Abstract
Background
Oesophageal squamous cell carcinoma (OSCC) often arises from preceding dysplastic lesions in the oesophageal epithelium. However, the molecular changes occurring in premalignant lesions are not well understood. An epigenetic change is an example of OSCC that may occur within the epithelium.
Aim
To investigate the methylation status of multiple promoters in cancer‐derived DNA, as well as in the background epithelium of OSCC, including dysplastic lesions and non‐neoplastic mucosa. The normal epithelium from patients without cancer was also examined. The findings were correlated with the mutational status of p53.
Patients and methods
56 patients with advanced OSCC, 21 patients with intraepithelial neoplasia (IEN), 56 patients with a background of non‐neoplastic epithelium, adjacent to the OSCC, and 42 normal control epithelia from healthy volunteers were studied. The promoter methylation status of SFRP1, SFRP2, DCC, APC, p16INK4a, p14ARF, MINT1, MINT2, MINT31, CACNA1G, COX2, DAPK, hMLH1 and MGMT was examined by methylation‐specific single polymerase chain reaction or combined bisulphite restriction analysis. The mutation of p53 by direct sequencing was assessed.
Results
DNA methylation was observed in OSCC and in its background epithelium. The frequency of CpG island methylation increased from a baseline level in the background non‐neoplastic epithelium, through IEN, to advanced OSCC. However, mutations in p53 were almost exclusively observed in IEN and OSCC. More extensive DNA methylation was seen in the neoplastic lesions (OSCC or IEN) having a p53 mutation than in those with wild‐type p53.
Conclusion
DNA methylation is present at low levels in the non‐neoplastic oesophageal epithelium and appears to contribute to the progression of the dysplasia–carcinoma sequence in OSCC carcinogenesis.
Despite the increased incidence of oesophageal adenocarcinoma in the Western World, oesophageal squamous cell carcinoma (OSCC) remains a common type of malignancy worldwide. Tumorigenesis of OSCC is a multistep process and OSCC often develops multifocally within the oesophageal epithelium. Environmental and dietary factors, such as alcohol, tobacco, and high levels of nitrates in the soil and drinking water, have been associated with the aetiology of OSCC.1,2 Mutations in the p53 gene are one of the most frequent genetic changes observed in oesophageal cancer and dysplasia.3,4,5 A varying p53 mutational status is often observed in multiple lesions from the same patient with OSCC.6 Wide areas within the oesophageal mucosa become simultaneously genetically unstable after a prolonged exposure to carcinogens, leading to a pattern of neoplastic transformation described as “field carcinogenesis”.7,8,9
Epigenetic changes in DNA without concomitant changes in the underlying genetic code are now known to occur often in human cancers.10,11,12 Promoter hypermethylation and resulting transcriptional repression of functionally important cancer‐related tumour suppressor genes appear to drive tumorigenesis. The promoter CpG island, in normal tissues, is generally protected from aberrant hypermethylation, but this protection may be lost in the early phase of tumorigenesis. We therefore studied the extent of promoter methylation in normal epithelium from patients without cancer and in the oesophageal epithelium, ranging from the background non‐neoplastic epithelium to OSCC, and compared the results with the p53 mutational status.
Materials and methods
Tissue samples
Tumours and the corresponding background non‐neoplastic epithelia were obtained from 56 patients with OSCC who underwent curative surgery without prior chemotherapy or radiotherapy between 1996 and 2004 at the Okayama University Medical Hospital, Okayama, Japan. The stage of OSCC was classified according to the TNM classification. Among 56 patients with OSCC, 21 intraepithelial neoplastic (IEN) lesions were identified in the background epithelial specimens with OSCC and sampled for further analysis. IEN lesions were identified and collected using 1.5% Lugol solution sprayed over the resected oesophageal mucosa. The intraepithelial lesion was confirmed histopathologically. IEN was diagnosed when atypical cell proliferation was seen in the upper one third of the epithelium. In situ carcinoma was also included in IEN. Controls of normal oesophageal epithelium were obtained by biopsy using a video endoscope (model Q240, Olympus Optical, Tokyo, Japan) from 42 healthy age‐matched volunteers. The mean (standard deviation (SD)) age of the healthy volunteers was 68.0 (13.4) years, 17 (40%) being women. Informed consent was obtained in writing from all patients before recruitment or enrolment into the study. Tissue samples were collected and stored at −80°C.
