Irrespective of initial stimuli, increased production of type I collagen is a common hallmark of fibrotic diseases in various organs including the liver. A dynamic balance between the production and degradation of collagen is seen, which is rigorously controlled by several growth factors and cytokines. Of these, transforming growth factor β (TGFβ) is the most potent factor in stimulating type I collagen gene transcription. It also regulates expression of matrix metalloproteinases and their inhibitors, and modulates inflammatory reactions by influencing T cell functions. Therefore, TGFβ is considered to be the major factor accelerating liver fibrosis. Identification and characterisation of Smad proteins, intracellular mediators of the signal transduction of TGFβ, have led to a better understanding of the precise mechanisms of TGFβ functions from the viewpoint of its intracellular signalling pathway and crosstalk with other factors. Several studies have focused on the suppression of TGFβ activation and intervention of the TGFβ/Smad signalling pathways to treat liver fibrosis. However, as generalised blockade of TGFβ activity may result in the promotion of carcinogenesis and excessive immune reactions, much attention has to be paid to selective intervention of the TGFβ/Smad signal specifically in collagen‐producing cells in the fibrotic tissue. TGFβ also affects the growth and differentiation of stem and progenitor cells. From this point of view, a new concept of the treatment for liver fibrosis may arise from the discipline of stem cell biology by modulating TGFβ and Smad signalling in stem/progenitor cells.
Background
TGFβ is a member of a large family of pleiotropic cytokines that includes bone morphogenetic proteins (BMPs), activins and other related factors. Mammals have three different forms of TGFβ (β1, β2 and β3). Although the three isoforms of TGFβ are encoded by distinct genes located on different chromosomes, they have approximately 80% homology at the level of amino acid sequence. Of these, TGFβ1 has been most extensively studied for its roles in various pathophysiological conditions.
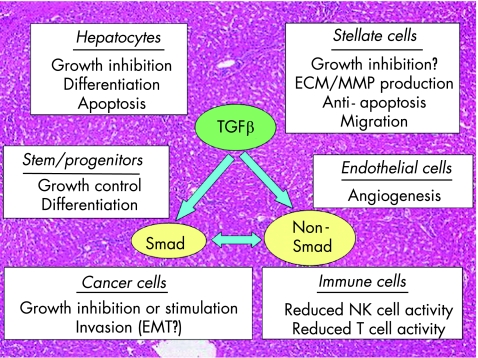
TGFβ1 (henceforth referred to as TGFβ) is a homodimeric polypeptide with a molecular weight of 25 kDa. It was first identified in the culture medium conditioned by transformed cells, and was named for its ability to induce anchorage‐independent growth of rodent fibroblasts.1 However, subsequent studies have shown that it is a potent growth inhibitor for most epithelial types of cells including parenchymal hepatocytes. In addition, TGFβ is multifunctional and regulates cell survival, differentiation, migration, adhesion and synthesis of extracellular matrix (ECM) components. It is therefore involved in many biological and pathological processes including embryonic development, tissue remodelling, inflammation, angiogenesis, atherosclerosis, fibrosis and carcinogenesis. Through these functions, TGFβ plays an important part in both normal and diseased conditions in the liver2,3 and in other organs (fig 1). During the past decade, identification and characterisation of Smad proteins, intracellular mediators of the signal transduction of TGFβ superfamily members, have led to a better understanding of the precise mechanisms of TGFβ functions from the viewpoint of its intracellular signalling pathway and crosstalk with other factors.4
Figure 1 Diverse effects of transforming growth factor β (TGFβ) and Smad signal in liver biology and pathobiology. TGFβ and Smad play important parts in both normal and diseased situations in the liver by exerting differential effects on each cell population. Non‐Smad signalling pathways of TGFβ have also been implicated in various pathophysiological conditions either independently of or through crosstalk with Smad signals. ECM, extracellular matrix; MMP, matrix metalloproteinase; EMT, epithelial‐to‐mesenchymal transition, NK, natural killer.
This review summarises recent advances in our understanding of the pathological roles of TGF‐β and Smad signalling in hepatic fibrogenesis, with an emphasis on the transcriptional regulation of type I collagen gene expression. It also highlights the current attempts to treat liver fibrosis by targeting TGFβ/Smad signalling.
Overview of the TGFβ/Smad signalling pathway
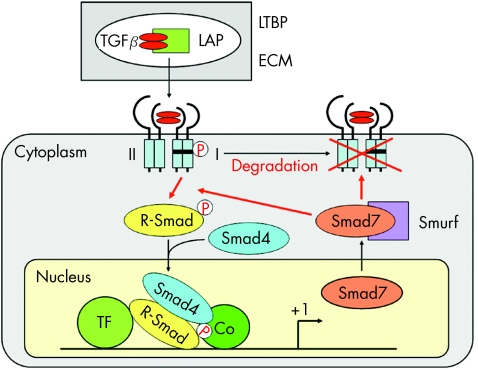
TGFβ protein is synthesised and secreted as a precursor that includes a latency‐associated peptide (LAP). This complex is often bound by one of the several latent TGFβ‐binding proteins, which act as an important safeguard against inadvertent activation of TGFβ. In addition, they assemble with several ECM components such as type IV collagen, fibronectin and fibrillin. Thus, ECM represents a reservoir from which active TGFβ can readily be recruited without de novo synthesis (fig 2). Several factors including matrix metalloproteinases (MMPs), plasmin, plasminogen activators, αvβ6 integrin and thrombospondins have been reported to accelerate the release of active TGFβ from the latent complex. Of these, thrombospondin 1 expression in hepatic stellate cells (HSC) is induced by platelet‐derived growth factor (PDGF) and promotes the effects of TGFβ.5 A peptide derived from the amino terminus of LAP blocks the activation of TGFβ by thrombospondin 1, and suppresses the progression of experimental liver fibrosis in rats.6 On the other hand, a protease inhibitor, camostat mesilate, prevents progression of rat liver fibrosis by reducing plasmin activity and suppressing TGFβ activation.7
Figure 2 Signal transduction pathways of transforming growth factor β (TGFβ) and its intracellular mediators, Smad proteins. Active TGFβ released from the latent TGFβ‐binding proteins (LTBP) and latency‐associated peptide (LAP) binds to the specific receptors on the cell surface. Receptor‐activated Smad (R‐Smad) and Smad4 transduce the signal from the cytoplasm to the nucleus, where they regulate target gene transcription usually in association with DNA‐binding transcription factors (TF) and coactivators (Co). By contrast, inhibitory Smad7 is strongly and rapidly induced by TGFβ itself, and acts in an autoregulatory negative feedback loop of the TGFβ/Smad signal. ECM, extracellular matrix.
Activated TGFβ first binds to the TGFβ type II receptor on the cell surface. It subsequently recruits the TGFβ type I receptor, thus forming a heterotetrameric complex of these two types of receptors (fig 2). The type II receptor kinase phosphorylates the type I receptor in the GS region, which is located immediately upstream of the kinase domain. Type I receptor in turn phosphorylates Smad2 and Smad3, so‐called receptor‐activated Smad (R‐Smad), which form hetero‐oligomers with a common mediator, Smad4 (Co‐Smad). They translocate from the cytoplasm to the nucleus, where they regulate the transcription of target genes.4 In addition to the weak DNA‐binding activity of Smad3 and Smad4 (but not Smad2), they usually associate with other DNA‐binding transcription factors (fig 2), which specify the promoter binding and transcriptional regulation of target genes. Smad2 and Smad3 are also linked to the general transcriptional machinery through a direct interaction with transcriptional coactivators p300 or CBP (fig 2).
