro a molecular pathway triggered by HMGB1 to inhibit apoptosis via c‐IAP2 induction. Our data indicate a strong correlation between upregulation of the apoptosis repressing HMGB1 and c‐IAP2 proteins in the pathogenesis of colon carcinoma.
Keywords: apoptosis inhibitors, nuclear factor κB, colorectal cancer, high mobility group box 1 protein, c‐IAP2
High mobility group box 1 protein (HMGB1; formerly HMG1) is a nuclear non‐histone protein which binds unspecifically to the minor groove of DNA and facilitates the assembly of site specific DNA binding proteins such as P53 at their cognate binding sites within chromatin.1 As an architectural chromatin binding factor it bends DNA and promotes protein assembly on specific DNA targets.2 HMGB1 interacts with cellular and viral proteins to increase their ability to interact with DNA. The protein consists of 215 amino acids and is remarkably conserved among species.3 Together with two other nuclear factors, HMGB2 and HMGB3, HMGB1 belongs to the HMGB protein family. All members share a bipolar structure of two L‐shaped DNA binding elements (HMG box A and B), each made up of 80 amino acids arranged in three positively charged α helices, and a C terminal, extremely negatively charged domain, which has recently been linked to the regulation of HMGB1 acetylation.4
While expression of HMGB2 and HMGB3 is more restricted, HMGB1 has been considered as ubiquitously expressed in all vertebrate nuclei.3 However, recent reports indicate that its expression can be developmentally regulated and induced by environmental stimuli, arguing that HMGB1 is not a housekeeping protein.5
HMGB1 has been implicated in a variety of biologically important processes, including transcription, DNA repair, V(D)J recombination,2,6 differentiation, development, and extracellular signalling.7,8 In certain cell types, the protein is observed at the cell surface where it has been reported to contribute to cellular migration and tumour invasion.9
Recently, a further function of HMGB1 has been identified. The molecule can be secreted by particular cells such as activated monocytes and macrophages,10 thereby playing important roles in inflammation, cell migration, and tumour invasion. The receptor for advanced glycation end products (RAGE) is the only cellular receptor identified for extracellular HMGB1 until now.11 Interestingly, HMGB1 is released by necrotic, but not by apoptotic, cells.12 In apoptotic cells, HMGB1 is bound firmly and irreversibly to chromatin because of histone underacetylation, while HMGB1 released from necrotic cells triggers inflammation due to its function as a cytokine.
Increased expression of HMGB1 has been reported for several different tumour types, including breast carcinoma,13 melanoma,14 gastrointestinal stromal tumours,15 and acute myeloblastic leukaemia.16 Furthermore, overexpression of another structurally distinct family of the high mobility group proteins, the HMGA group (consisting of HMGA‐1a, b, c, ‐2 (formerly HMG I(Y), HMG I‐C)), has been strongly correlated with tumorigenesis (Fedele and colleagues17 and references herein).
Different models exist which would explain the beneficial influence of HMGB1 on tumour development. As an extracellular molecule, HMGB1 induces several intracellular signalling pathways via binding to its receptor receptor for advanced glycation end products (RAGE), including one involving the small GTPases Rac and Cdc42, and a second one that implies the Ras‐mitogen activated protein (MAP) kinase pathway.18 Consequently, blockade of the RAGE‐HMGB1 interaction suppresses activation of p44/p42, p38, and SAP/JNK MAP kinases, and inhibits tumour growth.9 In addition, constant release of HMGB1 as a proinflammatory cytokine from necrotic tumour cells would create a microenvironment similar to chronic inflammations, a condition known to contribute to the development of epithelial malignancies (for review see Mignogna and colleagues19 and references herein).
We have recently identified HMGB1 as a potent inhibitor of yeast cell death induced by overexpression of the mammalian proapoptotic Bcl‐2 family member Bak, a finding most likely not associated with HMGB1 secretion.13 In the same study, we also showed that HMGB1 overexpression suppressed different apoptotic stimuli in mammalian cells. Inhibition of apoptosis is a necessary prerequisite for tumour development.20 During transformation and tumour growth, a cancer cell has to withstand numerous apoptotic triggers. For example, activation of oncogenes such as c‐myc, c‐fos, or c‐jun,21 hypoxia within the tumour mass,22 or anoikis of a metastasising cell detached from the primary tumour,23 are common apoptotic triggers within tumours. However, mutation of the proto‐cancer cells by deactivation of proapoptotic molecules essential for the cell death machinery (loss of function; for example, p5324) or by overexpression of antiapoptotic proteins (gain of function; for example, Bcl‐225) renders the cell resistant to apoptosis. Therefore, HMGB1 upregulation may contribute to tumorigenesis by inhibiting apoptosis in transformed cells. The molecular basis for the antiapoptotic potential of intracellular HMGB1 has not been analysed to date.
Nuclear HMGB1 not only binds to and bends DNA, but by simultaneous interaction with several transcription factors modulates their transactivation or repression capacity (for example, steroid hormone receptors,26 p53,27 or nuclear factor κB (NFκB)).28,29 Although NFκB can promote programmed cell death in response to certain death inducing signals and in certain cell types, it is most commonly involved in suppressing apoptosis by transactivating expression of antiapoptotic genes.30 A prominent example is suppression of tumour necrosis factor α induced cell death by the NFκB target gene product inhibitor of apoptosis c‐IAP2.31,32 The inhibitor of apoptosis protein (IAP) family members bind and inhibit uncleaved as well as activated caspases, thereby preventing cell death downstream of cytochrome c (Cyt c) release within the mitochondrial apoptosis pathway.33 Overexpression of antiapoptotic IAP proteins has been linked to tumorigenesis (see Liston and colleagues33 for references).
In this paper, we describe frequent elevation of both HMGB1 and c‐IAP2 in primary human colon carcinomas. In vitro, HMGB1 mediated enhancement of NFκB activity leads to upregulation of the NFκB target c‐IAP2, and this pathway may result in inhibition of apoptosis in tumour cells.
Materials and methods
DNA constructs
bak‐gfp and pBabe Hmgb1 have been described previously.13pCMV‐IκBαM is an IκBα dominant negative construct from Clontech (Heidelberg, Germany) which contains two mutations that prevent IκBα phosphorylation and degradation.
