Abstract
The pattern of circadian behavioral rhythms is photoperiod-dependent, highlighted by the conservation of a phase relation between the behavioral rhythm and photoperiod. A model of two separate, but mutually coupled, circadian oscillators has been proposed to explain photoperiodic responses of behavioral rhythm in nocturnal rodents: an evening oscillator, which drives the activity onset and entrains to dusk, and a morning oscillator, which drives the end of activity and entrains to dawn. Continuous measurement of circadian rhythms in clock gene Per1 expression by a bioluminescence reporter enabled us to identify the separate oscillating cell groups in the mouse suprachiasmatic nucleus (SCN), which composed circadian oscillations of different phases and responded to photoperiods differentially. The circadian oscillation in the posterior SCN was phase-locked to the end of activity under three photoperiods examined. On the other hand, the oscillation in the anterior SCN was phase-locked to the onset of activity but showed a bimodal pattern under a long photoperiod [light–dark cycle (LD)18:6]. The bimodality in the anterior SCN reflected two circadian oscillatory cell groups of early and late phases. The anterior oscillation was unimodal under intermediate (LD12:12) and short (LD6:18) photoperiods, which was always phase-lagged behind the posterior oscillation when the late phase in LD18:6 was taken. The phase difference was largest in LD18:6 and smallest in LD6:18. These findings indicate that three oscillating cell groups in the SCN constitute regionally specific circadian oscillations, and at least two of them are involved in photoperiodic response of behavioral rhythm.
Keywords: bioluminescence reporter, circadian rhythm, clock gene, photoperiod, behavioral rhythm
Adaptation to seasonal changes in environment is critical to the survival of many organisms. Photoperiodic time measurement by the circadian clock is one of the strategies by which they conserve the phase relation between behavioral events such as the activity onset and dawn or dusk (1). A dramatic change induced by photoperiod is in the length of an activity band, the duration of activity in behavioral rhythms. Nocturnal rodents such as rats and mice exhibit compressed activity bands in long photoperiods and decompressed bands in short photoperiods. A long-standing hypothesis for the photoperiodic time measurement assumes two separate, but mutually coupled, circadian oscillators that drive the activity onset and end of activity, respectively, and respond to dawn and dusk differentially. Therefore, their phase-relationship encodes day lengths and changes the length of an activity band (1).
The circadian clock in mammals is located in the suprachiasmatic nucleus (SCN) of the hypothalamus; it entrains to a light–dark cycle (LD) and determines the phases of overt circadian rhythms in behavior and physiology (2). Over the last decade, our understanding of the circadian clock in the SCN has advanced tremendously (3–5). The SCN consists of a number of oscillating cells in the circadian domain, which are independent but coupled with each other to produce coherent SCN output rhythms (4–8). The intracellular molecular machinery of circadian oscillation consists of interlocked transcriptional and translational autofeedback loops, in which at least six clock genes are involved (3). Clock and Bmal1 are positive elements of the feedback loop, and Per1, Per2, Cry1, and Cry2 are negative elements.
There is substantial evidence that the SCN contains at least two independent suboscillators. Circadian rhythms in two major neuropeptides in the SCN, arginine vasopressin (AVP) and vasoactive intestinal peptide (VIP), free-ran in vitro with different periods and showed internal desynchronization (9). More recently, two distinct peaks of electrophysiological activity in the cultured SCN were observed in the Syrian hamster (10,11), and the peaks responded differentially to photoperiods (10). In addition, circadian rhythms in clock gene expression and their protein products were reported to change in different photoperiods (12–17). Generally, the interval of high Per1 and Per2 expression is extended in long photoperiods, whereas the rhythm amplitudes are increased in short photoperiods. Johnston and colleagues (18, 19) observed differential phasing of clock gene expression rhythms in the rostral and caudal regions of the hamster SCN and proposed a model in which the evening (E) oscillator is located in the rostral and the morning (M) oscillator in the caudal SCN (19).