In contrast with those signal‐transducing R‐Smads and Co‐Smads, Smad7 is known as an inhibitory Smad (I‐Smad): it inhibits TGFβ signalling by interfering with the phosphorylation of Smad2 and Smad3 by the type I TGFβ receptor kinase.8 Smad7 expression is strongly and rapidly induced by TGFβ itself. In addition, Smad7 interacts with a group of ubiquitin ligases, termed Smurf. After recruitment of Smad7–Smurf1 complex to the activated TGFβ receptors, Smurf1 degrades the receptors through proteasomal and lysosomal pathways.9 Thus, Smad7 acts in an autoregulatory negative feedback loop of the TGFβ/Smad signal (fig 2). In addition, several corepressors have been reported, including c‐Ski, SnoN, transforming growth inhibiting factor and smad nuclear‐interacting protein 1. They interact with Smad2 or Smad3 and inhibit TGFβ responses.
TGFβ/Smad signal in hepatic fibrogenesis
Type I collagen, the major ECM component of the fibrotic tissue, is a heterotrimer composed of two α1 chains and one α2 chain. They are encoded by two distinct genes, COL1A1 and COL1A2, respectively. Increased production of type I collagen is a common hallmark of fibrotic diseases in various organs including the liver. A dynamic balance between the production and degradation of collagen is rigorously controlled by several growth factors and cytokines, and a disruption of this equilibrium results in organ fibrosis. TGFβ is the most potent factor in stimulating type I collagen gene transcription. It also regulates expression of MMPs and their inhibitors, and modulates inflammatory reactions by influencing T cell functions. Therefore, TGFβ is considered to be a major factor accelerating the progression of organ fibrosis.
In the normal liver, sinusoidal endothelial cells and Kupffer cells contain relatively high levels of TGFβ mRNA, whereas HSC express little amounts of TGFβ. However, once stimulated by fibrogenic stimuli, HSC are the only cells that respond by expressing increased amounts of all three different isoforms of TGFβ. As activated HSC are the principal cells to produce type I collagen in fibrotic liver, they contribute to the development of liver fibrosis through autocrine and paracrine loops of TGFβ‐stimulated collagen production.
During the past decade, intensive studies have been focusing on transcriptional regulation of type I collagen expression, especially on the molecular mechanisms responsible for the TGFβ‐elicited COL1A2 stimulation and its pathological roles during the fibrogenic process in liver.10
Transcriptional regulation of collagen gene expression by transforming growth factor β and Smad proteins
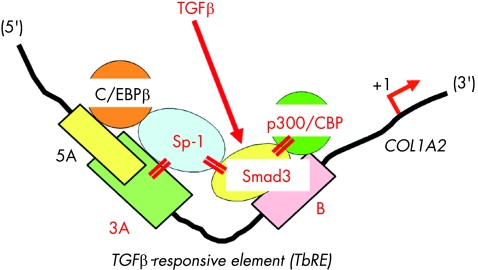
Our initial studies using primary cultures of skin fibroblasts have shown that the −313 to −183 upstream sequence of the COL1A2 promoter is essential for basal and TGFβ‐stimulated transcription.11 Within this segment are three DNA–protein binding sites, Box 5A, Box 3A and Box B (fig 3). Among them, Box 3A is the binding site for a ubiquitous trans‐activator Sp1, whereas unknown cofactors bind to the downstream Box B.11,12 Treatment with TGFβ modified the interaction between Sp1 and the cofactors and subsequently enhanced their binding to the cognate DNA sequences. We therefore designated the 3A plus B region the TGFβ‐responsive element, TbRE.11 Subsequently, Smad3 was shown to bind to Box B,13 and its interaction with Sp1 has been implicated in TGFβ‐elicited COL1A2 stimulation in embryonic fibroblasts (fig 3).14 TGFβ also enhances functional cooperation between Smad3 and p300/CBP coactivators.15 It is therefore suggested that Sp1, Smad proteins and p300/CBP coactivators form a multimeric complex to mediate the stimulatory effect of TGFβ on COL1A2 transcription (fig 3).
Figure 3 Schematic representation of the transforming growth factor β‐responsive element (TbRE) located on the α2(I) collagen (COL1A2) gene promoter. TbRE consists of Box 3A and Box B, which are separated 14 bases from each other. TGFβ treatment enhances the interaction between Box 3A‐bound Sp1, Box B‐bound Smad3 and p300/CBP coactivators, and subsequently stimulates COL1A2 transcription. On the other hand, Box 5A, located immediately upstream of and partly overlapping Box 3A, is a binding site for a COL1A2 repressor, CAAT/enhancer binding protein β (C/EBPβ).
Synergistic cooperation between Sp1 and Smad3 also mediates the stimulatory effect of TGFβ on COL1A2 transcription in activated HSC.16,17 However, the intracellular localisation of R‐Smads and their responses to exogenous TGFβ differ depending on the activation status of HSC. TGFβ induces phosphorylation and nuclear translocation of Smad2 in quiescent HSC and those of Smad3 in transdifferentiated HSC.18 More importantly, ligand‐independent nuclear localisation of Smad2, but not Smad3, was observed in culture‐activated HSC18 and in myofibroblasts obtained from chronically CCl4‐treated fibrotic liver.19 By contrast, constitutive activation of the TbRE due to ligand‐independent phosphorylation and nuclear localisation of Smad3 has been shown in an activated HSC clone established from a carbon tetrachloride (CCl4)‐induced cirrhotic liver.20
In contrast with several studies on the molecular mechanisms regulating COL1A2 transcription, our knowledge regarding the regulation of coordinately expressed COL1A1 transcription is limited. Previously, it has been shown that hydrogen peroxide and TGFβ act on the same upstream sequence of COL1A1 promoter to stimulate gene transcription.21 More recently, both Smad and p38 mitogen‐activated protein kinase (MAPK) pathways have been implicated in transcriptional and post‐transcriptional regulation of COL1A1 expression.22
Molecular mechanisms responsible for cell type‐specific COL1A2 transcription
Type I collagen is produced predominantly in mesenchymal cells, but molecular mechanisms responsible for this cell lineage‐specific expression are not fully understood. A COL1A2 upstream sequence containing the TbRE was activated during the development of liver fibrosis.23 More importantly, the COL1A2 promoter was used only in non‐parenchymal cells but not in parenchymal hepatocytes. We therefore compared COL1A2 transcription and response to TGFβ between activated HSC and primary cultures of hepatocytes.17 Parenchymal hepatocytes exhibited low transcriptional activity of the TbRE and no response to TGFβ. In those cells, another GC box binding factor Sp3, rather than Sp1, bound to the TbRE. Transfection of HSC with an Sp3 expression plasmid abolished the COL1A2 response to TGFβ, whereas overexpression of Sp1 in parenchymal hepatocytes increased basal COL1A2 transcription and conferred TGFβ responsiveness.17 In addition, functional and physical interactions were observed between Sp1 and Smad3, but not between Sp3 and Smad3.17 These results indicate that interaction between GC box binding factors and Smad proteins modulates cell lineage‐specific COL1A2 transcription in the liver.