Haematoxylin and eosin or periodic acid schiff staining
Serial sections were cut from each paraffin wax block, and the sections were counterstained with periodic acid schiff (PAS) with and without diastase digestion, independently. Haematoxylin and eosin (H&E) staining was carried out for histopathological diagnosis.
DNA extraction
From frozen samples, including OSCC, non‐neoplastic tissue from patients with OSCC and normal control epithelia from healthy volunteers, DNA was extracted by a standard procedure involving digestion with proteinase K and phenol–chloroform extraction. For the IEN, paraffin‐wax‐embedded blocks were deparaffinised and 5–10‐µm samples were microdissected for unstained serial slides. The dissected samples, which were approximately 50 mm2, were incubated in 40 µl of lysis buffer (20 mM TRIS‐HCl (pH 8.0), 1 mM EDTA, 0.5% Tween 20 and 200 µg/ml proteinase K) at 37°C for 24 h and then at 95°C for 15 min to inactivate the proteinase K.
Bisulphite modification and DNA methylation analysis
Extracted DNA was bisulphite modified using an EZ DNA methylation kit (Zymo Research, Orange, California, USA). The methylation status of MINT1, MINT2, MINT31, CACNA1G, p16INK4a, p14ARF, COX2, DCC, APC1A, SFRP1, SFRP2 and DAPK was evaluated and determined by combined bisulphite restriction analysis (COBRA) as described previously.12,13,14,15,16,17,18 The methylation status of hMLH1 and MGMT was determined by our modified methylation‐specific polymerase chain reaction (PCR). Their primer sequences have been described previously.19
Mutation analysis of p53
We assessed the p53 mutation from exons 5–9 by direct sequencing of the 56 main oesophageal cancers, 21 IEN, 56 background non‐neoplastic epithelia and 42 normal control epithelia from healthy volunteers. PCR was carried out in a 25 µl reaction volume containing 50 ng of genomic DNA, 20 pmol of each primer, 0.8 mmol/l dNTPs, 1× reaction buffer, 1.5 mmol/l MgCl2 and 0.7 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, California, USA). Box 1 describes the primer sequences.
Box 1 Primer sequences
-
Exons 5 and 6
-
-
Forward: 5′‐TTTGCTGCCGTGTTCCAGTTG‐3′
-
-
Reverse: 5′‐TGGGAGGAGGGGTTAAGGG‐3′
-
-
-
Exon 7
-
-
Forward: 5′‐CTTGGGCCTGTGTTATCTCCT‐3′
-
-
Reverse: 5′‐TCAGGAGCCACTTGCCACCCT‐3′
-
-
-
Exons 8 and 9
-
-
Forward: 5′‐CCTTACTGCCTCTTGCTTCTC‐3′
-
-
Reverse: 5′‐CTAAGTCTTGGGACCTCTTATCAA‐3′.
-
-
All PCR products were purified using a PCR products presequencing kit (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK), reacted with the Big Dye Terminator FS Ready‐Reaction kit (Applied Biosystems) and analysed using an ABI PRISM3100 sequencer (Applied Biosystems). Mutations found were confirmed by independent PCR amplification and by sequencing.
Statistical analysis
For binary data (methylated v non‐methylated), specific to patients, locus and tissue type were compared with those of controls (n = 42) and with background non‐neoplastic epithelium from patients (n = 56) using Mantel–Haenszel statistics. We compared the matched results for the 56 patients, for non‐neoplastic epithelium versus OSCC, using the Wilcoxon signed‐rank test. We also compared the non‐neoplastic epithelium, IEN and OSCC among the 21 patients with those in all of the three types of sample, using separate Wilcoxon's signed‐rank tests for two‐way comparisons and using a logistic regression. Differences in the frequencies of DNA methylation were evaluated by Pearson's χ2 test. The number of methylated genes per patient was analysed using the non‐parametric Kruskal–Wallis test and a similar non‐parametric test for a trend (procedure “nptrend”, Stata Corporation). Differences with age were tested using the Student's t test. All reported p values are two‐sided and a p value <0.05 was considered significant.
