Abstract
A classification of fusiform neocortical interneurons (n = 60) was performed with an unsupervised cluster analysis based on the comparison of multiple electrophysiological and molecular parameters studied by patch-clamp and single-cell multiplex reverse transcription–PCR in rat neocortical acute slices. The multiplex reverse transcription–PCR protocol was designed to detect simultaneously the expression of GAD65, GAD67, calbindin, parvalbumin, calretinin, neuropeptide Y, vasoactive intestinal peptide (VIP), somatostatin (SS), cholecystokinin, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid, kainate, N-methyl-d-aspartate, and metabotropic glutamate receptor subtypes. Three groups of fusiform interneurons with distinctive features were disclosed by the cluster analysis. The first type of fusiform neuron (n = 12), termed regular spiking nonpyramidal (RSNP)-SS cluster, was characterized by a firing pattern of RSNP cells and by a high occurrence of SS. The second type of fusiform neuron (n = 32), termed RSNP-VIP cluster, predominantly expressed VIP and also showed firing properties of RSNP neurons with accommodation profiles different from those of RSNP-SS cells. Finally, the last type of fusiform neuron (n = 16) contained a majority of irregular spiking-VIPergic neurons. In addition, the analysis of glutamate receptors revealed cell-type-specific expression profiles. This study shows that combinations of multiple independent criteria define distinct neocortical populations of interneurons potentially involved in specific functions.
Because, in part, of their diversity, the function of neuron subtypes in the physiology of the neocortex is still poorly understood. A better knowledge of the different neuronal populations that compose this heterogeneous brain structure may therefore contribute to elucidating their specific role.
Attempts to classify neurons rely on several independent criteria (morphological, physiological, and molecular). In the neocortex, neurons are classified as pyramidal cells or nonpyramidal cells according to their morphology. Pyramidal cells accumulate glutamate and constitute the main class of excitatory projecting neurons (1). In contrast, nonpyramidal cells, also termed interneurons, are mainly inhibitory γ-aminobutyric acid-ergic neurons with a short axon involved in local circuits (2) and have a large diversity of morphology (3). The expression of biochemical markers has been used to define different classes of nonpyramidal cells. The distribution of three calcium binding proteins, calbindin (CB), parvalbumin (PV), and calretinin (CR), and four neuropeptides, neuropeptide Y (NPY), vasoactive intestinal peptide (VIP), somatostatin (SS), and cholecystokinin (CCK), defines partially overlapping groups of interneurons (4–12). In addition to this morphological and molecular diversity, nonpyramidal cells also have a large repertoire of firing behaviors (13–18), such as fast spiking (FS), regular spiking nonpyramidal (RSNP), or irregular spiking (IS).
Some types of interneurons have been defined based on a correlation between the morphology, the pattern of expression of molecular markers, and the firing properties (16–18). However, for the majority of the interneurons, no clear correlation could be established between morphological, molecular, and electrophysiological properties (14–17).
Among other morphological types, fusiform interneurons have a large diversity of expression pattern of the biochemical markers and of firing properties (16–18). This diversity presumably reflects the fact that, although morphologically homogeneous, this group of cells is composed of more than one neuronal subtype.
This question was addressed by using a cluster analysis of fusiform interneurons for which multiple electrophysiological and molecular parameters were determined by patch-clamp and single-cell multiplex reverse transcription–PCR (RT-mPCR). This type of analysis groups together cells with similar properties and segregates cells that are very different. Pyramidal (n = 9) and FS (n = 16) cells were included as controls to validate the RT-mPCR protocol and to select the parameters of the cluster analysis. By using this approach, different subtypes of fusiform cells with distinctive properties were disclosed. In conjunction, the expression patterns of glutamate receptors of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid, kainate, N-methyl-d-aspartate, and metabotropic subtypes were determined for all cell types.
Materials and Methods
Recordings.
Parasagittal acute slices (300-μm-thick) were obtained from frontoparietal cortices of 12- to 34-day-old Wistar rats. Patch pipettes (3–5 MΩ) pulled from borosilicate glass were filled with 8 μl of autoclaved RT-PCR internal solution. Whole-cell recordings were made from layer II, III, and V neurons selected under visual control by using Nomarski optics and infrared videomicroscopy (19). The cell content was harvested under visual control and expelled into a test tube; RT was performed overnight at 37°C as described (20).
Solutions.
The bath solution contained (in mM): 121 NaCl, 2.5 KCl, 1.25 NaH2PO4, 2 CaCl2, 1 MgCl2, 26 NaHCO3, 20 glucose, and 5 pyruvate, bubbled with a mixture of 95% O2/5% CO2. Pipettes were filled with solution containing (in mM): 144 potassium gluconate, 3 MgCl2, 0.2 EGTA, and 10 Hepes (pH 7.2; 285/295 mosmol) adjusted with KOH.
mPCR.