Differential roles of Smad2 and Smad3 in HSC activation and collagen expression
Despite their high degree of homology, Smad2 and Smad3 have distinct roles in various biological and pathological situations. This has been clearly shown by the comparison of genetically engineered Smad2‐null and Smad3‐null mice. Smad2‐null mice are embryonic lethal, indicating its critical role in prenatal development.24 By contrast, mice lacking Smad3 are viable, although they exhibit small stature and impaired mucosal immunity.25 A previous study using embryonic fibroblasts from Smad2‐null and Smad3‐null mice has shown that Smad2 plays an important part in TGFβ‐induced MMP2 expression, whereas Smad3 is required for the induction of c‐fos, Smad7 and TGFβ.26 On the other hand, lack of either Smad2 or Smad3 results in markedly suppressed plasminogen activator inhibitor 1 (PAI) expression, indicating that both Smad2 and Smad3 are necessary to mediate TGFβ‐elicited PAI‐1 gene transcription.26 Another study using HSC isolated from Smad3‐null mice has clearly shown that Smad3 plays an important part in stimulating type I collagen expression in activated HSC.27 However, because of the lack of adult Smad2‐null mouse, the relative levels and functions of Smad2 and Smad3 in specific liver cell types and their relevance to pathophysiological conditions in the liver remained unknown. Recently, differential roles of Smad2 and Smad3 in HSC function have been reported by infecting cultured rat HSC with adenoviruses expressing wild‐type and dominant‐negative Smad2 or Smad3.28 Smad3‐overexpressing cells exhibited increased expression of type I collagen and fibronectin, increased chemotaxis and decreased proliferation. It was also shown that Smad3, but not Smad2, increased α‐smooth‐muscle actin organisation in stress fibres. From these findings, it is suggested that Smad3 plays a more important part than Smad2 in the morphological and functional maturation of myofibroblasts, and thus in the development of liver fibrosis.28 Several small molecules inhibiting kinase activity of the TGFβ/activin type I receptors have been developed for their possible therapeutic application.29 However, they suppress phosphorylation of both Smad2 and Smad3. Development of specific inhibitors for each of Smad2 and Smad3 not only shows the differential roles of these two R‐Smads, but is also useful for more specific regulation of TGFβ target gene expression.
Crosstalk between TGFβ/Smad and other signalling pathways
Signal crosstalk with TGFβ/Smad pathway
Recent studies have shown that several factors interact with the TGFβ/Smad signal. Physical interactions and functional cooperation of Smad proteins with transcriptional factors allow signal crosstalk with other signalling pathways. Crosstalk with the Smad signal may also result from the ability of TGFβ to activate signalling pathways independently of Smad. Among many factors interacting with the TGFβ/Smad signal, signal crosstalk with MAPK and BMP7 has been shown, owing to its relevance to the regulation of type I collagen gene expression.
MAPK
Several mitogenic or stress stimuli activate members of the MAPK family of proteins and induce diverse cellular responses, including proliferation, differentiation and regulation of specific metabolic pathways. TGFβ can induce the activation of all three known MAPK pathways, extracellular signal‐regulated kinase (ERK), p38 MAPK and c‐Jun N‐terminal kinase (JNK). One of the first demonstrated signal crosstalk between Smad and MAPK pathways was the ERK‐elicited inhibition of nuclear translocation of Smad1.30 ERK phosphorylates the specific serine residues in the linker domain of Smad1 and counter‐represses BMP/Smad1‐induced transcription. Similarly, negative regulation of TGFβ/Smad signal by the activated ERK pathway has been reported.31 On the other hand, p38 MAPK usually acts as an alternative pathway to mediate TGFβ signal,32 and it has been recently shown that p38 MAPK promotes the effects of Smad3 by enhancing its association with p300/CBP coactivators.33 TGFβ‐stimulated JNK can also phosphorylate Smad3 and induce its nuclear translocation.34 However, recent studies using phosphorylation site‐specific anti‐Smad2 and anti‐Smad3 antibodies have shown that p38 MAPK35 and JNK36 phosphorylate Smad3 at the linker region in activated HSC. Apart from the fact that each experiment used different types of cells, the reason why the same phosphorylation at the liker region of R‐Smads by ERK and p38 MAPK/JNK either stimulates or suppresses their nuclear localisation remains an interesting open question. In addition, the effects of MAPK signals on type I collagen gene expression have also been a matter of controversy. Although most studies have shown that specific inhibition of the p38 MAPK pathway results in a decrease in collagen gene expression,22,35,37 ERK signal either increases38 or decreases39 collagen gene expression depending on the cell types examined. Although there is no doubt that MAPK pathways greatly influence the TGFβ/Smad signals, the precise mechanisms of their crosstalk need to be elucidated in each cellular or pathophysiological context.
BMP‐7
BMPs, as members of the TGFβ superfamily, are structurally related to the prototype molecule TGFβ. However, they bind to distinct type II receptors and subsequently activate specific type I serine–threonine kinase receptors.4 While TGFβ receptors phosphorylate Smad2 and Smad3, the BMP signal is transmitted by phosphorylation of Smad1, Smad5 and Smad8. Recent studies have shown that BMP7 opposes the fibrogenic activity of TGFβ by suppressing the nuclear translocation of Smad3 in glomerular mesangial cells40 and pulmonary myofibroblasts.41 In the latter study on pulmonary myofibroblasts using transgenic COL1A2 reporter mice, BMP7 increased the expression of inhibitors of differentiation (Id)2 and 3, and ectopic expression of Id2 and Id3 was found to suppress COL1A2 transcription.41 These results therefore indicate that BMP7 antagonises TGFβ‐dependent fibrogenic activity of mouse pulmonary myofibroblasts not only by inhibiting the TGFβ/Smad3 signal but also by inducing Id2 and Id3. It is worth examining whether overexpression of BMP7 suppresses hepatic fibrogenesis through the same mechanisms. BMP7 and TGFβ are also reported to antagonise each other in epithelial‐to‐mesenchymal transition (EMT),42 which plays an important part in fibrogenesis43 as well as in carcinogenesis. To date, the presence and functional roles of EMT have not been established in the liver.
Antagonistic factors suppressing TGFβ/Smad‐stimulated COL1A2 transcription
As TGF‐β is a key player in stimulating collagen gene transcription during the progression of liver fibrosis, counter‐repression of the TGFβ‐stimulated collagen expression would be a potent therapeutic means for preventing organ fibrosis by suppressing excessive collagen deposition in the liver.3,44 Among several growth factors and cytokines, tumour necrosis factor α (TNFα) and interferon (IFN)γ are well known to suppress TGFβ‐elicited COL1A2 stimulation. More recently, hepatocyte growth factor (HGF) has also been reported to suppress experimental liver fibrosis. Most of these antagonistic actions are exerted via crosstalk between TGFβ/Smad and other signalling pathways.