Results
Clinical and pathological features
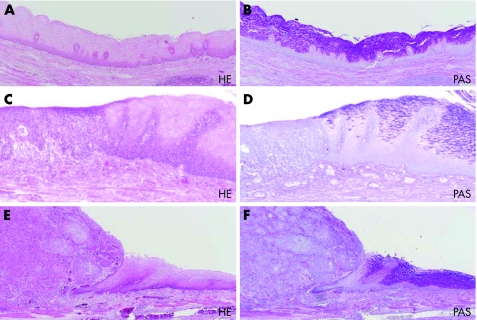
Table 1 shows the demographics of the patients with OSCC, with regard to the adjacent background non‐neoplastic epithelium, along with those for the patients with IEN and those with normal control oesophageal epithelium (healthy volunteers). The mean age of patients with OSCC was 65.8 years (median age 67 years) and the male:female ratio was 47:9. The mean age of the healthy volunteers was 67.9 years (median age 71 years) and the male:female ratio was 25:17. From a total of 56 oesophageal specimens, 21 with IEN were collected. Patients with OSCC with multiple neoplastic lesions and those with a single neoplastic lesion (main advanced OSCC) were not significantly different in age, sex, pathology of main cancer or stage (data not shown). Figure 1 shows the histopathological features of the advanced OSCC, IEN and background non‐neoplastic epithelium. IEN lesions were initially screened as Lugol‐negative lesions, and the collected lesions were histopathologically confirmed by H&E and PAS staining. All of the IEN lesions were glycogen negative, as shown by the negative PAS diastase staining.
Table 1 Pathological features of the samples examined.
| OSCC (n = 56) | Intraepithelial neoplasia (n = 21) | Background non‐neoplastic epithelium (n = 56) | Normal control epithelium (n = 42) | p Value | |
|---|---|---|---|---|---|
| Age | |||||
| Mean age | 65.8 | 64 | 65.8 | 67.9 | 0.6* |
| Median age | 67 | 67 | 67 | 71 | |
| Sex | |||||
| Male | 47 | 18 | 47 | 25 | 0.006† |
| Female | 9 | 3 | 9 | 17 | |
| Lesion | |||||
| Single | 36 | 7 | 36 | ||
| Multiple | 20 | 14 | 20 | ||
| Stage | |||||
| I | 4 | 1 | 4 | ||
| II | 16 | 7 | 16 | ||
| III | 23 | 6 | 23 | ||
| IV | 13 | 7 | 13 | ||
| Differentiation | |||||
| Well | 9 | 5 | 9 | ||
| Moderate | 30 | 10 | 30 | ||
| Poor | 17 | 6 | 17 | ||
| Smoke | |||||
| Smoker | 43 | 19 | 43 | ||
| Non‐smoker | 13 | 2 | 13 | ||
| Alcohol | |||||
| Drinker | 45 | 20 | 45 | ||
| Non‐drinker | 11 | 1 | 11 |
OSCC, oesophageal squamous cell carcinoma.
*p Value (two‐sided) was calculated using the Student's t test between OSCC and normal epithelium.
†p Value (two‐sided) was calculated using Pearson's χ2 test between OSCC and normal epithelium.
Figure 1 Histopathological features of advanced oesophageal squamous cell carcinoma (OSCC), intraepithelial lesions and background non‐neoplastic epithelium. Microscopic results of: (A, B) background non‐neoplastic epithelium of OSCC; (C, D) intraepithelial neoplasia (IEN); (E, F) OSCC. Left panels show haematoxylin and eosin (H&E) staining (A, C, E) and right panels show periodic acid schiff (PAS) staining (B, D, F). PAS staining showed the boundary of normal mucosa (right) and IEN (left; ×100). The background non‐neoplastic epithelium shows glycogen‐positive cells in the upper four fifths region (B), whereas IEN (D) and OSCC (F) contain no glycogen‐positive cells.
DNA methylation profiles
Using a panel of 14 promoter loci, the DNA methylation status was assessed using methylation‐specific PCR or COBRA in the 56 advanced OSCC, 56 corresponding background non‐neoplastic epithelia and 21 IEN lesions from patients with OSCC. The DNA methylation status was also assessed in control oesophageal epithelia from 42 OSCC healthy volunteers without OSCC. Minor promoter hypermethylation was observed in the epithelium of some of the healthy volunteers. Nearly 25% of the controls already possessed methylation in MGMT and SFRP1 (table 2). Compared with the 42 normal control epithelia, the background non‐neoplastic epithelium from the 56 patients was significantly more methylated, according to the Mantel–Haenszel test (χ12 = 43.5, p<0.001). In a matched comparison of tumour tissue and background non‐neoplastic epithelium from the 56 patients, there was significantly higher methylation in the tumour samples (p<0.001) and in the 21 patients with OSCC, who also had IEN. All three two‐way comparisons showed significantly increased methylation in the tissue with the worse disease (IEN>non‐neoplastic epithelium, p = 0.008; OSCC>non‐neoplastic epithelium, p<0.001; and OSCC>IEN, p = 0.012). We also compared all three types of sample in these 21 patients using a logistic regression, which showed that there was significant heterogeneity in the methylation status of individual loci among patients and of multiple loci within individual patients.