Comparative genomic hybridisation (CGH) database
The CGH database used for this study was described by Berrar and colleagues34 and contains CGH data from 1645 different tumours, including 538 lymphoma, 137 leukaemia, 426 carcinomas, 218 sarcoma, and 326 CNS tumours.
Colon tissue microarray
Performance of fluorescence in situ hybridisation (FISH) experiments has been described elsewhere.35 Briefly, bacterial artificial chromosome clone RP11‐550P23 containing the human HMGB1 gene (www.ensemble.org) was prepared from bacteria cultures using a Qiagen‐Plasmid‐Kit (Qiagen, Hilden, Germany) and labelled by nick translation with cyanine‐3‐dUTP (Perkin Elmer Life Science, Boston, Massachusetts, USA). Using this probe, a tissue microarray (CB2c, BioCat) containing 29 colon carcinoma biopsies and 29 biopsies of corresponding normal colon tissue were analysed. The average signal number per cell was 1.8 (40 cells evaluated) with an standard deviation of 0.13. The cut off value for copy number gains was defined as the average signal number per cell from normal tissues plus 2× the standard deviation (that is, 2.0).
Expression of HMGB1 and c‐IAP2 in human colon carcinomas was studied using the same tissue microarray as for FISH. HMGB1 levels were determined using a rabbit antibody (BD PharMingen Biosciences, Heidelberg, Germany) which was detected using biotinylated antirabbit IgG, followed by streptavidin‐Cy5 (Dianova, Hamburg, Germany). c‐IAP2 was recognised with a rabbit antibody from R&D Systems (Niesbaden, Germany). For quantification purposes, 16 bit Gray scale images were acquired from each biopsy of the microarray, and intensity values (I biopsy) were calculated according to:
Ibiospsy = ((%Pixelbiopsy > BGlocal + 1 SD)/100%) × (I mean,biospsy − BGlocal)
where BG = background intensity, SD = standard deviation, and I mean,biospsy = intensity of a single biopsy on the microarray. If either c‐IAP2 or HMGB1 were expressed at higher levels in a tumour compared with the highest expression in any of the normal tissues, this was considered as increased expression level.
Western blot analysis
Western blot analysis was performed according to standard protocols. Briefly, tumour and normal tissue samples were frozen in a 15 ml falcon tube and homogenised with a polytron in 2 ml of homogenisation buffer (50 mM Tris HCl, pH 7.4, 1 mM EGTA, 5 mM MgCl2 plus protease inhibitor cocktail). Also added were 400 μl 5×Laemmli's sample buffer (62.5 mM Tris base, 10% (w/v) glycerol, 2.3% sodium dodecyl sulphate, 5% 2‐mercaptoethanol). Protein lysates were transferred into TL100 tubes and centrifuged at 85 000 rpm for 20 minutes in a Beckman TL‐100 ultracentrifuge. The upper phase was boiled for five minutes and used for western blot analysis. Protein lysate (10 μg) was separated in a sodium dodecyl sulphate‐13% polyacrylamide gel. Proteins were transferred to PVDF membranes (Millipore, Schwalbach, Germany), and the blot was blocked and probed with a rabbit anti‐HMGB1 antibody (BD PharMingen Biosciences) and with anti‐β‐actin (C11; Santa Cruz, Heidelberg, Germany) in 5% non‐fat milk in phosphate buffered saline‐Tween 20 (0.1%). Immmunoreactive HMGB1 and β‐actin were visualised with horseradish peroxidase labelled secondary antibodies (GE Health Care, Freiburg, Germany) and ECL blotting substrate (GE Health Care).
Immunohistochemistry
Immunohistochemical analysis of colon carcinoma tissues was performed using standard protocols. After deparaffinising, 3 μm sections from formalin fixed paraffin embedded tissue blocks were boiled in 10 mM citrate buffer for antigen retrieval. A polyclonal rabbit anti‐HMGB1 antiserum (1:200; BD PharMingen Biosciences) or a rabbit anti‐c‐IAP2 antiserum (1:20; R&D Systems) were used as primary antibodies.
Apoptosis assays
Normal rat kidney cells (NRK1) were transfected with pBabe Hmgb1 using Fugene (Roche, Mannheim, Germany) according to the manufacturer's protocol, and apoptosis was induced by cotransfection with bak‐gfp. Cell death was quantified using the LIVE/DEAD viability/cytotoxicity kit from Molecular Probes (Leiden, the Netherlands) according to the supplier's protocol. For each experiment, at least 200 green and red cells were counted using a Nikon (Düsseldorf, Germany) fluorescence microscope (TE300).
RKO cells (3×105 per well) were transfected in six well plates with Hmgb1 or IκBαM alone or in combination using Lipofectamine (Invitrogen, Karlsruhe, Germany) according to the supplier's protocol. Cells were treated with 4 μg/ml mitomycin C (Roche) for 24 hours and fixed in 70% ice cold ethanol. Apoptosis was quantified using propidium iodide and FACS analysis (FACS Calilbur; Becton Dickinson, Karlsruhe, Germany) following standard protocols.
Quantification of caspase activities
Caspase activity was quantified using the Caspase Fluorometric Protease Assay Kit (BioVision, Heidelberg, Germany), according to the supplier's protocol. The assay is based on cleavage of the fluorogenic peptides Ac‐LEHD‐AFC (Casp‐9) and Ac‐DEVD‐AFC (Casp‐3), which are added to cell lysates prepared from apoptotic cells and non‐apoptotic controls.