TNFα
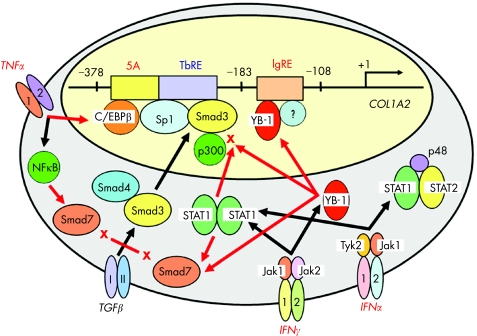
TGFβ and TNFα exert opposing effects on both synthesis and degradation of type I collagen. Our initial study has revealed that the counter‐repression of TGFβ‐stimulated COL1A2 transcription by TNFα is mediated through the same TbRE as well as by increasing the amount of a repressor protein bound to the immediately upstream sequence, Box 5A (fig 3).45 It has also been shown that CAAT/enhancer‐binding protein β is the major component of the Box 5A‐bound complex and mediates the TNFα‐elicited COL1A2 repression.46 A subsequent study showed that TNFα‐activated nuclear factor κB induced inhibitory Smad7 and suppressed TGFβ/Smad signalling (fig 4).47 On the other hand, others have proposed that TNFα prevents TGFβ/Smad‐induced gene transcription through the induction of c‐Jun/Jun B interacting with Smad3 and p300/CBP coactivators.48 The latter study showed a good example of a crosstalk between TGFβ/Smad and JNK/Jun signals.
Figure 4 Antagonistic actions of tumour necrosis factor α (TNFα) and interferons (IFNs) on transforming growth factor β (TGFβ)‐stimulated ×2(I) collagen gene transcription. Several antagonistic factors including TNFα and IFNs (IFNγ and IFNα) counter‐repress TGFβ‐stimulated COL1A2 transcription via crosstalk between TGFβ/Smad and other signalling pathways. IgRE, IFNγ‐responsive element; NF‐κB, nuclear factor κB; Stat, signal transducer and activator of transcription.
IFNγ
There have been several studies showing the inhibitory effects of IFNγ on collagen expression in both in vitro49,50 and in vivo51,52 experimental systems. With regard to the molecular mechanisms responsible for the inhibitory effects of IFNγ, several studies have shown crosstalk between the TGFβ and IFNγ signalling pathways (fig 4). The IFNγ‐responsive element (IgRE) has been mapped to the −161 to −150 sequence of the COL1A2 promoter,53 downstream of the TbRE (fig 4). A Y box binding protein, YB‐1, binds to the IgRE and mediates the inhibitory effects of IFNγ on COL1A2 transcription.54 Although IFNγ‐induced Smad7 expression via Jak1/Stat1 activation was initially proposed,55 others have reported that Stat1 activated by IFNγ interacts with p300/CBP coactivators and suppresses Smad3/p300‐stimulated COL1A2 transcription.56 We have shown that YB‐1 counter‐represses TGFβ‐stimulated COL1A2 transcription by interfering with the Smad3–p300 interaction through its preferential binding to p300.57 More recently, it has been shown that YB‐1 activates Smad7 gene transcription.58 Taken together, it is suggested that IFNγ‐induced YB‐1 inhibits COL1A2 transcription through several different mechanisms and plays a critical role in counter‐repressing TGFβ/Smad‐stimulated COL1A2 transcription (fig 4).
IFNα
IFNα is now widely used for the treatment of chronic hepatitis C. It has been reported that IFNα treatment results in an improvement in the serum levels of fibrotic markers, not only in the patients who responded to the treatment but also in those who did not respond.59,60 In addition, quantitative histopathological analyses of paired liver biopsy specimens have shown some improvement in the degree of fibrosis after IFNα treatment irrespective of the initial virological response.61,62 These results may suggest that IFNα has direct antifibrotic effects in addition to its antiviral activity. It has been shown that IFNα, as well as IFNγ, inhibits proliferation and collagen synthesis of cultured HSC,50 and that they suppress activation of the COL1A2 promoter induced by repeated CCl4 injections in vivo.63 IFNα and IFNγ inhibit COL1A2 transcription through the same mechanism that interferes with the interaction between phosphorylated Stat1 and p300 (fig 4).63
HGF
HGF was originally identified as a potent mitogen for adult rat hepatocytes, but subsequent studies have shown that it exhibits mitogenic, motogenic and morphogenic activities for a variety of cells. HGF administration not only stimulates liver regeneration but also prevents the occurrence of liver fibrosis and accelerates the recovery from the pre‐existing fibrosis.64,65 Moreover, HGF gene treatment has been shown to suppress TGFβ expression, inhibit hepatocyte apoptosis, and produce the complete resolution of fibrosis in rat cirrhotic liver.66 In addition to its inhibitory effect on TGFβ expression, other possible mechanisms responsible for the antifibrotic actions of HGF are stimulation of collagenase expression64,67 and its effect on growth inhibition and apoptosis of activated HSC.68
Cell type‐specific intervention of TGFβ/Smad signal to treat liver fibrosis
On the basis of the significant roles of TGFβ and Smad proteins in regulating collagen expression, there have been several studies attempting to block the TGFβ/Smad signal and suppress organ fibrosis by using the adenoviral gene transfer techniques. One of them blocked the TGFβ signalling by expressing a dominant‐negative TGFβ type II receptor, and prevented liver fibrosis and dysfunction in dimethylnitrosamine‐treated rats.69 It also prevented the progression of pre‐existing liver fibrosis and enhanced hepatocyte regeneration.70 Similarly, a soluble form of TGFβ type II receptor consisting of only the extracellular domain suppressed experimental liver fibrosis induced by common bile duct ligation71 or dimethylnitrosamine administration.72 Others injected a recombinant adenovirus carrying inhibitory Smad7 cDNA into rats and succeeded in treating liver fibrosis induced by common bile duct ligation.73
All of these adenoviral gene treatments have a great potential in treating liver fibrosis in humans. However, there are several concerns arising before their clinical application. Firstly, it should be seriously considered that blocking of the TGFβ/Smad signal may result in the promotion of carcinogenesis. As already described, TGFβ has an antiproliferative effect on most epithelial types of cells including parenchymal hepatocytes, and Smad4 was originally identified as the product of a tumour suppressor gene. It is especially important in the case of advanced liver fibrosis in humans, as most hepatocellular carcinomas originate from the underlying cirrhotic liver tissue. Owing to its critical roles in immune suppression, generalised blockade of TGFβ activity may also lead to excessive immune reactions and extensive inflammation. In relation to the first problem, the second concern is the limitation in tissue specificity and cell type specificity of gene delivery. All of the above experiments used strong enhancer/promoter elements such as the one derived from cytomegalovirus. These strong expression units exhibit no tissue specificity, which may cause severe adverse effects in the non‐target organs. Considering that collagens play critical parts in the maintenance of organ architecture and tissue integrity as well as in various physiological processes, it is more logical to use a tissue‐specific enhancer/promoter sequence for the fibrotic tissue‐specific regulation of collagen metabolism.
Several promoter sequences have been tested for their ability to mediate cell‐specific expression. One of them, the promoter of tissue inhibitor of metalloproteinase 1 gene, which is activated during hepatic fibrogenesis, induced cell death in culture activated HSC by expressing the herpes simplex virus thymidine kinase.74 We have recently shown in vivo the cell type‐specific gene expression by using a recombinant adenovirus carrying the −17 kilobase tissue‐specific COL1A2 enhancer sequence.75 Under the control of this COL1A2 enhancer, enhanced green fluorescent protein was expressed only in CCl4‐treated liver but not in untreated normal liver. It was expressed mainly in activated HSC in the fibrotic liver, but not in any other organs including kidney, lung and skin (fig 5). Furthermore, adenovirus‐mediated overexpression of YB‐1 driven by the COL1A2 enhancer inhibited COL1A2 promoter activation after CCl4 injections, and subsequently suppressed the progression of liver fibrosis.75 These results validate a new concept of the treatment for liver fibrosis by suppressing excessive collagen deposition in fibrotic tissue without affecting non‐target normal organs.