Table 2 Frequency of methylated promoters in each pathological oesophageal lesion.
| Gene/loci | Number of patients with methylation in designated loci (%) | |||
|---|---|---|---|---|
| Normal control epithelium (n = 42) | Background non‐neoplastic epithelium (n = 56) | Intraepithelial neoplasia (n = 21) | OSCC (n = 56) | |
| MGMT | 10 (23.8) | 42 (75.0) | 17 (81.0) | 45 (80.4) |
| SFRP1 | 9 (21.4) | 25 (44.6) | 13 (61.9) | 36 (64.3) |
| p16INK4a | 0 (0) | 18 (32.1) | 8 (38.1) | 22 (39.3) |
| DCC | 0 (0) | 3 (5.4) | 8 (38.1) | 26 (46.4) |
| MINT1 | 0 (0) | 1 (1.8) | 3 (14.3) | 14 (25.0) |
| MINT2 | 4 (9.5) | 2 (3.6) | 1 (4.8) | 8 (14.3) |
| MINT31 | 1 (2.4) | 4 (7.1) | 7 (33.3) | 18 (32.1) |
| CACNA1G | 0 (0) | 6 (10.7) | 5 (23.8) | 13 (23.2) |
| DAPK | 2 (4.8) | 3 (5.4) | 5 (23.8) | 15 (26.8) |
| p14ARF | 0 (0) | 2 (3.6) | 2 (9.5) | 7 (12.5) |
| SFRP2 | 2 (4.8) | 5 (8.9) | 3 (14.3) | 11 (19.6) |
| APC1A | 4 (9.5) | 10 (17.9) | 4 (19.0) | 15 (26.7) |
| COX2 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| hMLH1 | 0 (0) | 1 (1.8) | 0 (0) | 6 (10.7) |
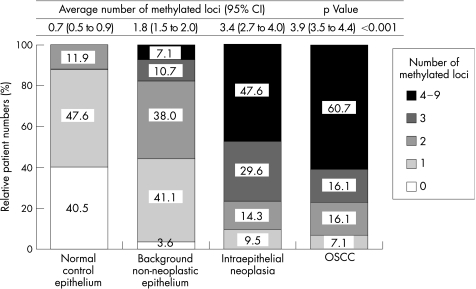
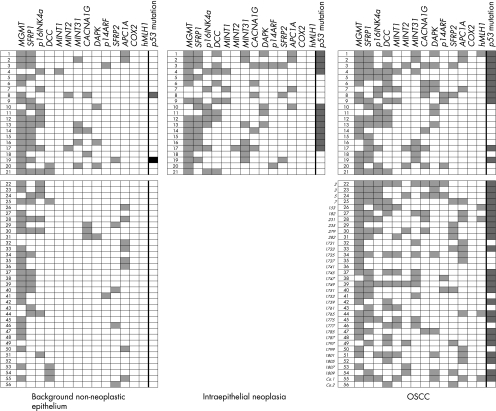
There was a gradual increase in the number of methylated loci from the normal epithelium (average number: 0.7, 95% CI 0.5 to 0.9), the OSCC‐background non‐neoplastic epithelium (average number 1.8, 95% CI 1.5 to 2.0), the IEN (average number 3.4, 95% CI 2.7 to 4.0), to the advanced OSCC (average number 3.9, 95% CI 3.5 to 4.4; fig 2). A non‐parametric test for a trend was performed on the three groups (background non‐neoplastic epithelium, IEN and OSCC) and a significant increase from the background epithelium to OSCC was shown (p = 0.03). Most of the individual markers, with the exception of MINT2, SFRP2 and APC1A, showed a significant increase in the frequency of methylation, within the range of normal epithelium, through to OSCC (table 2). The OSCC non‐neoplastic epithelium showed more extensive methylation than for the normal (control) epithelium (fig 2). Figure 3 shows the methylation status of each promoter occurring in the sequence from background non‐neoplastic epithelium, to IEN and to OSCC, in individual patients. Fifty six patients with OSCC, with or without IEN lesions (IEN lesions were identified in 21 of the patients with OSCC), are shown in the upper panel. OSCCSimilar clusters of promoters are shown to have methylation in the same patients on comparing the IEN lesions and the OSCC. Methylation profiles tend to accumulate along with the development of cancer from background non‐neoplastic epithelium through to OSCC, within the same patients.