The RT-mPCR protocol was designed to detect simultaneously and qualitatively at the single-cell level the expression of GAD65, GAD67, CB, CR, PV, NPY, VIP, SS, CCK, GluR1–7, KA1, KA2, NR2A–D, and metabotropic glutamate receptor (mGluR)1–8 mRNAs. The two amplification steps were performed essentially as described (17, 21). The PCR primers used for GAD65, GAD67, CB, CR, PV, NPY, VIP, SS, and CCK amplification were described in ref. 17, and those for GluR1–7, KA1, KA2, and NR2A-D were described in ref. 18. The following sets of PCR primers were used for mGluRs detection (from 5′ to 3′): CCAGATGAACAAAAGCGGAATG (sense mGluR1), CAAGAAAAGGCGATGGCTATGA (antisense mGluR1), TCTGTCCTGATGGCTCCTATGC (sense mGluR2), ACACGGGCACTGGGTTTCTG (antisense mGluR2), CAAGAAAAC-ATCCCACTGCTCA (sense mGluR3), TAGGACTTGCGG-ATGTTGGAGC (antisense mGluR3), GGACACTTGACCC-CCGCTTT (sense mGluR4), CCCATTGGGCCTGAAGTTGC-(antisense mGluR4), AATGGATGATGACGAAGTGTGG (sense mGluR5), AGACCGAGGCAGGCAAACAC (antisense mGluR5), GCTCCGGTGGCCAGTCAGAT (sense mGluR6), TCGCCTGACCACCGTAGAAC (antisense mGluR6), GTCTTCATTTGGTTTGGGGTTG (sense mGluR7), GGGCCTGTCACTGGGTTTGT (antisense mGluR7), TCGGAGTGTTTGTGTGGTTTGT (sense mGluR8), and TGGTCTGTCATTTCCCTTTTGG (antisense mGluR8), generating PCR fragments of 266, 435, 364, 451, 293, 414, 503, and 508 bp for mGluR1–8, respectively. Each individual second step PCR (10 μl) was then run on a 1.5% agarose gel with Φ×174 cut by HaeIII as molecular mass markers and stained with ethidium bromide. All PCR primers used in the present work were designed to amplify cDNA regions spanning at least one intron. The sensitivity of the RT-mPCR procedure was tested on 500 pg of total cortical RNAs (not shown).
Identification of the PCR Products.
PCR-generated fragments obtained from each cell were dot blotted. The Southern blots were probed with the specific oligonucleotides described in refs. 17 and 18; for mGluR, the following oligoprobes were used: GCCTTGCTTAAAGGGTCAGATT (mGluR1), CCCACAGTGATGCTCCTACAGCT (mGluR2), CAGGAAGCACGGCTGCGT (mGluR3), CCACCATGTCCAACAAGTTCAC (mGluR4), ACCATGTGAGAAAGGCCAAATAA (mGluR5), CCAATGGGCAGAGGCCCT (mGluR6), ATCAAGGCTGTCACACAAACCC (mGluR7), and AGCTGCTACCATGCAAAGCAAA (mGluR8).
Electrophysiological Analysis.
The following electrophysiological parameters were measured as described (17). Those parameters were (i) input resistance, (ii and iii) amplitude of the first and the second spike, respectively, (iv) spike amplitude reduction, (v and vi) duration of the first and the second spike, respectively, (vii) spike duration increase, (viii and ix) amplitude of the afterpotential (AHP) of the first and the second spike, respectively, and (x) frequency adaptation occurring during the first 200 ms of discharge (early adaptation), and (xi) frequency adaptation occurring after the first 200 ms of discharge (late adaptation). The input resistance was determined for small intensities of hyperpolarizing current eliciting no or very weak sag. In some neurons, a pronounced reduction of the amplitude of the action potentials was followed by an increase of the spike amplitude during an 800-ms depolarization protocol. This firing property was measured as the difference between the peak of the smallest action potential and the peak of the following biggest action potential and was termed accommodative hump (xii). Two additional parameters—the minimal discharge frequency (xiii) observed during 800 ms of depolarization for a current step eliciting an initial firing frequency of 100 Hz and the time at which this minimal frequency occurred (xiv)—took into account irregularities in the discharge.
Statistical Analysis.
To classify the cells, unsupervised clustering was performed by using Ward's method (22). Briefly, this analysis consists in first grouping the closest individuals (each of them being represented by a point in a multidimensional space) by using the matrix of their Euclidean distances. Then, at each stage, the number of groups is reduced by one (or more) through merger of two groups (or individuals) whose combination gives the least possible increase in the within-group sum of squared deviation. This method is usually implemented through updating a stored matrix of Euclidean distances between cluster centroids (23). Average within-cluster distance shown in Fig. 1 represents the Euclidean distance between the centroids of the merged clusters (24).
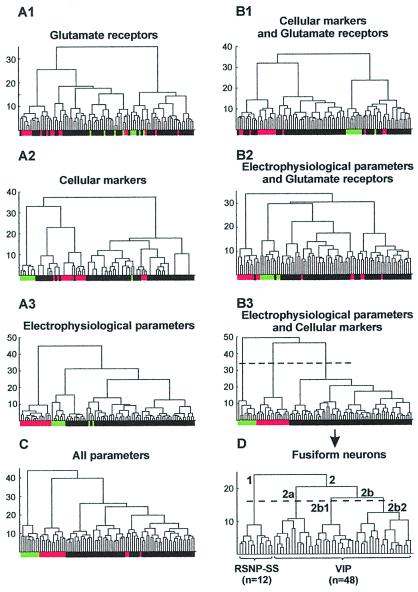
Figure 1.