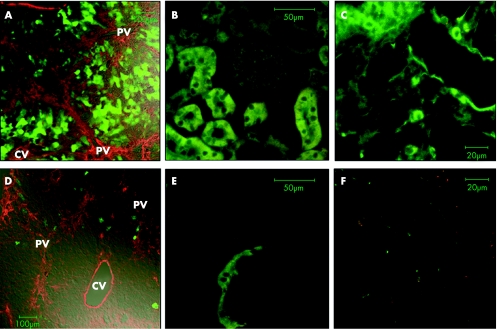
Figure 5 Cell type‐specific gene expression in the fibrotic liver tissue by using a tissue‐specific ×2(I) collagen gene enhancer sequence.75 When using the CAG expression unit as a control, enhanced green fluorescent protein (EGFP) fluorescence (green) was expressed in several parenchymal hepatocytes within the regenerating nodules (A) as well as in renal tubules (B) and pulmonary epithelium (C). By contrast, EGFP expression driven by the COL1A2 enhancer was observed mainly in activated hepatic stellate cells (HSCs) along the fibrotic septa but seldom in parenchymal hepatocytes (D). It was not expressed in kidney (E) or lung tissue (F) either. In panels A and D, expression of α‐smooth‐muscle actin (red), a marker for activated HSC, was examined by immunofluorescence staining. Note the weak autofluorescence signals observed in the kidney (panels B and E). CV, central vein; PV, portal vein.
On the other hand, the adenoviral system still has several limitations and problems, despite the cell type‐specific gene expression described above. Firstly, adenovirus is immunogenic and rapidly induces neutralising antibodies. Thus, it is not suitable for repeated injections in vivo. Secondly, the virus has dose‐related cytotoxicity, which raises safety concerns on clinical application. From this point of view, development of better delivery systems is needed to treat chronic diseases such as organ fibrosis more effectively. Another attempt for cell type‐specific fibrosis treatment uses modified human albumin as a selective carrier of antifibrotic reagents. For example, mannose‐6‐phosphate76 and a PDGF receptor‐recognising macromolecule77 have been tested for their binding capacity, specifically to activated HSC.
Problems and future perspectives
Despite the great advances in understanding the significant roles of TGFβ/Smad signals in the liver, there still remain several important issues to be dealt with. One of them is the differential roles of the three TGFβ isoforms in liver biology and pathobiology. These three isoforms exhibit essentially the same biological activities in vitro, but exert different biological effects in vivo. Most of the findings described above are observed with TGFβ1, and it is not clear whether they are also true for TGFβ2 and TGFβ3. It should be noted that there are two TGFβ type III receptor proteins, betaglycan and endoglin, both of which are expressed in HSC.78,79 As they bind to the different isoforms of TGFβ with different affinities and modulate the ligand binding to the type I and II receptors, they may play a critical role in modifying the TGFβ/Smad signals in various pathophysiological conditions in the liver.
The second issue is related to the heterogeneity of HSC and other collagen‐producing cells in the liver.80,81 In addition to HSC and myofibroblasts, portal fibroblasts have been implicated in the pathogenesis of biliary fibrosis. It has been shown that portal fibroblasts express a considerable amount of TGFβ2 and, unlike activated HSC, their growth is inhibited by TGFβ1 and TGFβ2, but not stimulated by PDGF treatment.82 Thus, TGFβ functions may be different depending on its isoforms, cellular context and the causes of liver injury. Another population of collagen‐producing cells in the liver is derived from bone marrow, which is described below.
In contrast with the expanding knowledge of molecular mechanisms regulating collagen gene transcription, the role of the TGFβ/Smad signal in the regulation of MMP gene expression has been much less understood. Most MMP genes possess a TGFβ inhibitory element in their promoter sequences. It has been shown that TGFβ modulates MMP13 expression in HSC by complex mechanisms involving p38 MAPK and other signalling pathways.83 In addition, reciprocal regulation of collagen and MMP expression by TGFβ is obviously an important but poorly understood area of investigation.84
With regard to the treatment of liver fibrosis, we may consider using endogenous factors or cells that can reverse fibrosis, rather than introducing exogenous genes and reagents. One such candidate is bone marrow‐derived cells expressing MMPs. It has recently been reported that infusion of bone marrow cells from syngeneic mice into cirrhotic animals resulted in migration of those cells into fibrotic liver.85 They not only stimulated liver regeneration by differentiating into parenchymal hepatocytes85 but also suppressed the progression of fibrosis by expressing MMP9.86 These results indicate that bone marrow transplantation could be a choice of treatment for advanced liver cirrhosis in humans. However, if the therapeutic derivation of autologous bone marrow cells and their differentiation into MMP‐expressing cells can be effectively achieved, it will be obviously a safer and less invasive approach for the treatment of liver fibrosis. On the other hand, others have reported that bone marrow‐derived cells contribute to the progression of liver fibrosis by expressing type I collagen in human87 and murine liver.88 As TGFβ, as well as other cytokines, is also involved in growth control and differentiation of stem/progenitor cells,89,90 a new concept of the treatment for liver fibrosis may arise from the discipline of stem cell biology by modulating TGFβ and Smad signalling in stem/progenitor cells.
Acknowledgements
We thank Dr Tomoyuki Nemoto at University of Fukui Faculty of Medical Sciences, Dr Atsuhito Nakao at University of Yamanashi Faculty of Medicine, Dr Kiyoshi Higashi at Sumitomo Chemical, Dr Kazuo Ikeda at Osaka City University Graduate School of Medicine and Ms Miwa Kushida at Tokai University School of Medicine for their contribution. We also thank Dr Francesco Ramirez at Child Health Institute of New Jersey for his kind cooperation and continuous encouragement throughout the work.
Abbreviations
BMP - bone morphogenetic protein
ECM - extracellular matrix
ERK - extracellular signal‐regulated kinase
HGF - hepatocyte growth factor
HSC - hepatic stellate cells
IFN - interferon
IgRE - IFN γ responsive element
JNK - c‐Jun N‐terminal kinase
LAP - latency‐associated peptide
MAPK - mitogen‐activated protein kinase
MMP - matrix metalloproteinase
PDGF - platelet‐derived growth factor
TbRE - TGF‐β‐responsive element
TGF - transforming growth factor
TNF - tumour necrosis factor
Footnotes
Competing interests: None declared.