Quantification of mRNA by real time PCR
c‐IAP2 and β‐actin mRNA levels were measured with an iCycler (BioRad, Munich, Germany) using quantitative polymerase chain reaction (PCR) analysis according to standard protocols. RKO cells (1×106) were transfected in 10 cm plates using polyethylenimine with 5 μg of total DNA of either empty vector, Hmgb1, IκBαM, or with Hmgb1 together with IκBαM. Forty eight hours later, cells were harvested, and RNA was isolated using RNeasy spin columns from Qiagen. For quantitative reverse transcription‐PCR, 1 μg of total RNA was converted to cDNA using iCript reverse transcriptase (BioRad) according to the manufacturer's specifications. PCR reaction mixtures consisted of 12.5 μl of 2× iQ SYBER Green Supermix (BioRad), 0.25 μl of each target primer (10 μM), and 5 μl diluted cDNA template (1:50) in a reaction volume of 25 μl. Thermal cycling conditions on the iCycler (BioRad) comprised an initial denaturation step of three min at 95°C, 40 cycles of 30 seconds at 95°C, and 30 seconds variable annealing/elongation temperature, depending on the respective set of target primers. dsDNA specific fluorescence was measured at the end of each extension phase. c‐IAP2 mRNA expression was normalised to the non‐regulated housekeeping gene β‐actin.
Oligonucleotide primers for c‐IAP2 and for β‐actin were as follows:
c‐IAP2‐forward: 5′‐TCC GTC AAG TTC AAG CCA GTT‐3′
c‐IAP2‐reverse: 5′‐TCT CCT GGG CTG TCT GAT GTG‐3′
β‐actin‐forward: 5′‐GCG GGA AAT CGT GCG TGA CAT T‐3′
β‐actin‐reverse: 5′‐GAT GGA GTT GAA GGT AGT TTC GTG‐3′.
Statistical analysis
For statistical analyses of FISH and protein expression results of the colon carcinoma tissue microarray, unpaired Student's t tests were applied.
Results
HMGB1 protein levels are increased in human colon carcinomas
When we hybridised a human cancer cDNA profiling array (Clontech) with a 32[P]‐labelled Hmgb1 cDNA probe, we observed strong upregulation of HMGB1 mRNA in carcinomas derived from the colon, uterus, and stomach in comparison with corresponding normal tissue from the same patients (see supplementary fig 1; supplementary fig 1 can be viewed on the Gut website at http://www.gutjnl.com/supplemental). In particular, 14 of the 35 colon carcinomas (40%) showed enhanced HMGB‐1 mRNA levels while 18 of 40 carcinomas of the uterus and nine of 20 stomach tumours expressed increased amounts of HMGB1 message.
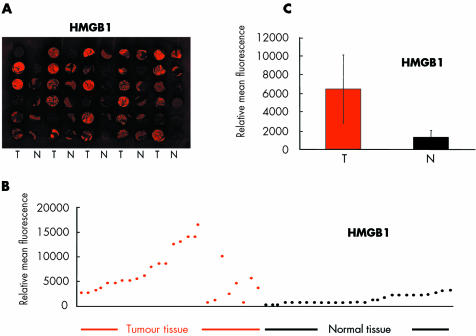
Figure 1 Enhanced protein levels of high mobility group box 1 protein (HMGB1) in colon carcinomas compared with normal tissue from the same patients, as revealed by immunohistochemical analysis of a colon tissue microarray. (A) HMGB1 expression in colon carcinomas (T) and normal tissue (N) biopsies visualised by anti‐HMGB1 antibody and Cy5 detection. (B) Relative mean intensities of the anti‐HMGB1 antibody signals in different colon cancer biopsies, as well as in normal tissue. (C) Mean (SD) intensity of anti‐HMGB1 antibody signals in colon cancers (T) versus normal tissue samples (N) (p<0.001). HMGB1 protein expression was on average four times higher in colon carcinoma samples than in corresponding normal tissue. For this calculation, the average value for all tumour samples was compared with the mean value for all normal tissues. When comparing individual pairs, HMGB1 expression in the tumour was on average 32.9 times higher than in matching normal tissue (see supplementary table 1; supplementary table 1 can be viewed on the Gut website at http://www.gutjnl.com/supplemental).
To screen a larger panel of colon carcinomas for HMGB1 expression, we analysed a human colon tissue microarray comprising fixed tissue sections from 29 different colon carcinomas and normal colon samples from the same patients. The intensity of HMGB1 staining was quantified, and the results are shown in fig 1 and supplementary table 1 (supplementary table 1 can be viewed on the Gut website at http://www.gutjnl.com/supplemental). Using this immunohistochemical method, we detected significantly higher HMGB‐1 fluorescence intensity in the colon carcinoma biopsies than in corresponding normal colon tissue (fig 1C; p<0.001). In all individual pairs of tumour and normal tissues, HMGB1 expression was elevated in tumour tissue (supplementary table 1; supplementary table 1 can be viewed on the Gut website at http://www.gutjnl.com/supplemental) and the average value of HMGB1 expression in the tumours was about four times higher than the mean HMGB1 expression in matching normal tissues (fig 1C). When comparing individual pairs, HMGB1 expression in the tumour was on average 32.9 times higher than in matching normal tissue.
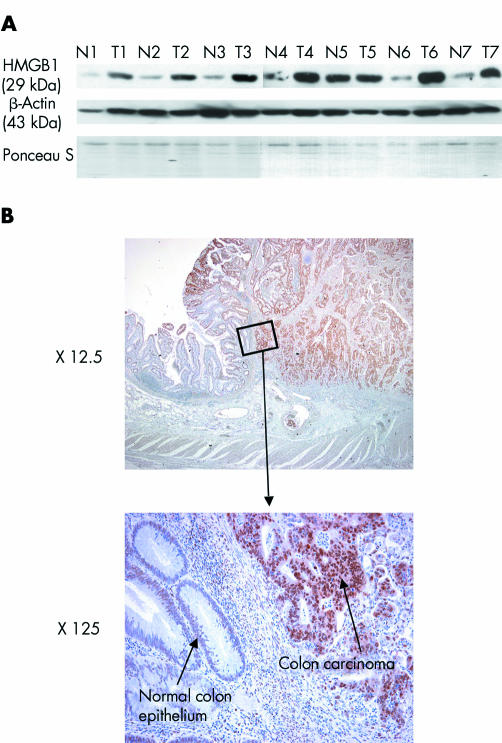
Our results obtained with the colon tissue microarray are consistent with western blot analysis on seven pairs of whole cell lysates of human colon carcinomas and normal colon tissue isolated from the same patients (fig 2A). In six out of seven pairs (86%), HMGB1 protein levels were increased in tumour samples compared with non‐cancerous colon mucosa.