Figure 2 Histopathological status of oesophageal lesions and DNA methylation profiles. A gradual increase in the number of methylated loci was observed from the normal control epithelium through the background non‐neoplastic epithelium of OSCC and intraepithelial neoplasia to advanced OSCC. p Value was calculated by the non‐parametric Kruskal–Wallis test.
Figure 3 The methylation status of individual promoter methylation in background non‐neoplastic epithelium, intraepithelial neoplasia and in oesophageal squamous cell cancer tissue from 56 patients with oesophageal squamous cell cancer (OSCC). Promoter methylation and p53 mutational status in 21 patients with OSCC accompanying intraepithelial neoplasia (IEN) are shown in the upper panel. Promoter methylation and p53 mutational status in 25 patients with OSCC, not accompanying IEN, are shown in the lower panel. The number of methylated loci and the frequency of p53‐mutated patients were similar in IEN and OSCC. Methylated loci (closed boxes in the first 14 columns); non‐methylated loci (open boxes); p53 mutant (closed boxes in the last column); p53 wild (open boxes).
Analysis of the p53 mutation
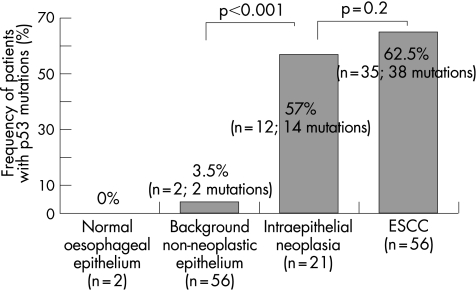
Exons 5–9 of the p53 gene were amplified and sequenced. Most of the changes found in p53 were missense mutations. A total of 35 of 56 OSCC tumour samples (62.5%, 38 mutations) and 12 of 21 samples of IEN (57%, 14 mutations) harboured p53 mutations (fig 4). However, only 2 of 56 background non‐neoplastic mucosa showed p53 mutations (3.5%, 2 mutations), and no mutation was found in the normal mucosa (fig 4). The p53 mutational profiles in the IEN were different from those of the corresponding main advanced OSCC, within the same patient, there being one exception (patient 17; table 3).
Figure 4 Mutational analysis of p53. The frequency of p53 mutation among patients with normal oesophageal epithelium, background non‐neoplastic epithelium, intraepithelial neoplasia and, oesophageal squamous cell carcinoma (OSCC). Exons 5–9 of p53 were amplified and sequenced. A total of 35 of 56 main patients with OSCC (62.5%, 38 mutations) and 12 of 21 patients with intraepithelial neoplasia (57%, 14 mutations) showed p53 mutations. However, only 2 of 56 background non‐neoplastic epithelia and none of the normal control epithelia showed p53 mutations. p Value was calculated by Pearson's χ2 test.
Table 3 Comparison of p53 mutation and methylation frequency between intraepithelial neoplasia of the oesophagus and advanced oesophageal squamous cell carcinoma.
| Patient number | Sex | Age | Differentiation | Stage | Number of methylation | p53 mutation spectrum | ||
|---|---|---|---|---|---|---|---|---|
| IEN | OSCC | IEN | OSCC | |||||
| 1 | M | 51 | Moderate | IV | 6 | 6 | C580T | G422A |
| 2 | M | 67 | Poor | III | 4 | 4 | C449T | C535T |
| 3 | M | 53 | Poor | III | 7 | 7 | G437A/A659G | G818T |
| 4 | M | 62 | Poor | IV | 4 | 6 | Deletion | A659G |
| 5 | M | 70 | Moderate | IV | 2 | 4 | w | T400G |
| 6 | M | 76 | Poor | III | 3 | 4 | w | C637T |
| 7 | M | 54 | Well | III | 3 | 6 | w | A659G |
| 8 | F | 73 | Well | II | 2 | 5 | w | G824A |
| 9 | M | 70 | Well | III | 4 | 5 | w | G743A |
| 10 | F | 69 | Well | II | 4 | 3 | A659G | w |
| 11 | M | 74 | Moderate | II | 3 | 3 | A671G | w |
| 12 | M | 61 | Moderate | II | 4 | 4 | A659G | w |
| 13 | M | 70 | Moderate | II | 5 | 5 | A659G | w |
| 14 | M | 69 | Poor | I | 3 | 2 | T755C | w |
| 15 | F | 54 | Moderate | III | 2 | 2 | T421C/A659G | w |
| 16 | M | 68 | Well | II | 3 | 2 | A659G | w |
| 17 | M | 46 | Moderate | IV | 6 | 5 | A659G | A659G |
| 18 | M | 63 | Moderate | IV | 2 | 3 | w | w |
| 19 | M | 60 | Moderate | II | 4 | 3 | w | w |
| 20 | M | 66 | Moderate | IV | 2 | 5 | w | w |
| 21 | M | 69 | Well | IV | 3 | 4 | w | w |
OSCC, oesophageal squamous cell carcinoma; F, female; IEN, intraepithelial neoplasia; M, male; w, wild‐type p53.