References
- 1.Roberts A B, Lamb L C, Newton D L.et al Transforming growth factors: isolation of polypeptides from virally and chemically transformed cells by acid/ethanol extraction. Proc Natl Acad Sci USA 1980773494–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bissell D M, Roulot D, George J. Transforming growth factor‐β and the liver. Hepatology 200134859–867. [DOI] [PubMed] [Google Scholar]
- 3.Gressner A M, Weiskirchen R. Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF‐β as major players and therapeutic targets. J Cell Mol Med 20051076–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Massague J, Seoane J, Wotton D. Smad transcription factors. Gene Dev 2005192783–2810. [DOI] [PubMed] [Google Scholar]
- 5.Breitkopf K, Sawitza I, Westhoff J H.et al Thrombospondin 1 acts as a strong promoter of transforming growth factor β effects via two distinct mechanisms in hepatic stellate cells. Gut 200554673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondou H, Mushiake S, Etani Y.et al A blocking peptide for transforming growth factor‐β1 activation prevents hepatic fibrosis in vivo. J Hepatol 200339742–748. [DOI] [PubMed] [Google Scholar]
- 7.Okuno M, Akita K, Moriwaki H.et al Prevention of rat hepatic fibrosis by the protease inhibitor, camostat mesilate, via reduced generation of active TGF‐β. Gastroenterology 20011201784–1800. [DOI] [PubMed] [Google Scholar]
- 8.Nakao A, Afrakhte M, Moren A.et al Identification of Smad7, a TGFβ‐inducible antagonist of TGF‐β signalling. Nature 1997389631–635. [DOI] [PubMed] [Google Scholar]
- 9.Ebisawa T, Fukuchi M, Murakami G.et al Smurf1 interacts with transforming growth factor‐β type I receptor through Smad7 and induce receptor degradation. J Biol Chem 200127612477–12480. [DOI] [PubMed] [Google Scholar]
- 10.Inagaki Y, Nemoto T, Nakao A. Transcriptional activation of type I collagen gene during hepatic fibrogenesis. In: Okazaki I, Ninomiya Y, Friedman SL, Tanikawa K, eds. Extracellular matrix and the liver‐approach to gene therapy. New York: Academic Press, 2003233–248.
- 11.Inagaki Y, Truter S, Ramirez F. Transforming growth factor‐β stimulates α2(I) collagen gene expression through a cis‐acting element that contains an Sp1 binding site. J Biol Chem 199426914828–14834. [PubMed] [Google Scholar]
- 12.Greenwel P, Inagaki Y, Hu W.et al Sp1 is required for the early response of α2(I) collagen to transforming factor‐β1. J Biol Chem 199727219738–19745. [DOI] [PubMed] [Google Scholar]
- 13.Chen S ‐ J, Yuan W, Lo S.et al Interaction of Smad3 with a proximal Smad‐binding element of the human α2(I) procollagen gene promoter required for transcriptional activation by TGF‐β. J Cell Physiol 2000183381–392. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Oul J, Inagaki Y.et al Synergistic cooperation between Sp1 and Smad3/Smad4 mediates transforming growth factor β1 stimulation of α2(I)‐collagen (COL1A2) transcription. J Biol Chem 200027539237–39245. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh A K, Yuan W, Mori Y.et al Smad‐dependent stimulation of type I collagen gene expression in human skin fibroblasts by TGF‐β involves functional cooperation with p300/CBP transcriptional coactivators. Oncogene 2000193546–3555. [DOI] [PubMed] [Google Scholar]
- 16.Inagaki Y, Truter S, Greenwel P.et al Regulation of the α2(I) collagen gene transcription in fat‐storing cells derived from a cirrhotic liver. Hepatology 199522573–579. [PubMed] [Google Scholar]
- 17.Inagaki Y, Nemoto T, Nakao A.et al Interaction between GC box binding factors and Smad proteins modulates cell lineage‐specific α2(I) collagen gene transcription. J Biol Chem 200127616573–16579. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Gaca M D A, Swenson E S.et al Smad2 and Smad3 are differentially activated by TGF‐β in quiescent and activated hepatic stellate cells: constitutive nuclear localization of Smads in activated cells is TGF‐β independent. J Biol Chem 200327811721–11728. [DOI] [PubMed] [Google Scholar]
- 19.Tahashi Y, Matsuzaki K, Date M.et al Differential regulation of TGF‐β signal in hepatic stellate cells between acute and chronic rat liver injury. Hepatology 20023549–61. [DOI] [PubMed] [Google Scholar]
- 20.Inagaki Y, Mamura M, Kanamaru Y.et al Constitutive phosphorylation and nuclear localization of Smad3 are correlated with increased collagen gene transcription in activated hepatic stellate cells. J Cell Physiol 2001187117–123. [DOI] [PubMed] [Google Scholar]
- 21.Garcia‐Trevijano E R, Iraburu M J, Fontana L.et al Transforming growth factor β1 induces the expression of α1(I) procollagen mRNA by a hydrogen peroxide‐C/EBPβ‐dependent mechanism in rat hepatic stellate cells. Hepatology 199929960–970. [DOI] [PubMed] [Google Scholar]
- 22.Tsukada S, Westwick J K, Ikejima K.et al SMAD and p38 MAPK signaling pathways independently regulate α1(I) collagen gene expression in unstimulated and transforming growth factor‐β‐stimulated hepatic stellate cells. J Biol Chem 200528010055–10064. [DOI] [PubMed] [Google Scholar]
- 23.Inagaki Y, Truter S, Bou‐Gharios G.et al Activation of proα2(I) collagen promoter during hepatic fibrogenesis in transgenic mice. Biochem Biophys Res Commun 1998250606–611. [DOI] [PubMed] [Google Scholar]
- 24.Nomura M, Li E. Smad2 role in mesoderm formation, left‐right patterning and craniofacial development. Nature 1998393786–790. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Letterio J J, Lechleider R J.et al Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF‐β. EMBO J 1999181280–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piek E, Jun Ju W, Heyer J.et al Functional characterization of transforming growth factor β signaling in Smad2‐ and Smad3‐deficient fibroblasts. J Biol Chem 200127619945–19953. [DOI] [PubMed] [Google Scholar]
- 27.Schnabl B, Kweon Y O, Frederick J P.et al The role of Smad3 in mediating mouse hepatic stellate cell activation. Hepatology 20013489–100. [DOI] [PubMed] [Google Scholar]
- 28.Uemura M, Swenson E S, Gaca M D A.et al Smad2 and Smad3 play different roles in rat hepatic stellate cell function and α‐smooth muscle actin organization. Mol Biol Cell 2005164214–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inman G J, Nicolas F J, Callahan J F.et al SB‐431542 is a potent and specific inhibitor of transforming growth factor‐β superfamily type I activin receptor‐like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol 20026265–74. [DOI] [PubMed] [Google Scholar]
- 30.Kretzschmar M, Doody J, Massague J. Opposing BMP and EGF signalling pathways converge on the TGF‐β family mediator Smad1. Nature 1997389618–622. [DOI] [PubMed] [Google Scholar]