Figure 2 Western blot and immunohistochemical analysis of paraffin embedded tumour tissue demonstrate increased protein expression of high mobility group box 1 protein (HMGB1) in colon carcinomas. (A) Western blot analysis illustrates elevated HMGB1 protein levels in six of seven different human colon carcinomas compared with normal colon mucosa (prepared from the same patients). In tumour No 5, no upregulated HMGB1 levels were detected (T5 v N5). Tumour samples displayed 70–80% tumour cell content, as estimated by histological analysis. Staining of the membrane with Ponceau S and incubation with an anti‐β‐actin antibody are shown as protein loading controls. (B) Immunohistochemical staining of HMGB1 in one representative colorectal carcinoma. Note the strong nuclear expression of HMGB1 in tumour cells and weaker expression in adjacent normal colon epithelial cells (magnification ×125, specific HMGB1 staining in brown, nuclear counterstaining in blue). Twenty of 24 colon carcinomas displayed overexpressed HMGB1 levels in tumour cells when analysed by immunohistochemistry. Tumour samples investigated by colon tissue microarray (29 tumours, see fig 1), western blot (seven tumours), and immunohistochemistry (24 tumours) were all from different patients.
As an alternative method of detecting HMGB1 protein expression in tumour tissue, we performed immunohistochemical staining of sections derived from 24 different colon carcinomas. Figure 2B shows one representative example. Of the 24 carcinomas analysed by microscope, 20 (83%) showed profound HMGB1 protein upregulation in tumour cells compared with normal colon epithelium of the same patient. Tumour samples used for colon tissue microarray (29 tumours), western blot (seven tumours) and immunohistochemical analysis (24 tumours) were all derived from different patients. These studies confirm the correlation between elevated HMGB1 protein expression and neoplastic transformation in human colon carcinomas.
The chromosomal subregion containing the HMGB1 locus is overrepresented in human colon carcinomas
CGH is a technique used in cytogenetic diagnosis to analyse genomic imbalances (amplifications and deletions) in tumour DNA. We have implemented a CGH database which comprises 1645 cases from different human tumour types.34 This CGH database allows identification of cytogenetic changes at a particular chromosomal subregion with a resolution of approximately 10 Mb. When we analysed the genomic subregion corresponding to the HMGB1 locus (13q12), we detected an increase in copy numbers in 35 of 96 colon carcinomas contained in our database (36.5%; data not shown). Correspondingly, when we performed a FISH analysis of the same 29 colon carcinomas on the human colon tissue microarray which had been used for immunohistochemical analysis of HMGB1 protein expression shown in fig 1, copy number gains (see material and methods for definition) of the HMGB‐1 locus were found in 13 of 27 tumours which revealed evaluable FISH signals. The average number of FISH signals per cell was different in tumour versus the corresponding normal tissue (tumour: 2.2 (0.6); normal: 1.8 (0.1); p = 0.003). However, there was no correlation detected between the gene copy number of the individual tumours and expression levels of HMGB1. Possible reasons for this will be discussed below.
HMGB1 overexpression inhibits caspase activation
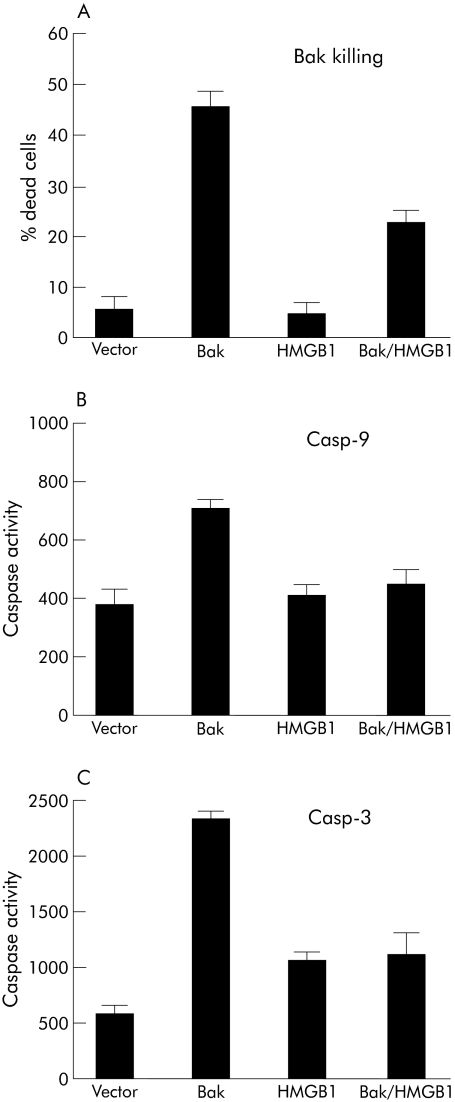
We have shown previously that HMGB1 overexpression inhibits Bak induced cell death in the human colon carcinoma cell line RKO.13 Likewise, Bak mediated killing of the NRK1 rat kidney cell line is inhibited by cotransfection of full length Hmgb1 (fig 3A). Proapoptotic Bak expression leads to release of mitochondrial Cyt c into the cytoplasm and to activation of the intrinsic apoptosis pathway.36 On apoptosome formation, Casp‐9 becomes activated which in turn leads to proteolytic activation of effector caspases such as Casp‐3.37 Using fluorescent substrate peptides, we measured caspase activities in cell lysates prepared from NRK1 cells which were transfected with bak alone or cotransfected with bak and Hmgb1. As shown in fig 3B and 3C, HMGB1 coexpression inhibited Casp‐9 and Casp‐3 activity in bak transfected cells, and this caspase inhibition correlated with suppression of Bak induced apoptosis by HMGB1. We observed a similar reduction in caspase activity by HMGB1 following UV radiation (data not shown). As Bak overexpression induces Cyt c release from mitochondria and as Bak induced apoptosis is inhibited by HMGB1, interference of HMGB1 with the apoptotic programme must occur between Cyt c release and Casp‐9 activation after apoptosome formation.