Comparison of different cluster analyses applied to neocortical fusiform interneurons (black boxes, n = 60) as well as to pyramidal (green boxes, n = 9) and FS (red boxes, n = 16) cells taken as controls. For each diagram, the x axis represents the individuals, and the y axis represents the average within-cluster linkage distance. Dotted lines in B3 and D indicate the limits between clusters as suggested by the Thorndike procedure (see Materials and Methods). (A) Analyses based on the expression profiles of glutamate receptors (A1), of cellular markers (A2), or on electrophysiological properties (A3). (B and C) Analyses based on the combination of two of three sets of parameters (B1, B2, and B3) or on all parameters (C). Note that the control pyramidal and FS cells were segregated only by the combination of electrophysiological parameters and of cellular markers and that the expression of glutamate receptors blurs the classification. (D) Cluster analysis with the same parameters as in B3 restricted to the fusiform cell population. This analysis disclosed three groups of fusiform RSNP cells mainly expressing SS (RSNP-SS; branch labeled 1; n = 12) or mainly expressing VIP (branch labeled 2; n = 48). Within the group of VIP expressing cells, most of neurons of branch 2a were IS interneurons, and the majority of cells in branch 2b were RSNP cells. Within branch 2b, two subpopulations of RSNP-VIP (branches 2b1 and 2b2) were suggested by the Thorndike procedure.
Three sets of parameters were used independently or in different combinations to perform different cluster analyses that were compared by their ability to discriminate pyramidal and FS cells taken as controls. The first set consisted in 14 electrophysiological parameters (listed in Table 1 and above). The second set of parameters grouped nine interneurons' selective cellular markers (GAD65, GAD67, CB, PV, CR, NPY, VIP, SS, and CCK). Finally, the last set of parameters included the glutamate receptors [25 parameters: GluR1–4 (flip and flop), GluR5–7, KA1, KA2, NR2A–D, and mGluR1–8]. For the molecular data, the presence of a given PCR product was digitized by 1, and its absence was digitized by 0. Molecular and electrophysiological parameters were standardized by centering and reducing all of the values. The cluster analyses were run on statistica software (Statsoft, Tulsa, OK).
Table 1.
Electrophysiological properties of five types of cortical neurons
| Electrophysiological parameters | Pyr (n = 9) | FS (n = 16) | RSNP-SS (n = 12) | RSNP-VIP (n = 32) | IS (n = 16) |
|---|---|---|---|---|---|
| (i) Input resistance, MΩ | 247 ± 88 | 203 ± 85 | 298 ± 108 | 430 ± 139 | 395 ± 157 |
| FS, Pyr, RSNP-SS ≪ IS, RSNP-VIP | |||||
| (ii) First spike amplitude, mV | 89.9 ± 10.2 | 80.8 ± 9.6 | 88.1 ± 6.0 | 91.6 ± 6.6 | 83.4 ± 10.0 |
| FS, IS < Pyr, RSNP-SS, RSNP-VIP | |||||
| (iii) Second spike amplitude, mV | 80.3 ± 11.4 | 76.6 ± 8.6 | 85.8 ± 5.6 | 84.6 ± 6.5 | 71.2 ± 10.2 |
| FS, IS ≪ RSNP-VIP, RSNP-SS | |||||
| (iv) Amplitude reduction, % | 10.7 ± 7.7 | 5.0 ± 3.5 | 2.5 ± 3.4 | 7.5 ± 6.1 | 14.5 ± 8.1 |
| RSNP-SS ≪ RSNP-VIP, Pyr, IS | |||||
| (v) First spike duration, ms | 1.27 ± 0.33 | 0.43 ± 0.12 | 0.48 ± 0.15 | 0.69 ± 0.23 | 0.54 ± 0.23 |
| FS, RSNP-SS, IS ≪ RSNP-VIP ≪ Pyr | |||||
| (vi) Second spike duration, ms | 2.01 ± 0.93 | 0.43 ± 0.15 | 0.51 ± 0.15 | 0.78 ± 0.31 | 0.61 ± 0.26 |
| FS, RSNP-SS ≪ RSNP-VIP ≪ Pyr and IS ≪ Pyr | |||||
| (vii) Duration increase, % | 58.3 ± 48.2 | −2.4 ± 12.7 | 6.6 ± 10.1 | 10.0 ± 9.7 | 15.0 ± 15.6 |
| FS, RSNP-SS, RSNP-VIP, IS ≪ Pyr and FS ≪ RSNP-VIP, IS | |||||
| (viii) First spike AHP, mV | −2.6 ± 2.6 | −21.5 ± 3.1 | −12.2 ± 4.8 | −13.6 ± 4.0 | −11.5 ± 3.5 |
| FS < RSNP-VIP, RSNP-SS, IS ≪ Pyr | |||||
| (ix) Second spike AHP, mV | −8.2 ± 4.4 | −22.8 ± 2.5 | −11.8 ± 4.2 | −14.2 ± 4.3 | −14.4 ± 4.2 |
| FS ≪ IS, RSNP-VIP ≪ Pyr and FS < RSNP-SS | |||||
| (x) Early adaptation, % | 77.1 ± 7.9 | 19.6 ± 18.0 | 58.3 ± 18.0 | 61.1 ± 15.4 | 85.4 ± 14.5 |
| FS < RSNP-SS, RSNP-VIP ≪ Pyr, IS | |||||
| (xi) Late adaptation, % | 4.2 ± 2.3 | 7.3 ± 8.7 | 10.8 ± 9.4 | 8.8 ± 7.0 | 8.1 ± 12.8 |
| n.s. | |||||
| (xii) Accommodative hump, mV | 12.0 ± 4.0 | 3.0 ± 3.7 | 1.8 ± 2.8 | 7.1 ± 4.8 | 3.2 ± 3.8 |
| FS, RSNP-SS, IS ≪ RSNP-VIP ≪ Pyr | |||||
| (xiii) Minimal frequency, Hz | 18.5 ± 5.4 | 57.4 ± 24.2 | 28.0 ± 8.6 | 27.4 ± 12.3 | 14.7 ± 22.3 |
| IS, Pyr < RSNP-SS, RSNP-VIP ≪ FS | |||||
| (xiv) Time for minimal frequency, ms | 774 ± 66 | 749 ± 128 | 735 ± 113 | 726 ± 112 | 464 ± 140 |
| IS ≪ Pyr, FS, RSNP-SS, RSNP-VIP | |||||
Values are means ± SD. n, number of cells; Pyr, pyramidal cells; <, significantly smaller with P = 0.05; ≪, significantly smaller with P = 0.01; n.s., not statistically different. Statistically significant differences were determined by using the Mann–Whitney U test. Electrophysiological parameters were measured as described in Materials and Methods.