- 31.Kretzschmar M, Doody J, Timokhina I.et al A mechanism of repression of TGFβ/Smad signaling by oncogenic Ras. Gene Dev 199913804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanafusa H, Ninomiya‐Tsuji J, Masuyama N.et al Involvement of the p38 mitogen‐activated protein kinase pathway in transforming growth factor‐β‐induced gene expression. J Biol Chem 199927427161–27167. [DOI] [PubMed] [Google Scholar]
- 33.Abecassis L, Rogier E, Vazquez A.et al Evidence for a role of MSK1 in transforming growth factor‐β‐mediated responses through p38α and Smad signaling pathways. J Biol Chem 200427930474–30479. [DOI] [PubMed] [Google Scholar]
- 34.Engel M E, McDonnell M A, Law B K.et al Independent SMAD and JNK signaling in transforming growth factor‐β‐mediated transcription. J Biol Chem 199927437413–37420. [DOI] [PubMed] [Google Scholar]
- 35.Furukawa F, Matsuzaki K, Mori S.et al p38 MAPK mediates fibrogenic signal through Smad3 phosphorylation in rat myofibroblasts. Hepatology 200338879–889. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida K, Matsuzaki K, Mori S.et al Transforming growth factor‐β and platelet‐derived growth factor signal via c‐Jun N‐terminal kinase‐dependent Smad2/Smad3 phosphorylation in rat hepatic stellate cells after acute liver injury. Am J Pathol 20051661029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez‐Barbero A, Obreo J, Yuste L.et al Transforming growth factor‐β1 induces collagen synthesis and accumulation via p38 mitogen‐activated protein kinase (MAPK) pathway in cultured L6E9 myoblasts. FEBS Lett 2002513282–288. [DOI] [PubMed] [Google Scholar]
- 38.Hayashida T, Poncelet A ‐ C, Hubchak S C.et al TGF‐β1 activates MAP kinase in human mesangial cells: a possible role in collagen expression. Kidney Int 19991710–1720. [DOI] [PubMed]
- 39.Reunanen N, Foschi M, Han J.et al Activation of extracellular signal‐regulated kinase 1/2 inhibits type I collagen expression by human skin fibroblasts. J Biol Chem 200027534634–34639. [DOI] [PubMed] [Google Scholar]
- 40.Wang S, Hirschberg R. BMP7 antagonizes TGF‐beta‐dependent fibrogenesis in mesangial cells. Am J Physiol Renal Physiol 2003284F1006–F1013. [DOI] [PubMed] [Google Scholar]
- 41.Izumi N, Mizuguchi S, Inagaki Y.et al BMP‐7 opposes TGFβ1‐mediated collagen induction in mouse pulmonary myofibroblasts through Id2. Am J Physiol Lung Cell Mol Physiol 2006290L120–L126. [DOI] [PubMed] [Google Scholar]
- 42.Zeisberg M, Hanai J, Sugimoto H.et al BMP‐7 counteracts TGF‐beta1‐induced epithelial‐to‐mesenchymal transition and reverses chronic renal injury. Nat Med 20039964–968. [DOI] [PubMed] [Google Scholar]
- 43.Zeisberg M, Kalluri R. The role of epithelial‐to‐mesenchymal transition in renal fibrosis. J Mol Med 200482175–181. [DOI] [PubMed] [Google Scholar]
- 44.Rockey D C. Antifibrotic therapy in chronic liver disease. Clin Gastroenterol Hepatol 2005395–107. [DOI] [PubMed] [Google Scholar]
- 45.Inagaki Y, Truter S, Tanaka S.et al Overlapping pathways mediate the opposing actions of tumor necrosis factor‐α and transforming growth factor‐β on α2(I) collagen gene transcription. J Biol Chem 19952703353–3358. [DOI] [PubMed] [Google Scholar]
- 46.Greenwel P, Tanaka S, Penkov D.et al Tumor necrosis factor alpha inhibits type I collagen synthesis through repressive CCAAT/enhancer‐binding proteins. Mol Cell Biol 200020912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bitzer M, von Gersdorff G, Liang D.et al A mechanism of suppression of TGF‐β/SMAD signaling by NF‐κB/RelA. Genes Dev 200014187–197. [PMC free article] [PubMed] [Google Scholar]
- 48.Verrecchia F, Pessah M, Atfi A.et al Tumor necrosis factor‐α inhibits transforming growth factor‐β/Smad signaling in human dermal fibroblasts via AP‐1 activation. J Biol Chem 200027530226–30231. [DOI] [PubMed] [Google Scholar]
- 49.Rockey D C, Maher J J, Jarnagin W R.et al Inhibition of rat hepatic lipocyte activation in culture by interferon‐γ. Hepatology 199216776–784. [DOI] [PubMed] [Google Scholar]
- 50.Mallat A, Preaux A ‐ M, Blazejewski S.et al Interferon alpha and gamma inhibit proliferation and collagen synthesis of human Ito cells in culture. Hepatology 1995211003–1010. [PubMed] [Google Scholar]
- 51.Czaja M J, Weiner F R, Takahashi S.et al γ‐interferon treatment inhibits collagen deposition in murine Schistosomiasis. Hepatology 198910795–800. [DOI] [PubMed] [Google Scholar]
- 52.Baroni G S, D'Ambrosio L, Curto P.et al Interferon gamma decreases hepatic stellate cell activation and extracellular matrix deposition in rat liver fibrosis. Hepatology 1996231189–1199. [DOI] [PubMed] [Google Scholar]
- 53.Higashi K, Kouba D J, Song Y ‐ J.et al A proximal element within the human α2(I) collagen (COL1A2) promoter, distinct from the tumor necrosis factor‐α response element, mediates transcriptional repression by interferon‐γ. Matrix Biol 199816447–456. [DOI] [PubMed] [Google Scholar]
- 54.Higashi K, Inagaki Y, Suzuki N.et al Y‐box‐binding protein YB‐1 mediates transcriptional repression of human α2(I) collagen gene expression by interferon‐γ. J Biol Chem 20032785156–5162. [DOI] [PubMed] [Google Scholar]
- 55.Ulloa L, Doody J, Massague J. Inhibition of transforming growth factor‐β/Smad signalling by the interferon‐γ/STAT pathway. Nature 1999397710–713. [DOI] [PubMed] [Google Scholar]
- 56.Ghosh A K, Yuan W, Mori Y.et al Antagonistic regulation of type I collagen gene expression by interferon‐γ and transforming growth factor‐β. Integration at the level of p300/CBP transcriptional coactivators. J Biol Chem 200127611041–11048. [DOI] [PubMed] [Google Scholar]
- 57.Higashi K, Inagaki Y, Fujimori K.et al Interferon‐γ interferes with transforming growth factor‐β signaling through direct interaction of YB‐1 with Smad3. J Biol Chem 200327843470–43479. [DOI] [PubMed] [Google Scholar]
- 58.Dooley S, Said H M, Gressner A M.et al Y‐box protein‐1 is the crucial mediator of antifibrotic interferon‐γ effects. J Biol Chem 20062811784–1795. [DOI] [PubMed] [Google Scholar]
- 59.Hiramatsu N, Hayashi N, Kasahara A.et al Improvement of liver fibrosis in chronic hepatitis patients treated with natural interferon alpha. Hepatology 199522135–142. [DOI] [PubMed] [Google Scholar]
- 60.Suou T, Hosho K, Kishimoto Y.et al Long‐term decrease in serum N‐terminal propeptide of type III procollagen in patients with chronic hepatitis C treated with interferon alpha. Hepatology 199522426–431. [PubMed] [Google Scholar]