Figure 3 High mobility group box 1 protein (HMGB1) expression inhibits Bak induced apoptosis and caspase (Casp)‐9 and ‐3 activation. NRK1 cells were seeded in six well plates at 3×105 cells/well and transfected with either 0.5 μg bak‐gfp cDNA or with 2.0 μg of pBabe‐Hmgb1 alone or in combination. Twenty four hours later, cells were harvested, and the amount of dead cells was measured using the Live/Dead assay from Molecular Probes (A). In parallel, the activities of Casp‐9 (B) and Casp‐3 (C) were determined in a fluorometric cleavage assay. Mean (SD) values of three different experiments are shown.
HMGB1 increases NFκB activity and leads to c‐IAP2 upregulation in RKO cells
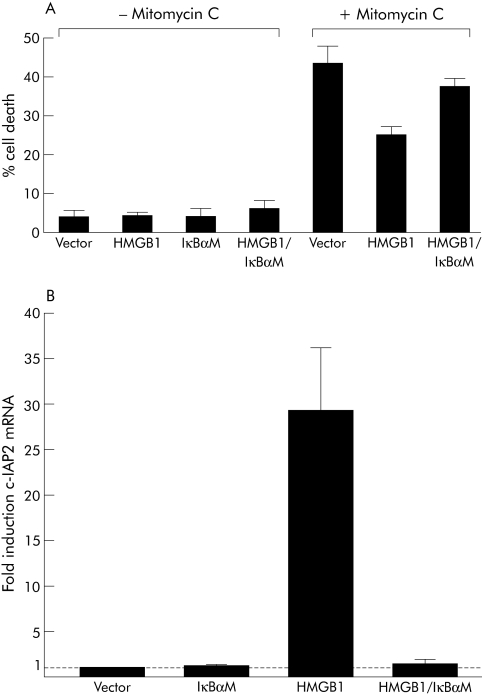
Results from the caspase activity assays suggest that HMGB1 interferes with the apoptotic machinery between Bak induced Cyt c release and apoptosomal activation of Casp‐9. This antiapoptotic effect could be mediated via inhibitor of apoptosis proteins (IAPs) which directly bind and inhibit caspases.33 IAPs belong to the target genes of the transcription factor NFκB,31,32,38 and overexpression of HMGB1 enhances NFκB activity in mouse embryonic fibroblasts28 and RKO cells (see supplementary fig 2; supplementary fig 2 can be viewed on the Gut website at http://www.gutjnl.com/supplemental). We therefore used a dominant negative non‐phosphorylated IκBα construct (IκBαM) in transfected RKO cells to test whether interfering with NFκB activity would diminish inhibition of apoptosis by HMGB1 overexpression. As shown in fig 4A, HMGB1 transfection indeed protected RKO cells from mitomycin C induced apoptosis, and this protection was abolished on cotransfection with IκBαM.
Figure 4 High mobility group box 1 protein (HMGB1) overexpression leads to nuclear factor κB (NFκB) dependent inhibition of mitomycin C induced apoptosis in RKO cells and to upregulation of c‐IAP2 mRNA. (A) RKO cells were transfected in six well plates with Hmgb1 or with the NFκB inhibitor IκBαM, or with both cDNAs in combination. Forty eight hours later, cells were treated for 24 hours with mitomycin C. Apoptotic cells with hypoploid DNA content were quantified by FACS analysis after ethanol fixation and PI staining. Values are mean (SD). (B) Overexpression of HMGB1 in RKO colon carcinoma cells caused NFκB dependent upregulation of c‐IAP2 mRNA. RKO cells were transfected with empty vector, the NFκB inhibitor IκBαM or Hmgb1 alone, or with Hmgb1 together with IκBαM. Forty eight hours later, c‐IAP2 mRNA levels were quantified by real time polymerase chain reaction (PCR). Three independent quantitative PCR experiments were performed. Values are mean (SD).
We then analysed whether activation of NFκB in RKO cells on HMGB1 overexpression would upregulate the antiapoptotic IAP family member c‐IAP2. Transfection of RKO cells with Hmgb1 increased c‐IAP2 mRNA severalfold, which was completely inhibited on cotransfection with the NFκB inhibitor IkBαM (fig 4B).
Upregulated HMGB1 protein levels correlate with increased amounts of c‐IAP2 in colon carcinomas
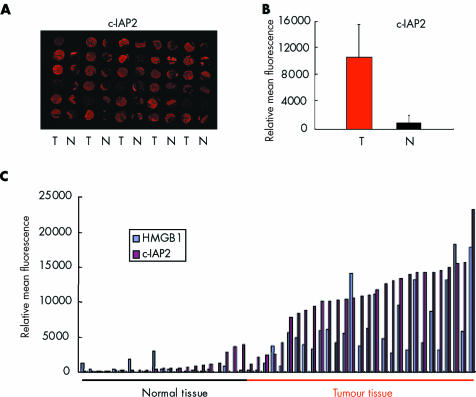
In the next experiment we tested the human colon tissue microarray with 29 different human colon carcinomas and normal colon samples from the same patients (which had been used for immunohistochemical HMGB1 (fig 1) and FISH analysis before) for simultaneous expression of HMGB1 and c‐IAP2 (fig 5). The array slides were incubated with appropriate antibodies, and upcoming fluorescence intensities were quantified. Results were obtained from all 29 colon tumours and 21 of 29 normal tissue biopsies. c‐IAP‐2 protein levels were upregulated in 25 of 29 tumours (86%). On average, eight times more c‐IAP2 protein was measured in tumour samples than in corresponding normal tissue (fig 5B). Twenty two of 29 tumours (76%) showed increased amounts of HMGB1 protein, and in 21 of these 22 carcinomas c‐IAP2 was simultaneously upregulated (fig 5C). Only one tumour displayed enhanced HMGB1 levels without elevated c‐IAP2 amounts, and only four tumours overexpressed c‐IAP2 without increased amounts of HMGB1 protein. Three of the 29 carcinomas showed no enhancement of either HMGB1 or c‐IAP2 protein levels. These data confirm the strong correlation between elevated HMGB1 and c‐IAP2 protein expression in human colon carcinomas.