The final number of clusters was suggested by the Thorndike procedure (25). Briefly, the average within-cluster distance is plotted at each stage of the amalgamation schedule, resulting in a decrease in the average within-cluster distance as the number of clusters increases. The final number of classes (or cell types) is determined at the stage where the maximal decrease is reached in this plot.
Results
Classification of Neocortical Neurons.
The aim of the present study was the identification of possible subtypes of fusiform interneurons. To validate our analysis method, pyramidal neurons and FS cells were also included as controls.
Fusiform neurons (n = 60) in layers II, III, and V of the neocortex were selected according to their morphology as seen with infrared videomicroscopy. Neurons with a vertically oriented soma were considered for further analysis when their vertical axis was at least twice their horizontal axis. Pyramidal neurons (n = 9) were selected according to the triangular shape of their soma and their firing properties (17). FS cells (n = 16) were identified by their firing properties.
Neocortical neurons (n = 85) were electrophysiologically characterized and subsequently analyzed by single-cell RT-mPCR for the expression of 30 different genes that included cellular markers and glutamate receptors (see Materials and Methods). The classification of neocortical neurons was performed by using unsupervised cluster analyses based on different combinations of parameters (see Materials and Methods).
Fig. 1 shows the tree diagrams of cluster analyses applied to the sample of 85 neocortical neurons, based on the expression profile of glutamate receptors (Fig. 1A1) or of cellular markers (Fig. 1A2), on electrophysiological parameters (Fig. 1A3), or on different combinations of these parameters (Fig. 1B1–3 and C). Only the combination of cellular markers and electrophysiological parameters (Fig. 1B3) allowed the segregation of pyramidal and FS cells from fusiform interneurons. Therefore, this combination of parameters was used to perform a subsequent analysis restricted to the 60 fusiform interneurons (Fig. 1D). The Thorndike procedure (see Materials and Methods) suggested four clusters of fusiform neurons, corresponding to branches 1, 2a, 2b1, and 2b2 in the tree diagram (Fig. 1D). The first cluster (Fig. 1D, branch 1) was composed of 12 RSNP neurons. Because 11 of 12 neurons expressed SS, this cluster was termed RSNP-SS cells. The three other clusters (Fig. 1D, branches 2a, 2b1, and 2b2 forming branch 2) contained the majority of fusiform neurons (48 of 60) characterized by a high occurrence of VIP (45 of 48 cells; see also Fig. 3). Cells segregated on branch 2a (n = 14) contained a majority of IS neurons (11 of 14; Fig. 1D); the 3 other neurons were RSNP cells. The remaining fusiform neurons (n = 34; branch 2b) had RSNP firing properties (except for 5 IS interneurons). Because the only significant differences (P = 0.05) observed between 2b1 and 2b2 RSNP cells were that 2b1 cells had slower action potential, smaller AHPs, more pronounced accommodative hump (see Materials and Methods), and lower and higher occurrence of CR and NPY, respectively, than 2b2 cells, these two cell clusters were merged and termed RSNP-VIP cluster.
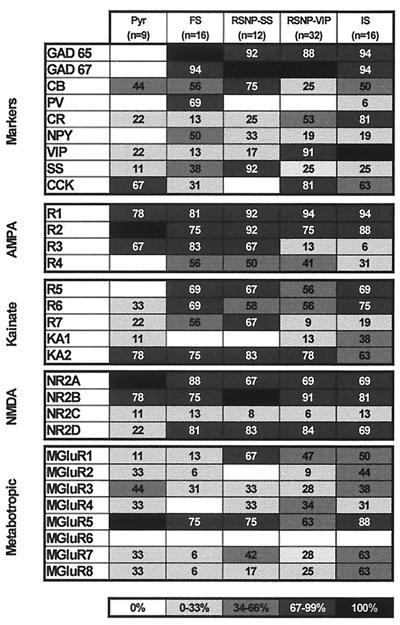
Figure 3.
Expression patterns of molecular markers and glutamate receptors in different classes of neocortical neurons. The percentage of neurons expressing a given mRNA is represented by a gray scale, as indicated on the bottom of the figure. Pyramidal cells (Pyr) did not express GADs in contrast to the other classes of neurons. FS cells consistently expressed PV and NPY. The majority of RSNP-SS cells expressed SS, whereas most of RSNP-VIP and IS cells expressed VIP. The distribution of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunits revealed that the five classes of neurons frequently expressed GluR1 and GluR2. GluR3 was very rare in RSNP-VIP and IS. Pyramidal cells lacked GluR4 subunits. The distribution of kainate receptor subunits showed that KA2 subunit was the most frequently observed subunit. GluR5 subunit was not expressed in pyramidal cells. Distribution of N-methyl-d-aspartate (NMDA) subunits indicated that NR2A and NR2B were detected frequently in the five classes of cells. NR2D was more frequent in interneurons than in pyramidal cells. The distribution of mGluRs showed that mGluR1 was frequent in fusiform cells. mGluR5 was the most frequently expressed mGluR. Group III mGluRs were rare in FS cells.