Comparison of DNA methylation and p53 mutation
The methylation status of the 14 promoter loci and the mutational status of p53 were compared for the 21 patients with IEN and the 56 patients with advanced OSCC (table 4). In both groups of patients, higher levels of methylation were observed in the lesions with p53 mutations (IEN; p<0.006, OSCC; p<0.001). No specific methylation marker or pattern was related to the p53 mutation.
Table 4 Methylation frequency in multiple promoter versus p53 mutational status in the intraepithelial neoplasia of the oesophagus or advanced oesophageal squamous cell carcinoma.
| Gene/loci | Intraepithelial neoplasia (n = 21) | p Value* | OSCC (n = 56) | p Value* | ||
|---|---|---|---|---|---|---|
| p53 wild (n = 9) | p53 mutant (n = 12) | p53 wild (n = 21) | p53 mutant (n = 35) | |||
| MGMT | 7 (77.8) | 10 (83.3) | 0.7 | 16 (76.2) | 29 (82.9) | 0.5 |
| SFRP1 | 3 (33.3) | 10 (83.3) | 0.01 | 11 (52.4) | 24 (68.6) | 0.09 |
| p16INK4a | 2 (22.2) | 6 (50.0) | 0.2 | 6 (28.6) | 16 (45.7) | 0.3 |
| DCC | 3 (33.3) | 5 (41.7) | 0.7 | 6 (28.6) | 19 (54.3) | 0.1 |
| MINT1 | 2 (22.2) | 1 (8.3) | 0.4 | 3 (14.3) | 11 (31.4) | 0.2 |
| MINT2 | 0 | 1 (8.3) | 0.4 | 2 (9.5) | 6 (17.1) | 0.5 |
| MINT31 | 2 (22.2) | 5 (41.7) | 0.1 | 6 (28.6) | 12 (34.3) | 0.8 |
| CACNA1G | 2 (22.2) | 3 (25.0) | 0.9 | 2 (9.5) | 12 (34.3) | 0.05 |
| DAPK | 0 | 5 (41.7) | 0.02 | 4 (19.0) | 11 (31.4) | 0.4 |
| p14ARF | 1 (11.1) | 1 (8.3) | 0.8 | 1 (4.8) | 6 (17.1) | 0.2 |
| SFRP2 | 2 (22.2) | 1 (8.3) | 0.4 | 1 (4.8) | 10 (28.6) | 0.04 |
| APC1A | 1 (11.1) | 3 (25.0) | 0.4 | 4 (19.0) | 11 (31.4) | 0.4 |
| COX2 | 0 | 0 | NA | 0 | 0 | NA |
| hMLH1 | 0 | 0 | NA | 0 | 5 (13.9) | 0.08 |
| Average number of | 2.4 | 4.1 | 0.006† | 2.9 | 4.9 | <0.001† |
| methylated loci (95% CI) | (1.6 to 3.3) | (3.3 to 4.9) | (2.1 to 3.5) | (4.1 to 5.2) | ||
*p Value was calculated by Pearson's χ2 test.
†p Value was calculated by the non‐parametric Kruskal–Wallis test.
OSCC, oesophageal squamous cell carcinoma;
Discussion
This study is the first to show that the non‐neoplastic oesophageal epithelium in patients with OSCC already accumulates a low level of DNA methylation in the gene promoters. The number of gene promoters with DNA methylation increased gradually from the normal healthy epithelium, through OSCC‐background non‐neoplastic epithelium and IEN identified in the epithelium of OSCC, to the main advanced form of OSCC.
Hiyama et al1 and Morita et al2 have reported that smoking or alcohol intake increases the risk of OSCC. Among 56 patients with OSCC in the present study, 43 (77%) had a history of smoking, whereas 45 (80%) had consumed alcohol (table 1). However, neither smoking nor alcohol was significantly associated with the methylation status of the examined promoters (data not shown).