Figure 5 c‐IAP2 protein levels are upregulated in colon carcinomas with increased amounts of high mobility group box 1 protein (HMGB1). (A) c‐IAP2 expression in sections of colon carcinomas (T) and corresponding normal tissue (N) from the same patient visualised by anti‐c‐IAP2 antibody and Cy5 detection. A slide from the same colon tissue microarray as shown in fig 1 was used. (B) Mean (SD) intensity of anti‐c‐IAP2 antibody signals in colon cancers versus normal tissue samples. c‐IAP2 protein expression was on average eight times higher in the colon carcinoma samples (T) than in corresponding normal tissue (N). (C) Relative mean intensities of the fluorescent anti‐HMGB1 and anti‐c‐IAP2 antibody signals from individual colon tumour and normal tissue biopsies represented on the tissue microarray shown in (A) (and in fig 1A). If either one of the proteins was expressed at a higher level in a tumour compared with the highest expression in any of the normal tissues, this was considered as increased expression level. When applying this stringent criterion, 22 of 29 carcinomas displayed elevated HMGB1 expression, and 21 of these 22 tumours simultaneously showed increased c‐IAP2 expression. Four tumours demonstrated upregulated c‐IAP2 levels without elevated HMGB1 quantities, while three tumours displayed normal HMGB1 and c‐IAP2 protein amounts. Only one carcinoma was found with upregulated HMGB1 and normal c‐IAP2 levels.
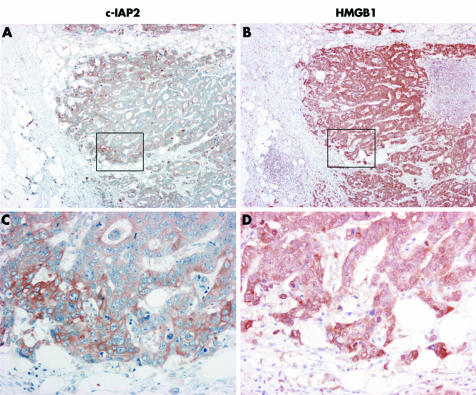
Figure 6 shows an example for immunohistochemical staining of a colon carcinoma overexpressing both HMGB1 and c‐IAP2. Interestingly, cytoplasmic and nuclear HMGB1 expression was even more pronounced in invading tumour cells at the tumour‐host interface, and in the same cells c‐IAP2 levels were also further upregulated (fig 6C).
Figure 6 Immunohistochemical staining of c‐IAP2 and high mobility group box 1 protein (HMGB1) demonstrates co‐upregulation of both proteins in colon carcinoma. Specific staining in brown and nuclear counterstaining in blue is shown in serial colon tissue sections. (C, D) Larger magnifications (×200) of the boxed regions in (A) and (B) (×40). Tumour cells express elevated protein levels of HMGB1 (B, D) and c‐IAP2 (A, C) in comparison with normal tissue. A further increase in nuclear and cytoplasmic HMGB1 expression was found in invading tumour cells at the tumour host interface, which was associated with co‐overexpression of c‐IAP‐2.
Discussion
In the present study, we showed strong upregulation of HMGB1 in primary human colon carcinomas. Using a cancer profiling array, elevated HMGB1 mRNA levels were detected in 40% of all colon carcinomas. Furthermore, increased HMGB1 protein levels were observed in 46 of 51 (90%) colon carcinoma samples analysed by tissue microarrrays, immunohistochemical staining of tissue sections, or western blot analysis. Search of a CGH database revealed an increase in HMGB1 copy number in 37% of colon cancers represented in the database. Correspondingly, we found a higher average number of HMGB‐1 signals in tumour samples compared with normal tissue by FISH analysis of the tissue microarray. No correlation between the gene copy number of the individual tumours and their expression level of HMGB1 was observed. However, it should be emphasised that tissue microarrays are suitable screening tools revealing information at the level of the whole tumour collections rather than individual tumour cases. This is mainly due to the fact that only small biopsies (not representative of the entire tumour specimen) are used for tissue microarrays. FISH and immunohistochemical analyses performed on different slices of tissue microarrays might therefore give different results with regard to individual tumours biopsies. Furthermore, copy number gains are most likely not the only molecular mechanism for increased HMGB1 expression.
Several publications have reported upregulated HMGB1 protein in different tumour types.5,13,14,15,16,39 In accordance with our data, an earlier report demonstrated higher HMGB1 levels in colorectal adenocarcinomas compared with corresponding non‐cancerous mucosa, albeit only at the mRNA level.40 Elevated HMGB1 expression (and of its receptor RAGE) has also been associated with cancer invasion and metastasis.9,41,42,43
HMGB1 may support tumour growth and metastasis by its ability as an extracellular ligand to induce proliferation,9,18,44 and/or by its proinflammatory properties as a cytokine.10,19 Alternatively (or in addition), HMGB1 could act as an oncoprotein because of its ability to suppress apoptosis.13 We were able to demonstrate that overexpression of HMGB1 inhibits Bak (and UV) induced cell death by preventing Casp‐9 (and consequently Casp‐3) activation. In vivo, caspase activity is regulated by IAP proteins, and our analysis of HMGB1 overexpressing colon cancers revealed co‐upregulation of c‐IAP2, which is known to directly bind to and inhibit Casp‐3 and Casp‐7 and to prevent activation of pro‐Casp‐9. IAPs are regulated by endogenous proteins (second mitochondrial activator of caspases (SMAC) and Omi) that are released from the mitochondria during apoptosis and which displace the IAPs from caspase molecules.33 Expression studies have manifested elevated IAP levels in cancer cell lines and primary tumour biopsy samples (for references see Liston and colleagues33). In addition, direct genetic evidence suggests an oncogenic role for the antiapoptotic IAPs.45,46,47,48,49 Our data show for the first time elevated expression of c‐IAP2 protein in human colon carcinomas, and correlation of this upregulation with increased HMGB1 levels.
HMGB1, in its nuclear function, has for a long time been known to enhance the activity of transcriptional activators and repressors by binding to transcription factors, and by simultaneously interacting with and distorting chromosomal DNA (McKinney and Prives27 and references herein). The phenotype of Hmgb1−/− knockout mice is in agreement with the role of HMGB1 as a regulator of transcription.50 One of the transcription factors whose activity is influenced by HMGB1 is NFκB.28 NFκB is a ubiquitous transcription factor which as a dimer is composed of two members of the Rel family (for review see Kucharczak and colleagues30). The mammalian Rel family consists of the c‐Rel, RelA, RelB, p105/ NFκB1, and p100/ NFκB2 proteins. On induction, the NFκB homo‐ or heterodimer translocates into the nucleus, binds to κB DNA binding motifs and activates certain target genes. Different members of the Rel family display specific DNA binding preferences.