Thus, this cluster analysis disclosed three main groups of fusiform neurons: RSNP-SS, IS, and RSNP-VIP cells.
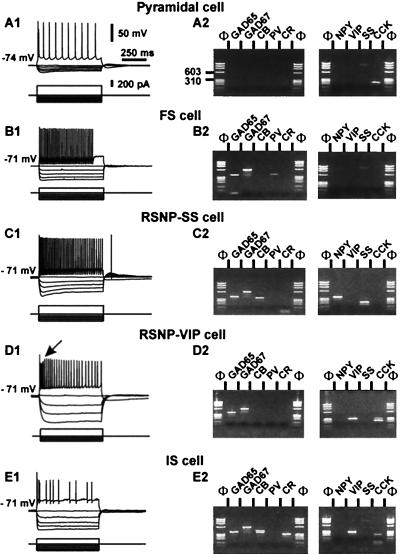
The molecular and electrophysiological analyses of members of these three neuronal classes and of pyramidal and FS cells are shown in Figs. 2 and 3. When depolarized, the cell shown in Fig. 2A1 showed distinctive long-duration action potentials of pyramidal cells (see Table 1). The molecular analysis of the pyramidal cell showed only CCK (Fig. 2A2) in agreement with previous reports (17, 26). Pyramidal cells did not express GADs, PV, or NPY (Fig. 3).
Figure 2.
Electrophysiological and biochemical analysis of five types of neocortical neurons. (Left) The firing properties of the five neurons. Membrane potentials were adjusted as indicated to the left of the recordings. (Right) The molecular analysis of the cells shown at Left. PCR products were resolved by agarose gel electrophoresis with Φ×174/HaeIII as a molecular mass marker. (A) Pyramidal neurons. (A1) In response to depolarizing current step (300 pA), the pyramidal cell fired action potentials with a marked frequency adaptation (Upper). (A2) RT-mPCR analysis showing only the expression of CCK mRNA. The faint band (≈800 bp) observed in the SS lane corresponds to the genomic amplification of SS. (B) FS interneuron. (B1) Note the high frequency, nonadapting discharge of fast action potentials with large hyperpolarized afterpotentials (Upper; 150 pA). (B2) The same FS cell expressed GAD65, GAD67, and PV. (C) RSNP-SS cell. (C1) Injection of a depolarizing current step produced an accommodating discharge of action potentials (Upper; 250 pA). Note the progressive reduction in the amplitude of the action potentials during the discharge. (C2) GAD65, GAD67, CB, NPY, and SS were detected in this neuron. (D) RSNP-VIP neuron. (D1) Injection of depolarizing current (Upper; 200 pA) elicited the firing of action potentials with a marked frequency adaptation. Note the augmentation after a marked reduction of spike amplitude (arrow). (D2) Agarose gel showing the expression of GAD65, GAD67, VIP, and CCK mRNAs. (E) IS interneuron. (E1) In response to depolarizing current, the neuron discharged an initial burst of action potentials followed by irregularly emitted action potentials. (E2) The agarose gel shows the expression of GAD65, GAD67, CB, CR, VIP, and CCK mRNAs.
The neuron shown in Fig. 2B showed fast action potentials with large AHPs and little frequency adaptation distinctive of FS cells (see Fig. 2B1 Upper and Table 1). The RT-mPCR analysis of the FS neuron revealed the expression of GAD65, GAD67, and PV mRNAs (Fig. 2B2 and Fig. 3). In the population of FS cells, GAD was always expressed (Fig. 3; refs. 15–17).
Fig. 2C illustrates the analysis of an RSNP-SS cell. This cell fired action potentials with a marked frequency adaptation (49.5%) distinctive of RSNP-SS neurons (Table 1) and no accommodative hump (Fig. 2C1 Upper). The molecular analysis of an RSNP-SS neuron (Fig. 2C2) showed the expression of GAD65, GAD67, CB, NPY, and SS. All RSNP-SS cells expressed GAD mRNAs (Fig. 3). SS was observed more frequently (P = 0.01) in RSNP-SS neurons than in other cell types (Fig. 3).
Fig. 2D shows an example of RSNP-VIP neuron firing action potentials with marked frequency adaptation (72.0%). This neuron had a pronounced accommodative hump (Fig. 2D1 Upper, arrow). This feature was a distinctive firing property of RSNP-VIP neurons in addition to their high input resistance (Table 1). The agarose gel of Fig. 2D2 shows that this RSNP-VIP neuron expressed GAD65, GAD67, VIP, and CCK. All RSNP-VIP neurons expressed GAD (Fig. 3). In addition, the expression of CCK was observed frequently in RSNP-VIP cells (26 of 32), confirming the frequent association of VIP and CCK in the neocortex (17, 27).
Fig. 2E shows analysis of an IS cell. When depolarized, IS cells fired an initial burst of action potentials, followed by irregularly emitted action potentials (Fig. 2E1). IS cells were characterized by a marked amplitude reduction of action potentials and by a short latency to reach the minimal discharge frequency during prolonged depolarization, which reflects the irregularity of their discharge (Table 1). The IS neuron shown in Fig. 2E2 expressed GAD65, GAD67, CB, CR, VIP, and CCK. All IS neurons expressed VIP and GAD mRNAs (Fig. 3). In addition, CR and CCK were expressed frequently in IS cells (Fig. 3), in good agreement with results obtained in previous studies (17, 18).
Comparison of Glutamate Receptor Expression in the Different Types of Neocortical Neurons.
