Ulcerative colitis is an inflammatory bowel disease (IBD) characterised by a chronic inflammation of the colorectal mucosa. The role of the gut microbiota in triggering or maintenance of the mucosal inflammation is strongly suspected.1 We may postulate that a localised dysbiosis could explain the localisation of the lesions. Many of the recent studies have been made possible by the development of culture‐independent techniques, which identify bacteria on the basis of comparisons of the nucleic acid sequence of rRNA molecules. Temporal temperature‐gradient gel electrophoresis (TTGE) showed that in healthy people dominant microbiota associated with colonic mucosa is similar from the caecum to the rectum.2 This has also been observed in Crohn's disease,2 and the dominant microbiota associated with ulcerated mucosa did not differ markedly from the microbiota associated with non‐ulcerated mucosa.3
The aim of this study was to compare the dominant microbiota associated with injured colonic mucosa versus healthy colonic mucosa, using TTGE in patients with active ulcerative colitis.
Ten patients with active ulcerative colitis were studied, after informed consent was obtained. Three patients had a proctitis and seven had a left‐sided colitis. None of them had received antibiotics or sulfasalazine within the previous 3 months. Four patients were receiving corticosteroids, four mesalazine and three azathioprine. Colonoscopy was performed during ulcerative colitis flare‐ups. Colonic or rectal biopsy samples were collected from both injured and healthy mucosa in each patient and were frozen immediately in liquid nitrogen, and stored at −80°C until analysis.
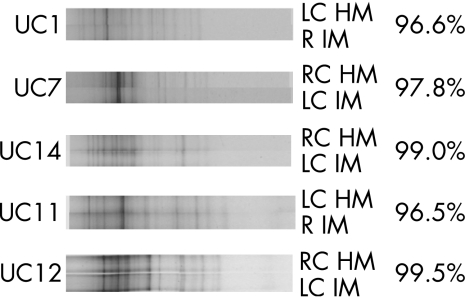
Global analysis of TTGE profiles showed a high biodiversity in both injured colonic mucosa and healthy colonic mucosa. Mean (standard deviation (SD)) numbers of bands were 11.5 (2.7) and 11.6 (2.6) in injured colonic mucosa and healthy colonic mucosa microbiota, respectively (no marked differences). Figure 1 shows the TTGE profiles of representative patients.
Figure 1 Temporal temperature‐gradient gel electrophoresis of 16S rDNA amplicons (obtained using primers for the V6–V8 regions) extracted from biopsy samples of healthy and injured mucosa from the colon or the rectum of five patients with ulcerative colitis (UC). HM, healthy mucosa; IM, injured mucosa; LC, left colon; R, rectum; RC, right colon; Right side, similarity indexes (expressed in %) paired samples.
Intraindividual analysis showed that in a given patient, the mean (SD) similarity index between injured colonic mucosa and healthy colonic mucosa TTGE profiles was 94.9 (4.6%). Similarity indexes of TTGE profiles of samples collected from a given segment in different patients ranged from 59.5 (15.4%) to 79.2 (8.2%) for injured colonic mucosa and from 62.4 (16.3%) to 71.2 (13.4%) for healthy colonic mucosa (table 1). The mucosa‐associated microbiota differed markedly between patients. However, these differences were comparable in injured and healthy mucosa and were within the range of interindividual variability. The interindividual similarity index of each healthy colonic mucosa or injured colonic mucosa segment was always significantly lower than the intraindividual similarity index between injured colonic mucosa and healthy colonic mucosa (table 1).