NFκB can promote programmed cell death in response to certain death inducing signals and in certain cell types whereas it is most commonly involved in suppressing apoptosis by transactivating the expression of antiapoptotic genes.30 One of these antiapoptotic NFκB target genes is c‐IAP2 which suppresses tumour necrosis factor induced cell death.31,32,38 In vitro, HMGB1 increases c‐IAP2 expression levels by enhancing NFκB activity. The data shown in the present study indicate that this is also true in vivo, as we observed constitutively high HMGB1 expression correlated with c‐IAP2 upregulation in colon carcinoma cells. Such a molecular mechanism would explain the antiapoptotic activity of nuclear HMGB1 and could well contribute to the development of colon cancer.
Supplementary figures 1 and 2 and supplementary table 1 can be viewed on the Gut website at http://www.gutjnl.com/supplemental.
Supplementary Material
Acknowledgements
The authors thank Sabina Solinas‐Toldo (Phase‐it AG) for help during CGH database analysis, Frauke Devens, Silke Deckert, and Annemarie Schimpf for excellent technical assistance, and Dr Trevor Littlewood and Dr Winfried Wels for critically reading the manuscript. This work is supported in part by the DFG (ZO 110/1‐1), the German National Genome Research Network (N1KR‐S12T23, 01 GR 0417 and KR‐S05T02), the Deutsche Krebshilfe (10‐1745‐Ho), and the Heinrich and Erna Schaufler‐Stiftung.
Abbreviations
HMGB1 - high mobility group box 1 protein
NFκB - nuclear factor κB
CGH - comparative genomic hybridisation
IAP - inhibitor of apoptosis protein
RAGE - receptor for advanced glycation end products
FISH - fluorescence in situ hybridisation
Casp - caspase
PCR - polymerase chain reaction
Cyt c - cytochrome c
Footnotes
Conflict of interest: None declared.
Supplementary figures 1 and 2 and supplementary table 1 can be viewed on the Gut website at http://www.gutjnl.com/supplemental.
References
- 1.Thomas J O, Travers A A. HMG1 and 2, and related ‘architectural' DNA‐binding proteins. Trends Biochem Sci 200126167–174. [DOI] [PubMed] [Google Scholar]
- 2.Bianchi M E, Beltrame M. Upwardly mobile proteins. Workshop: the role of HMG proteins in chromatin structure, gene expression and neoplasia, EMBO Rep 20001109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson U, Erlandsson‐Harris H, Yang H.et al HMGB1 as a DNA‐binding cytokine. J Leukoc Biol 2002721084–1091. [PubMed] [Google Scholar]
- 4.Pasheva E, Sarov M, Bidjekov K.et al In vitro acetylation of HMGB‐1 and ‐2 proteins by CBP: the role of the acidic tail. Biochemistry 2004432935–2940. [DOI] [PubMed] [Google Scholar]
- 5.Muller S, Ronfani L, Bianchi M E. Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J Intern Med 2004255332–343. [DOI] [PubMed] [Google Scholar]
- 6.Kwon J, Imbalzano A N, Matthews A.et al Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol Cell 19982829–839. [DOI] [PubMed] [Google Scholar]
- 7.Mitsouras K, Wong B, Arayata C.et al The DNA architectural protein HMGB1 displays two distinct modes of action that promote enhanceosome assembly. Mol Cell Biol 2002224390–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller S, Scaffidi P, Degryse B.et al New EMBO members' review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. Embo J 2001204337–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taguchi A, Blood D C, del Toro G.et al Blockade of RAGE‐amphoterin signalling suppresses tumour growth and metastases. Nature 2000405354–360. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Bloom O, Zhang M.et al HMG‐1 as a late mediator of endotoxin lethality in mice. Science 1999285248–251. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt A M, Vianna M, Gerlach M.et al Isolation and characterization of two binding proteins for advanced glycosylation end products from bovine lung which are present on the endothelial cell surface. J Biol Chem 199226714987–14997. [PubMed] [Google Scholar]
- 12.Scaffidi P, Misteli T, Bianchi M E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002418191–195. [DOI] [PubMed] [Google Scholar]
- 13.Brezniceanu M L, Volp K, Bosser S.et al HMGB1 inhibits cell death in yeast and mammalian cells and is abundantly expressed in human breast carcinoma. FASEB J 2003171295–1297. [DOI] [PubMed] [Google Scholar]
- 14.Poser I, Golob M, Buettner R.et al Upregulation of HMG1 leads to melanoma inhibitory activity expression in malignant melanoma cells and contributes to their malignancy phenotype. Mol Cell Biol 2003232991–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi Y R, Kim H, Kang H J.et al Overexpression of high mobility group box 1 in gastrointestinal stromal tumors with KIT mutation. Cancer Res 2003632188–2193. [PubMed] [Google Scholar]
- 16.Court E L, Ann Smith M, Avent N D.et al DNA microarray screening of differential gene expression in bone marrow samples from AML, non‐AML patients and AML cell lines. Leuk Res 200428743–753. [DOI] [PubMed] [Google Scholar]
- 17.Fedele M, Battista S, Kenyon L.et al Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene 2002213190–3198. [DOI] [PubMed] [Google Scholar]
- 18.Huttunen H J, Fages C, Rauvala H. Receptor for advanced glycation end products (RAGE)‐mediated neurite outgrowth and activation of NF‐kappaB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J Biol Chem 199927419919–19924. [DOI] [PubMed] [Google Scholar]
- 19.Mignogna M D, Fedele S, Lo Russo L.et al Immune activation and chronic inflammation as the cause of malignancy in oral lichen planus: is there any evidence? Oral Oncol 200440120–130. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg R A. The hallmarks of cancer. Cell 200010057–70. [DOI] [PubMed] [Google Scholar]
- 21.Hueber A O, Evan G I. Traps to catch unwary oncogenes. Trends Genet 199814364–367. [DOI] [PubMed] [Google Scholar]