The qualitative nature of the RT-mPCR amplification does not allow the quantification of the different glutamate receptors expressed for each recorded cell. Nevertheless, the comparison of glutamate receptors expression profiles of the different neuronal populations revealed some cell-type specificity.
Fig. 3 shows the distribution of the four α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor subunits (GluR1–4). GluR1 and GluR2 were the subunits detected most frequently for each cell type. We found a higher occurrence of the flop variants (not shown), except for GluR3 in pyramidal cells (67% of pyramidal cells expressed the flip variant versus 11% for the flop). GluR4 was absent in our sample of pyramidal neurons. Both IS and RSNP-VIP cells had a low occurrence of GluR3 (P = 0.01).
The pattern of expression of kainate receptor subunits in the five classes of neocortical neurons is shown in Fig. 3. The occurrence of low-affinity kainate receptor subunits (GluR5–7) was 55, 88, 92, 81, and 81% and that of high-affinity receptor subunits (KA1 and KA2) was 78, 75, 83, 78, and 69% in pyramidal, FS, RSNP-SS, RSNP-VIP, and IS cells, respectively. The overall occurrence of KA1 was much lower than that of KA2 (see Fig. 3). The expression of GluR5 was restricted to interneurons. RSNP-VIP and IS cells expressed GluR7 less frequently than did other interneurons (P = 0.01).
Fig. 3 shows the relative distribution of N-methyl-d-aspartate subunits in the different cell types. The occurrence of NR2A and NR2B was high in all populations. The occurrence of NR2C was low in all populations. The occurrence of NR2D was low in pyramids in marked contrast with interneurons (P = 0.05).
The distribution of mGluRs is presented in Fig. 3. mGluR1 occurred more frequently (P = 0.01) in fusiform interneurons, particularly in RSNP-SS cells, than in FS or pyramidal cells. mGluR2 was not found in RSNP-SS neurons and rarely occurred in FS and RSNP-VIP cells (P = 0.05). mGluR4 was not found in FS neurons. mGluR5 was the mGluR detected most frequently. mGluR6 was never detected, and mGluR7 and mGluR8 rarely occurred in FS cells. Group I mGluRs (mGluR1 and mGluR5) were the most frequently expressed in all subpopulations (100, 75, 83, 78, and 94% in pyramidal, FS, RSNP-SS, RSNP-VIP, and IS cells, respectively). Group II mGluRs (mGluR2 and mGluR3) were expressed frequently in pyramidal and IS neurons (67 and 63%, respectively) and had low occurrence in FS, RSNP-SS, and RSNP-VIP cells (38, 33, and 31%, respectively). Group III mGluRs (mGluR4, mGluR6, mGluR7, and mGluR8) had low occurrence (P = 0.05) in FS neurons (13%) in contrast to pyramidal, RSNP-SS, RSNP-VIP, and IS cells (56, 67, 69, and 75%, respectively).
Discussion
The aim of the present study was the identification and the characterization of different subpopulations of neocortical fusiform interneurons. Electrophysiological and molecular properties were investigated by combining patch-clamp recordings and single-cell RT-mPCR. Neuronal classes were defined by using cluster analysis of both molecular and electrophysiological properties. The reliability of this approach was supported by its ability to cluster pyramidal and FS cells taken as controls. It revealed three main groups in the heterogeneous population of fusiform cells (RSNP-SS, RSNP-VIP, and IS) with distinctive electrophysiological and molecular characteristics. In addition, the expression profiles of glutamate receptor genes showed cell-type-specific differences.
Different Subtypes of Neocortical Neurons.
The electrophysiological and molecular properties of pyramidal and FS neurons of the present study are consistent with numerous reports (1, 8, 14, 17, 26, 28–35). Interestingly, as observed in the hippocampus (36–38), we did not detect GluR5 in pyramidal neurons in contrast to interneurons. Similarly, NR2D was expressed preferentially by nonpyramidal cells, as reported (39). The present study also showed that FS cells rarely expressed group III mGluRs.
RSNP-SS neurons were characterized by a spike frequency adaptation with no accommodative hump, as observed for SS-expressing bitufted RSNP cells (40). The expression profile of RSNP-SS neurons revealed, in particular, a high occurrence of mGluR1 that has already been described (41, 42). We found that mGluR1 was also expressed frequently by neurons of the VIP cluster, indicating that the high occurrence of mGluR1 is a general feature of fusiform interneurons.
We found a high occurrence of VIP in fusiform neurons as already documented (10, 43). RSNP-VIP neurons can be differentiated from other interneurons based on the accommodative hump and input resistance. Molecular analysis confirmed that CCK and CR were expressed frequently in VIP-expressing neurons (7, 8, 17, 18, 27). In marked contrast to other clusters, VIP cells rarely expressed GluR3. In addition, VIP neurons were characterized by a low occurrence of GluR7 subunit, and IS neurons frequently expressed mGluR7 and mGluR8.
Although the firing pattern of IS cells has obvious and distinctive features (see Fig. 3 and refs. 17 and 18), IS interneurons and RSNP-VIP cells were not totally segregated with the present parameters. This result reflects the high overall similarity of IS and RSNP-VIP cells. Interestingly, we previously showed that both IS and RSNP-VIP cells responded to nicotinic agonists, whereas other types of neocortical neurons did not (44).
Cluster Analysis and Classification of Neocortical Neurons.