Mutation in the p53 gene has been shown to play an important role in oesophageal carcinogenesis. We therefore assessed p53 mutation in exons 5–9. We identified 38 mutations in 35 OSCC samples (three patients having double mutations), 14 mutations in 12 IEN (two patients having double mutations) and 2 mutations in 2 corresponding background non‐neoplastic mucosa. Most of the mutations identified were located in “hot spots” as shown in the IARC TP53 MUTATION database.20 The two background non‐malignant mucosal specimens with a p53 mutation may have come from micro‐malignant areas, which could not be easily identified by the routine microscopy used. Interestingly, these two areas also had frequent promoter methylation (data not shown). No significant difference was seen in p53 mutational status for sex or age (data not shown).
In this study, p53 mutations were almost exclusively observed in the IEN and OSCC samples and rarely in the background epithelium. Conversely, an average of two of 14 promoter loci examined had acquired methylation in the corresponding non‐neoplastic mucosa and 0.7 loci showed methylation in age‐matched normal oesophageal epithelium from healthy volunteers (fig 2). However, there was no significant association between the number of methylated genes or loci examined and the sex or age either in the OSCC or in the normal mucosa from healthy volunteers (data not shown). Interestingly, within 21 samples of IEN or 56 OSCC samples, a significantly higher number of loci showed methylation in samples with p53 mutation, compared with the samples without the mutation. These results suggest that the oesophageal epithelium, which has accumulated methylation, becomes highly susceptible to mutations in the p53 gene. The promoter methylation of MGMT has been shown to be associated with the G:C to A:T mutations in p53 in colorectal and other cancers.21,22,23,24 By contrast, MGMT methylation has not been shown to be associated with a p53 mutation in gastric carcinoma or OSCC.25,26 In our study, no association was found between aberrant MGMT methylation and the frequency of p53 mutations (table 4) or the presence of G:C to A:T transitions in p53 (data not shown).
We also compared the mutational status of the p53 and the pattern of DNA methylation in the IEN and the OSCC in the same patient (table 3 and fig 3). Interestingly, the frequency of methylation in the same individual was very similar. Moreover, almost the same promoter (loci) showed methylation both in the IEN and in the OSCC. The pattern of methylation in individual loci, shown in fig 3, indicates a progressive accumulation of methylations from non‐neoplastic epithelium to IEN to OSCC in individual patients. Thus, probably a field of methylation precedes and has some causative relationship with the development of intraepithelial lesions and cancer. By contrast, the specific mutations in p53 almost always differed between IEN and tumour lesions from the same patients. These results suggest that the oesophageal epithelium, affected by environmental carcinogens, accumulates methylation in increasing numbers of gene promoters over time and that these epigenetic changes eventually contribute to OSCC carcinogenesis. Also, epigenetic changes in a wide epithelial field appear to increase the probability of acquiring changes in genes such as p53, which may occur independently in multiple sites and may also contribute to carcinogenesis.
In summary, this study provides novel evidence for the field carcinogenesis in OSCC that is closely linked to the progressive methylation of multiple promoters and p53 mutations.
Abbreviations
COBRA - combined bisulphite restriction analysis
OSCC - oesophageal squamous cell carcinoma
H&E - haematoxylin and eosin
IEN - intraepithelial neoplasia
PAS - periodic acid schiff
PCR - polymerase chain reaction
Footnotes
OSCC, oesophageal squamous cell carcinoma.
Competing interests: None declared.