Transgenic mice carrying a luciferase reporter gene enable us to search for the E and M oscillators or even the E and M oscillating cells in the SCN. In the present study, we tried to separate oscillating cell populations that respond differentially to photoperiod by using transgenic mice with a Per1 luciferase reporter system. We found oscillating cell groups in the SCN, which were phase-locked separately to the activity onset and end of activity and whose phase relations were changed by photoperiod.
Results
Behavioral Rhythms in Different Photoperiods.
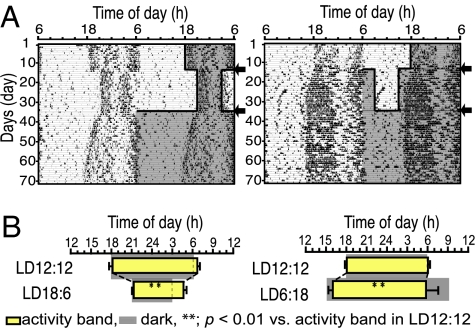
Photoperiodic responses of spontaneous activity rhythms were examined in the transgenic mice carrying a Per1-luciferase reporter gene (Per1-luc) initially kept in 12 h of light and 12 h of darkness (LD12:12) (Fig. 1A). Nocturnal activities of these mice were bimodal, showing the clear activity onset and end of activity in constant darkness (DD) as well as in LD. In steady-state entrainment, the length of activity band, an interval between the activity onset and the end of activity, was decreased (compression) by 5.26 h when the animals were transferred to LD18:6, and increased (decompression) by 1.49 h when transferred to LD6:18 (Table 1 and Fig. 1B). The compressed activity band in LD18:6 was decompressed again when released into constant darkness with a transient period of ≈2 weeks [supporting information (SI) Fig. 6]. The phase angle difference (ψ) between the activity onset and light off (ψonset) was basically unchanged in LD18:6 but became slightly but significantly more negative (P < 0.01) by −0.99 h in LD6:18. On the other hand, the phase angle difference between the end of activity and light on (ψend) was significantly changed (P < 0.01) in LD18:6 as well as LD6:18 by −0.96 h and by 3.53 h, respectively (Fig. 1B).
Fig. 1.
Circadian behavioral rhythms in different photoperiods. (A) Representative double-plotted circadian rhythms in spontaneous locomotor activity of a mouse transferred to LD18:6 (Left) and LD6:18 (Right) from LD12:12. The dark phase is shaded in the right-hand side of each actograph. Arrows indicate day of photoperiod transfer. (B) Phase relations of activity rhythms to different photoperiods. Yellow bars indicate activity bands (mean ± SD). Gray areas indicate dark phase.
Table 1.
Parameters of behavioral rhythms in different photoperiods
| Lighting (on–off) | Activity band, h |
ψ, h |
|||
|---|---|---|---|---|---|
| Activity time | Onset | End | ψonset | ψend | |
| LD12:12 (0600–1800) | 12.61 ± 0.77 | 18.16 ± 0.58 | 6.77 ± 0.34 | −0.16 ± 0.58 | −0.77 ± 0.34 |
| LD18:6 (0300–2100) | 7.35 ± 0.66** | 21.38 ± 0.34** | 4.73 ± 0.44** | −0.38 ± 0.34 | −1.73 ± 0.44** |
| LD12:12 (0600–1800) | 12.10 ± 0.47 | 18.19 ± 0.22 | 6.29 ± 0.48 | −0.19 ± 0.22 | −0.29 ± 0.48 |
| LD6:18 (0900–1500) | 13.59 ± 0.86** | 16.18 ± 0.60** | 5.76 ± 1.28 | −1.18 ± 0.60** | 3.24 ± 1.28** |
Phase angle differences (ψ) were calculated between the activity onset and light off (ψonset) and between the end of activity and light on (ψend). Paired Student's t test was used for comparing values of two different photoperiods. Values are mean ± SD (n = 9). **, P < 0.01 vs. respective LD12:12.
Per1-luc Expression Rhythms in the SCN Slices.