The general question addressed in this study relates to the existence and the definition of neuronal subtypes. It is accepted that excitatory pyramidal cells and inhibitory interneurons form two distinct types of neurons with distinct functions. The large diversity of interneurons suggests that they constitute different subtypes that might have specific functions. In this study, we selected a combination of electrophysiological and molecular parameters that discriminated subtypes of neocortical neurons and may be used to recognize these subtypes for future functional studies. Interestingly, pyramidal, FS, and fusiform neurons were segregated without including morphological parameters. In addition, the developmental modifications in the expression of some molecular markers that occur during the developmental stages analyzed in the present study (12 to 34 days) did not seem to alter the discriminative potency of the cluster analysis. For instance, PV neurons transiently expressing CB until the end of the 3rd postnatal week (32) were segregated in the FS cluster as expected (14, 17).
We found that the choice of parameters used in the cluster analysis was a critical point for the determination of neuronal subtypes. For instance, the addition of glutamate receptors in the cluster analysis reduced the discriminative properties of the other parameters. In that case, pyramidal and FS neurons were not segregated from interneurons by using an unsupervised procedure. This observation could be predicted, because, among the 21 different glutamate receptors analyzed, only a few have cell-type-specific expression (28, 34, 35), which is reminiscent of a study performed on hippocampal interneurons in which responses to mGluRs agonist could not be correlated with firing or morphological properties (45). Thus, definition of neuronal types strongly depends on the discriminative properties of the criteria used. It is expected that electrophysiological or molecular parameters with low occurrence will have high discriminative potency.
In summary, our study shows that the analysis of multiple parameters allowed the definition of subtypes of interneurons with distinctive properties. The subtype-specific distinctive properties of fusiform interneurons suggest that the different groups of fusiform neurons may be involved in specific functions.
Acknowledgments
We thank Aurélie Bridault and Mathieu Striker for their help in statistical analyses, and Samia Ben Ammou for technical assistance. This work was supported by European Union Biotech Grants 960382 and 960589 and by European Union Grant QLG3-CT-1999-00649. J.T.P. was supported by a Human Frontier Science Program Organization fellowship.
Abbreviations
- RT-mPCR
multiplex reverse transcription–PCR
- CB
calbindin
- PV
parvalbumin
- CR
calretinin
- NPY
neuropeptide Y
- VIP
vasoactive intestinal peptide
- SS
somatostatin
- CCK
cholecystokinin
- FS
fast spiking
- RSNP
regular spiking nonpyramidal
- IS
irregular spiking
- mGluR
metabotropic glutamate receptor
- AHP
afterpotential
References
- 1.Baughman R W, Gilbert C D. J Neurosci. 1981;1:427–439. doi: 10.1523/JNEUROSCI.01-04-00427.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houser C R, Hendry S H, Jones E G, Vaughn J E. J Neurocytol. 1983;12:617–638. doi: 10.1007/BF01181527. [DOI] [PubMed] [Google Scholar]
- 3.Ramón y Cajal S. General Structure of the Cerebral Cortex. 1995. trans. Swanson, N. & Swanson, R. W. (Oxford Univ. Press, New York), pp. 429–492. [Google Scholar]
- 4.Celio M R. Science. 1986;231:995–997. doi: 10.1126/science.3945815. [DOI] [PubMed] [Google Scholar]
- 5.van Brederode J F, Helliesen M K, Hendrickson A E. Neuroscience. 1991;44:157–171. doi: 10.1016/0306-4522(91)90258-p. [DOI] [PubMed] [Google Scholar]
- 6.Baimbridge K G, Celio M R, Rogers J H. Trends Neurosci. 1992;15:303–308. doi: 10.1016/0166-2236(92)90081-i. [DOI] [PubMed] [Google Scholar]
- 7.Rogers J H. Brain Res. 1992;587:147–157. doi: 10.1016/0006-8993(92)91439-l. [DOI] [PubMed] [Google Scholar]
- 8.Kubota Y, Hattori R, Yui Y. Brain Res. 1994;649:159–173. doi: 10.1016/0006-8993(94)91060-x. [DOI] [PubMed] [Google Scholar]
- 9.Hendry S H, Jones E G, DeFelipe J, Schmechel D, Brandon C, Emson P C. Proc Natl Acad Sci USA. 1984;81:6526–6530. doi: 10.1073/pnas.81.20.6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison J H, Magistretti P J, Benoit R, Bloom F E. Brain Res. 1984;292:269–282. doi: 10.1016/0006-8993(84)90763-7. [DOI] [PubMed] [Google Scholar]
- 11.Somogyi P, Hodgson A J, Smith A D, Nunzi M G, Gorio A, Wu J Y. J Neurosci. 1984;4:2590–2603. doi: 10.1523/JNEUROSCI.04-10-02590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demeulemeester H, Vandesande F, Orban G A, Brandon C, Vanderhaeghen J J. J Neurosci. 1988;8:988–1000. doi: 10.1523/JNEUROSCI.08-03-00988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connors B W, Gutnick M J. Trends Neurosci. 1990;13:99–104. doi: 10.1016/0166-2236(90)90185-d. [DOI] [PubMed] [Google Scholar]