References
- 1.Hiyama T, Sato T, Yoshino K.et al Second primary cancer following laryngeal cancer with special reference to smoking habits. Jpn J Cancer Res 199283334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morita M, Kusano H, Ohno S.et al Multiple occurrence of carcinoma in the upper aerodigestive tract associated with esophageal cancer: reference to smoking, drinking and family history. Int J Cancer 199458207–210. [DOI] [PubMed] [Google Scholar]
- 3.Bennett W P, Hollstein M C, He A.et al Archival analysis of p53 genetic and protein alterations in Chinese esophageal cancer. Oncogene 199161779–1784. [PubMed] [Google Scholar]
- 4.Wang L D, Hong J Y, Qiu S L.et al Accumulation of p53 protein in human esophageal precancerous lesions: a possible early biomarker for carcinogenesis. Cancer Res 1993531783–1787. [PubMed] [Google Scholar]
- 5.Gao H, Wang L D, Zhou Q.et al p53 tumor suppressor gene mutation in early esophageal precancerous lesions and carcinoma among high‐risk populations in Henan, China. Cancer Res 1994544342–4346. [PubMed] [Google Scholar]
- 6.Ito S, Ohga T, Saeki H.et al p53 mutation profiling of multiple esophageal carcinoma using laser capture microdissection to demonstrate field carcinogenesis. Int J Cancer 200511322–28. [DOI] [PubMed] [Google Scholar]
- 7.Sozzi G, Miozzo M, Pastorino U.et al Genetic evidence for an independent origin of multiple preneoplastic and neoplastic lung lesions. Cancer Res 199555135–140. [PubMed] [Google Scholar]
- 8.Izawa T, Obara T, Tanno S.et al Clonality and field cancerization in intraductal papillary‐mucinous tumors of the pancreas. Cancer 2001921807–1817. [DOI] [PubMed] [Google Scholar]
- 9.Kanjilal S, Strom S S, Claymon G L.et al p53 Mutations in nonmelanoma skin cancer of the head and neck: molecular evidence for field cancerization. Cancer Res 1995553604–3609. [PubMed] [Google Scholar]
- 10.Jones P A, Baylin S B. The fundamental role of epigenetic events in cancer. Nat Rev Genet 20023415–428. [DOI] [PubMed] [Google Scholar]
- 11.Jones P A, Laird P W. Cancer epigenetics comes of age. Nat Genet 199921163–167. [DOI] [PubMed] [Google Scholar]
- 12.Nagasaka T, Sharp G B, Notohara K.et al Hypermethylation of O6‐methylguanine‐DNA methyltransferase promoter may predict nonrecurrence after chemotherapy in colorectal cancer cases. Clin Cancer Res 200395306–5312. [PubMed] [Google Scholar]
- 13.Xiong Z, Laird P W. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 1997252532–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rashid A, Shen L, Morris J S.et al CpG island methylation in colorectal adenomas. Am J Pathol 20011591129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyota M, Kopecky K J, Toyota M O.et al Methylation profiling in acute myeloid leukemia. Blood 2001972823–2829. [DOI] [PubMed] [Google Scholar]
- 16.Shen L, Kondo Y, Hamilton S R.et al P14 methylation in human colon cancer is associated with microsatellite instability and wild‐type p53. Gastroenterology 2003124626–633. [DOI] [PubMed] [Google Scholar]
- 17.Toyota M, Shen L, Ohe‐Toyota M.et al Aberrant methylation of the cyclooxygenase 2 CpG island in colorectal tumors. Cancer Res 2000604044–4048. [PubMed] [Google Scholar]
- 18.Ogi K, Toyota M, Ohe‐Toyota M.et al Aberrant methylation of multiple genes and clinicopathological features in oral squamous cell carcinoma. Clin Cancer Res 200283164–3171. [PubMed] [Google Scholar]
- 19.Nagasaka T, Sasamoto H, Notohara K.et al Colorectal cancer with mutation in BRAF, KRAS, and wild‐type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol 2004224584–4594. [DOI] [PubMed] [Google Scholar]
- 20.Hainaut P, Hernandez T, Robinson A.et al IARC database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualization tools. Nucl Acids Res 199826205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf P, Hu Y C, Doffek K.et al O6‐methylguanine‐DNA methyltransferase promoter hypermethylation shifts the p53 mutational spectrum in non‐small cell lung cancer. Cancer Res 2001618113–8117. [PubMed] [Google Scholar]
- 22.Nakamura M, Watanabe T, Yonekawa Y.et al Promoter methylation of the DNA repair gene MGMT in astrocytomas is frequently associated with G:CA:T mutations of the TP 53 tumor suppressor gene. Carcinogenesis 2001221715–1719. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M, Toyota M, Sanchez‐Cespedes M.et al Inactivation of the DNA repair gene O6‐methylguanine‐DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K‐ras in colorectal tumorigenesis. Cancer Res 2000602368–2371. [PubMed] [Google Scholar]
- 24.Esteller M, Risques R ‐ A, Toyota M.et al Promoter hypermethylation of the DNA repair gene O6‐methylguanine‐DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res 2001614689–4692. [PubMed] [Google Scholar]
- 25.Oue N, Shigeishi H, Kuniyasu H.et al Promoter hypermethylation of MGMT is associated with protein loss in gastric carcinoma. Int J Cancer 200193805–809. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Lu W, Miao X.et al Inactivation of DNA repair gene O6‐methylguanine‐DNA methyltransferase by promoter hypermethylation and its relation to p53 mutations in esophageal squamous cell carcinoma. Carcinogenesis 2003241039–1044. [DOI] [PubMed] [Google Scholar]