- 61.Manabe N, Chevallier M, Chossegros P.et al Interferon‐α 2b therapy reduces liver fibrosis in chronic non‐A, non‐B hepatitis: a quantitative histological evaluation. Hepatology 1993181344–1349. [PubMed] [Google Scholar]
- 62.Sobesky R, Mathurin P, Charlotte F.et al Modeling the impact of interferon alpha treatment on liver fibrosis progression in chronic hepatitis C: a dynamic view. Gastroenterology 1999116378–386. [DOI] [PubMed] [Google Scholar]
- 63.Inagaki Y, Nemoto T, Kushida M.et al Interferon α down‐regulates collagen gene transcription and suppresses experimental hepatic fibrosis in mice. Hepatology 200338890–899. [DOI] [PubMed] [Google Scholar]
- 64.Matsuda Y, Matsumoto K, Ichida T.et al Hepatocyte growth factor suppresses the onset of liver cirrhosis and abrogates lethal hepatic dysfunction in rats. J Biochem 1995118643–649. [DOI] [PubMed] [Google Scholar]
- 65.Yasuda H, Imai E, Shiota A.et al Antifibrotic effect of a deletion variant of hepatocyte growth factor on liver fibrosis in rats. Hepatology 199624636–642. [DOI] [PubMed] [Google Scholar]
- 66.Ueki T, Kaneda Y, Tsutsui H.et al Hepatocyte growth factor gene therapy of liver cirrhosis in rats. Nat Med 19995226–230. [DOI] [PubMed] [Google Scholar]
- 67.Ozaki I, Zhao G, Mizuta T.et al Hepatocyte growth factor induces collagenase (matrix metalloproteinase‐1) via the transcription factor Ets‐1 in human hepatic stellate cells. J Hepatol 200236169–178. [DOI] [PubMed] [Google Scholar]
- 68.Kim W ‐ H, Matsumoto K, Bessho K.et al Growth inhibition and apoptosis in liver myofibroblasts promoted by hepatocyte growth factor leads to resolution from liver cirrhosis. Am J Pathol 20051661017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qi Z, Atsuchi N, Ooshima A.et al Blockade of type β transforming growth factor signaling prevents liver fibrosis and dysfunction in the rat. Proc Natl Acad Sci USA 1999962345–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakamura T, Sakata R, Ueno T.et al Inhibition of transforming growth factor‐β prevents progression of liver fibrosis and enhances hepatocyte regeneration in dimethylnitrosamine‐treated rats. Hepatology 200032247–255. [DOI] [PubMed] [Google Scholar]
- 71.George J, Roulot D, Koteliansky V E.et al In vivo inhibition of rat stellate cell activation by soluble transforming growth factor β type II receptor: a potential new therapy for hepatic fibrosis. Proc Natl Acad Sci USA 19999612719–12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ueno H, Sakamoto T, Nakamura T.et al A soluble transforming growth factor beta receptor expressed in muscle prevents liver fibrogenesis and dysfunction in rats. Hum Gene Ther 20001133–42. [DOI] [PubMed] [Google Scholar]
- 73.Dooley S, Hamzavi J, Breitkopf K.et al Smad7 prevents activation of hepatic stellate cells and liver fibrosis in rats. Gastroenterology 2003125178–191. [DOI] [PubMed] [Google Scholar]
- 74.Janoschek N, van de Leur E, Gressner A M.et al Induction of cell death in activated hepatic stellate cells by targeted gene expression of the thymidine kinase/ganciclovir system. Biochem Biophys Res Commun 20043161107–1115. [DOI] [PubMed] [Google Scholar]
- 75.Inagaki Y, Kushida M, Higashi K.et al Cell type‐specific intervention of TGF‐β/Smad signaling suppresses collagen gene expression and hepatic fibrosis in mice. Gastroenterology 2005129259–268. [DOI] [PubMed] [Google Scholar]
- 76.Beljaars L, Molema G, Weert B.et al Albumin modified with mannose 6‐phosphate: a potential carrier for selective delivery of antifibrotic drugs to rat and human hepatic stellate cells. Hepatology 1999291486–1493. [DOI] [PubMed] [Google Scholar]
- 77.Beljaars L, Weert B, Geerts A.et al The preferential homing of a platelet derived growth factor receptor‐recognizing macromolecule to fibroblast‐like cells in fibrotic tissue. Biochem Pharmacol 2003661307–1317. [DOI] [PubMed] [Google Scholar]
- 78.Friedman S L, Yamasaki G, Wong L. Modulation of transforming growth factor β receptors of rat lipocytes during the hepatic wound healing process. Enhanced binding and reduced gene expression accompany cellular activation in culture and in vivo. J Biol Chem 199426910551–10558. [PubMed] [Google Scholar]
- 79.Meurer S K, Tihaa L, Lahme B.et al Identification of endoglin in rat hepatic stellate cells: new insights into transforming growth factor β receptor signaling. J Biol Chem 20052803078–3087. [DOI] [PubMed] [Google Scholar]
- 80.Knittel T, Kobold D, Saile B.et al Rat liver myofibroblasts and hepatic stellate cells: different cell populations of the fibroblast lineage with fibrogenic potential. Gastroenterology 19991171205–1221. [DOI] [PubMed] [Google Scholar]
- 81.Magness S T, Bataller R, Yang L.et al A dual reporter gene transgenic mouse demonstrates heterogeneity in hepatic fibrogenic cell populations. Hepatology 2004401151–1159. [DOI] [PubMed] [Google Scholar]
- 82.Wells R G, Kruglov E, Dranoff J A. Autocrine release of TGF‐β by portal fibroblasts regulates cell growth. FEBS Lett 2004559107–110. [DOI] [PubMed] [Google Scholar]
- 83.Lechuga C G, Hernandes‐Nazara Z H, Dominguez J ‐ A.et al TGF‐β1 modulates matrix metalloproteinase‐13 expression in hepatic stellate cells by complex mechanisms involving p38MAPK, PI3‐kinase, Akt, and p70S6k. Am J Physiol Gastrointest Liver Physiol 2004287G974–G987. [DOI] [PubMed] [Google Scholar]
- 84.Shaefer B, Rivas‐Estilla A M, Meraz‐Cruz N.et al Reciprocal modulation of matrix metalloproteinase‐13 and type I collagen genes in rat hepatic stellate cells. Am J Pathol 20031621771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Terai S, Sakaida I, Yamamoto N.et al An in vivo model for monitoring trans‐differentiation of bone marrow cells into functional hepatocytes. J Biochem (Tokyo) 2003134551–558. [DOI] [PubMed] [Google Scholar]
- 86.Sakaida I, Terai S, Yamamoto N.et al Transplantation of bone marrow cells reduces CCl4‐induced liver fibrosis in mice. Hepatology 2004401304–1311. [DOI] [PubMed] [Google Scholar]
- 87.Forbes S J, Russo F P, Rey V.et al A significant proportion of myofibroblasts are of bone marrow origin in human liver fibrosis. Gastroenterology 2004126955–963. [DOI] [PubMed] [Google Scholar]
- 88.Russo F P, Alison M R, Bigger B W.et al The bone marrow functionally contribute to liver fibrosis. Gastroenterology 20061301807–1821. [DOI] [PubMed] [Google Scholar]
- 89.Lowes K N, Croager E J, Olynyk J K.et al Oval cell‐mediated liver regeneration: role of cytokines and growth factors. J Gastroenterol Hepatol 2003184–12. [DOI] [PubMed] [Google Scholar]
- 90.Mishra L, Derynck R, Mishra B. Transforming growth factor‐β signaling in stem cells and cancer. Science 200531068–71. [DOI] [PubMed] [Google Scholar]