Table 1 Comparison of interindividual similarity index of each healthy colonic mucosa or injured colonic mucosa segment versus mean intraindividual similarity index between injured colonic mucosa and healthy colonic mucosa.
| Healthy mucosa | Injured mucosa | |||
|---|---|---|---|---|
| Right colon | Left colon | Left colon | Rectum | |
| Number of samples | 6 | 4 | 7 | 3 |
| Mean (SD) similarity index (%) | 62.4 (16.3) | 71.2 (13.4) | 59.5 (15.4) | 79.2 (8.2) |
| Versus mean intra‐individual similarity (p value) | 0.001 | 0.004 | 0.001 | 0.001 |
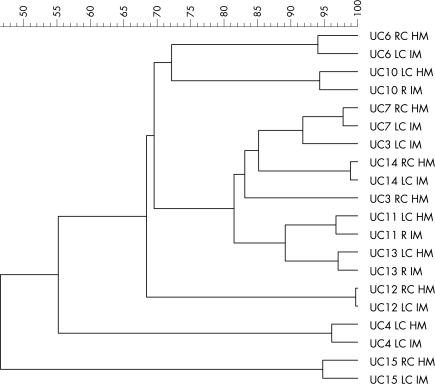
All TTGE profiles were compared in a single dendrogram. The branching distances between two samples shows their degree of relatedness. Except for patient 3, samples from the same patient clustered together (fig 2).
Figure 2 Dendrogram representing the temporal temperature‐gradient gel electrophoresis (TTGE) profiles of 16S rDNA amplicons (obtained using primers for the V6–V8 regions) extracted from biopsy samples of injured mucosa (IM) and healthy mucosa (HM) of 10 patients with ulcerative colitis (UC). The dendrogram represents a statistically optimal representation of the similarities between TTGE profiles on the basis of the matrix of Pearson's correlation coefficients and applying the unweighted pair group method using arithmetic averages. LC, left colon; R, rectum; RC, right colon.
The role of the intestinal microbiota in the pathogenesis of ulcerative colitis is suspected.1 This could involve a dysbiosis or an altered immune response to a normal microbial context.1 We showed, using TTGE, that the dominant mucosa‐associated microbiota in patients with ulcerative colitis does not differ between the injured and the healthy mucosa.
A previous study using the same method also showed no sign of localised dysbiosis in the dominant mucosal microbiota during Crohn's disease.3 However, these two studies cannot exclude quantitative differences in the microbiota or qualitative differences in the subdominant bacteria. Kleessen et al4 found more bacteria on the mucosal surface of patients with IBD than on those of controls without IBD. A decrease in the biodiversity of the active portion of the fecal microbiota has been reported in ulcerative colitis.5 Restricted biodiversity has also been observed in mucosa‐associated microbiota by Ott et al.6 However, in this study the dominant microbiota biodiversity, assessed by the number of bands on the TTGE, was similar in healthy colonic mucosa and in injured colonic mucosa. These results suggest that the dominant mucosa‐associated microbiota does not differ in ulcerated tissues during IBD; further studies searching for a hypothetical localised dysbiosis to explain the patchy distribution of the disease should thus focus on other targets, and probably to our view on usually subdominant bacteria such as Escherichia coli or sulphate reducing bacteria (which have been suspected to play a part in IBD in other studies).5,7
Footnotes
Competing interests: None.
References
- 1.Marteau P, Lepage P, Mangin I.et al Review article: gut flora and inflammatory bowel disease. Aliment Pharmacol Ther 20042018–23. [DOI] [PubMed] [Google Scholar]
- 2.Lepage P, Seksik P, Sutren M.et al Biodiversity of the mucosa‐associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm Bowel Dis 200511473–480. [DOI] [PubMed] [Google Scholar]
- 3.Seksik P, Lepage P, de la Cochetiere M F.et al Search for localized dysbiosis in Crohn's disease ulcerations by temporal temperature gradient gel electrophoresis of 16S rRNA. J Clin Microbiol 2005434654–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleessen B, Kroesen A J, Buhr H J.et al Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol 2002371034–1041. [DOI] [PubMed] [Google Scholar]
- 5.Sokol H, Lepage P, Seksik P.et al Temperature gradient gel electrophoresis of faecal 16S rRNA reveals active Escherichia coli in the microbiota of patients with ulcerative colitis. J Clin Microbiol 2006443172–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ott S J, Musfeldt M, Wenderoth D F.et al Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 200453685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pitcher M C, Beatty E R, Cummings J H. The contribution of sulphate reducing bacteria and 5‐aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 20004664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]