- 22.Kunz M, Ibrahim S M. Molecular responses to hypoxia in tumor cells. Mol Cancer 2003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng Q, McCauley L K, Wang C Y. Hepatocyte growth factor inhibits anoikis by induction of activator protein 1‐dependent cyclooxygenase‐2. Implication in head and neck squamous cell carcinoma progression. J Biol Chem 200227750137–50142. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt C A, Fridman J S, Yang M.et al Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 20021289–298. [DOI] [PubMed] [Google Scholar]
- 25.Kirkin V, Joos S, Zornig M. The role of Bcl‐2 family members in tumorigenesis. Biochim Biophys Acta 20041644229–249. [DOI] [PubMed] [Google Scholar]
- 26.Boonyaratanakornkit V, Melvin V, Prendergast P.et al High‐mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol 1998184471–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKinney K, Prives C. Efficient specific DNA binding by p53 requires both its central and C‐terminal domains as revealed by studies with high‐mobility group 1 protein. Mol Cell Biol 2002226797–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agresti A, Lupo R, Bianchi M E.et al HMGB1 interacts differentially with members of the Rel family of transcription factors. Biochem Biophys Res Commun 2003302421–426. [DOI] [PubMed] [Google Scholar]
- 29.Brickman J M, Adam M, Ptashne M. Interactions between an HMG‐1 protein and members of the Rel family. Proc Natl Acad Sci U S A 19999610679–10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kucharczak J, Simmons M J, Fan Y.et al To be, or not to be: NF‐kappaB is the answer—role of Rel/NF‐kappaB in the regulation of apoptosis. Oncogene 2003228961–8982. [DOI] [PubMed] [Google Scholar]
- 31.Wang C Y, Mayo M W, Korneluk R G.et al NF‐kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c‐IAP1 and c‐IAP2 to suppress caspase‐8 activation. Science 19982811680–1683. [DOI] [PubMed] [Google Scholar]
- 32.Chu Z L, McKinsey T A, Liu L.et al Suppression of tumor necrosis factor‐induced cell death by inhibitor of apoptosis c‐IAP2 is under NF‐kappaB control. Proc Natl Acad Sci U S A 19979410057–10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liston P, Fong W G, Korneluk R G. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene 2003228568–8580. [DOI] [PubMed] [Google Scholar]
- 34.Berrar D, Dubitzky W, Solinas‐Toldo S.et al A database system for comparative genomic hybridization analysis. IEEE Eng Med Biol Mag 20012075–83. [DOI] [PubMed] [Google Scholar]
- 35.Sticht C, Hofele C, Flechtenmacher C.et al Amplification of Cyclin L1 is associated with lymph node metastases in head and neck squamous cell carcinoma (HNSCC). Br J Cancer 200592770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scorrano L, Korsmeyer S J. Mechanisms of cytochrome c release by proapoptotic BCL‐2 family members. Biochem Biophys Res Commun 2003304437–444. [DOI] [PubMed] [Google Scholar]
- 37.Zou H, Yang R, Hao J.et al Regulation of the Apaf‐1/caspase‐9 apoptosome by caspase‐3 and XIAP. J Biol Chem 20032788091–8098. [DOI] [PubMed] [Google Scholar]
- 38.Deveraux Q L, Roy N, Stennicke H R.et al IAPs block apoptotic events induced by caspase‐8 and cytochrome c by direct inhibition of distinct caspases. EMBO J 1998172215–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lotze M T, DeMarco R A. Dealing with death: HMGB1 as a novel target for cancer therapy. Curr Opin Investig Drugs 200341405–1409. [PubMed] [Google Scholar]
- 40.Xiang Y Y, Wang D Y, Tanaka M.et al Expression of high‐mobility group‐1 mRNA in human gastrointestinal adenocarcinoma and corresponding non‐cancerous mucosa. Int J Cancer 1997741–6. [DOI] [PubMed] [Google Scholar]
- 41.Kuniyasu H, Chihara Y, Takahashi T. Co‐expression of receptor for advanced glycation end products and the ligand amphoterin associates closely with metastasis of colorectal cancer. Oncol Rep 200310445–448. [PubMed] [Google Scholar]
- 42.Nestl A, Von Stein O D, Zatloukal K.et al Gene expression patterns associated with the metastatic phenotype in rodent and human tumors. Cancer Res 2001611569–1577. [PubMed] [Google Scholar]
- 43.Kuniyasu H, Chihara Y, Kondo H.et al Amphoterin induction in prostatic stromal cells by androgen deprivation is associated with metastatic prostate cancer. Oncol Rep 2003101863–1868. [PubMed] [Google Scholar]
- 44.Palumbo R, Sampaolesi M, De Marchis F.et al Extracellular HMGB1, a signal of tissue damage, induces mesoangioblast migration and proliferation. J Cell Biol 2004164441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H, Ruskon‐Fourmestraux A, Lavergne‐Slove A.et al Resistance of t(11;18) positive gastric mucosa‐associated lymphoid tissue lymphoma to Helicobacter pylori eradication therapy. Lancet 200135739–40. [DOI] [PubMed] [Google Scholar]
- 46.Uren A G, O'Rourke K, Aravind L A.et al Identification of paracaspases and metacaspases: two ancient families of caspase‐like proteins, one of which plays a key role in MALT lymphoma. Mol Cell 20006961–967. [DOI] [PubMed] [Google Scholar]
- 47.Baens M, Maes B, Steyls A.et al The product of the t(11;18), an API2‐MLT fusion, marks nearly half of gastric MALT type lymphomas without large cell proliferation. Am J Pathol 20001561433–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai Z, Zhu W G, Morrison C D.et al A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum Mol Genet 200312791–801. [DOI] [PubMed] [Google Scholar]
- 49.Imoto I, Yang Z Q, Pimkhaokham A.et al Identification of cIAP1 as a candidate target gene within an amplicon at 11q22 in esophageal squamous cell carcinomas. Cancer Res 2001616629–6634. [PubMed] [Google Scholar]
- 50.Calogero S, Grassi F, Aguzzi A.et al The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet 199922276–280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.