- 14.Kawaguchi Y, Kubota Y. J Neurophysiol. 1993;70:387–396. doi: 10.1152/jn.1993.70.1.387. [DOI] [PubMed] [Google Scholar]
- 15.Kawaguchi Y, Kubota Y. J Neurosci. 1996;16:2701–2715. doi: 10.1523/JNEUROSCI.16-08-02701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaguchi Y. J Neurosci. 1995;15:2638–2655. doi: 10.1523/JNEUROSCI.15-04-02638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cauli B, Audinat E, Lambolez B, Angulo M C, Ropert N, Tsuzuki K, Hestrin S, Rossier J. J Neurosci. 1997;17:3894–3906. doi: 10.1523/JNEUROSCI.17-10-03894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porter J T, Cauli B, Staiger J F, Lambolez B, Rossier J, Audinat E. Eur J Neurosci. 1998;10:3617–3628. doi: 10.1046/j.1460-9568.1998.00367.x. [DOI] [PubMed] [Google Scholar]
- 19.Stuart G J, Dodt H U, Sakmann B. Pflügers Arch. 1993;423:511–518. doi: 10.1007/BF00374949. [DOI] [PubMed] [Google Scholar]
- 20.Lambolez B, Audinat E, Bochet P, Crepel F, Rossier J. Neuron. 1992;9:247–258. doi: 10.1016/0896-6273(92)90164-9. [DOI] [PubMed] [Google Scholar]
- 21.Ruano D, Lambolez B, Rossier J, Paternain A V, Lerma J. Neuron. 1995;14:1009–1017. doi: 10.1016/0896-6273(95)90339-9. [DOI] [PubMed] [Google Scholar]
- 22.Ward J H. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- 23.Wishart D. Biometrics. 1969;25:165–170. [Google Scholar]
- 24.Anderberg M R. Cluster Analysis for Applications. New York: Academic; 1973. [Google Scholar]
- 25.Thorndike R L. Psychometrika. 1953;18:267–276. [Google Scholar]
- 26.Schiffmann S N, Vanderhaeghen J J. J Comp Neurol. 1991;304:219–233. doi: 10.1002/cne.903040206. [DOI] [PubMed] [Google Scholar]
- 27.Papadopoulos G C, Parnavelas J G, Cavanagh M E. Brain Res. 1987;420:95–99. doi: 10.1016/0006-8993(87)90243-5. [DOI] [PubMed] [Google Scholar]
- 28.Jonas P, Racca C, Sakmann B, Seeburg P H, Monyer H. Neuron. 1994;12:1281–1289. doi: 10.1016/0896-6273(94)90444-8. [DOI] [PubMed] [Google Scholar]
- 29.Lambolez B, Ropert N, Perrais D, Rossier J, Hestrin S. Proc Natl Acad Sci USA. 1996;93:1797–1802. doi: 10.1073/pnas.93.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angulo M C, Lambolez B, Audinat E, Hestrin S, Rossier J. J Neurosci. 1997;17:6685–6696. doi: 10.1523/JNEUROSCI.17-17-06685.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geiger J R, Melcher T, Koh D S, Sakmann B, Seeburg P H, Jonas P, Monyer H. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 32.Alcantara S, de Lecea L, Del Rio J A, Ferrer I, Soriano E. Eur J Neurosci. 1996;8:1329–1339. doi: 10.1111/j.1460-9568.1996.tb01595.x. [DOI] [PubMed] [Google Scholar]
- 33.Flint A C, Maisch U S, Weishaupt J H, Kriegstein A R, Monyer H. J Neurosci. 1997;17:2469–2476. doi: 10.1523/JNEUROSCI.17-07-02469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wisden W, Seeburg P H. J Neurosci. 1993;13:3582–3598. doi: 10.1523/JNEUROSCI.13-08-03582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monyer H, Burnashev N, Laurie D J, Sakmann B, Seeburg P H. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 36.Clarke V R, Ballyk B A, Hoo K H, Mandelzys A, Pellizzari A, Bath C P, Thomas J, Sharpe E F, Davies C H, Ornstein P L, et al. Nature (London) 1997;389:599–603. doi: 10.1038/39315. [DOI] [PubMed] [Google Scholar]
- 37.Cossart R, Esclapez M, Hirsch J C, Bernard C, Ben-Ari Y. Nat Neurosci. 1998;1:470–478. doi: 10.1038/2185. [DOI] [PubMed] [Google Scholar]
- 38.Frerking M, Malenka R C, Nicoll R A. Nat Neurosci. 1998;1:479–486. doi: 10.1038/2194. [DOI] [PubMed] [Google Scholar]
- 39.Standaert D G, Landwehrmeyer G B, Kerner J A, Penney J B J, Young A B. Brain Res Mol Brain Res. 1996;42:89–102. doi: 10.1016/s0169-328x(96)00117-9. [DOI] [PubMed] [Google Scholar]
- 40.Reyes A, Lujan R, Rozov A, Burnashev N, Somogyi P, Sakmann B. Nat Neurosci. 1998;1:279–285. doi: 10.1038/1092. [DOI] [PubMed] [Google Scholar]
- 41.Baude A, Nusser Z, Roberts J D, Mulvihill E, McIlhinney R A, Somogyi P. Neuron. 1993;11:771–787. doi: 10.1016/0896-6273(93)90086-7. [DOI] [PubMed] [Google Scholar]
- 42.Kerner J A, Standaert D G, Penney J B J, Young A B, Landwehrmeyer G B. Brain Res Mol Brain Res. 1997;48:259–269. doi: 10.1016/s0169-328x(97)00102-2. [DOI] [PubMed] [Google Scholar]
- 43.Staiger J F, Zilles K, Freund T F. J Comp Neurol. 1996;367:194–204. doi: 10.1002/(SICI)1096-9861(19960401)367:2<194::AID-CNE3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 44.Porter J T, Cauli B, Tsuzuki K, Lambolez B, Rossier J, Audinat E. J Neurosci. 1999;19:5228–5235. doi: 10.1523/JNEUROSCI.19-13-05228.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parra P, Gulyas A I, Miles R. Neuron. 1998;20:983–993. doi: 10.1016/s0896-6273(00)80479-1. [DOI] [PubMed] [Google Scholar]