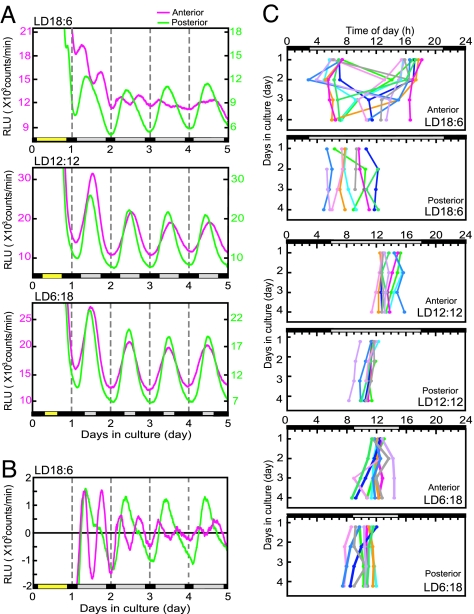
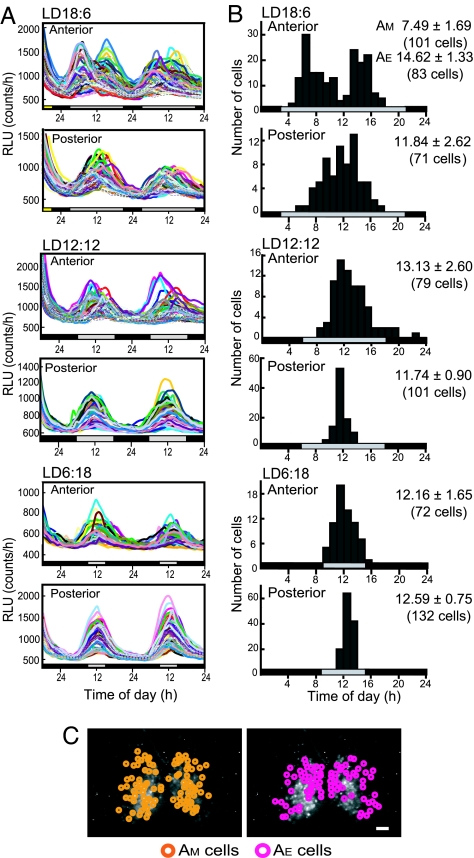
Circadian rhythms in Per1-luc were measured separately in the anterior and posterior SCN slices (SI Fig. 7 A and B). Robust circadian rhythms in Per1-luc activity were detected in all preparations, except for the anterior SCN in LD18:6, and the circadian peak phases on the first day of culture were located in the subjective day (Fig. 2A). The circadian peak on the first cycle in the anterior and posterior SCN were located at local time 13.69 ± 0.98 h (mean ± SD, n = 10) and 11.44 ± 0.68 h (n = 9) in LD12:12, which shifted to 12.20 ± 0.57 h (n = 9) and 10.45 ± 1.11 h in LD6:18 (n = 10), respectively. In the anterior SCN obtained from mice in LD18:6, the two antiphasic peaks were detected in the first few days of culture, which were located at the rising phase (6.75 ± 0.69 h) and falling phase (16.66 ± 0.95 h) (n = 10) of the posterior peak. The bimodal pattern became evident in the detrended curves (Fig. 2B). The posterior peak was located at 8.02 ± 1.75 h (n = 8) on the first day. In 9 of 10 anterior SCN slices from LD18:6, the two peaks merged into a single circadian peak on the course of culture (Fig. 2 C Top). Distribution of the posterior peaks was significantly wider in LD18:6 than in LD12:12 and LD6:18 (P < 0.01 and P < 0.05, respectively).
Fig. 2.
Per1-luc activity rhythms of the anterior and posterior SCN slices in different photoperiods. (A) Representative rhythms of Per1-luc activity in the anterior (pink) and posterior (green) SCN in LD18:6 (Top), LD12:12 (Middle), and LD6:18 (Bottom). Ordinate indicates strength of bioluminescence (relative light units, RLU). Abscissa indicates days in culture. Broken lines in the graph indicate 0000 hours. Yellow bars on the abscissa, light phase on the day of slice preparation. Gray and black bars, the subjective day and night on the following days, respectively. (B) Detrended waveforms of Per1-luc rhythms obtained by subtracting 12 h of moving average values from original luminescence curves. (C) Peak phases of Per1-luc rhythms from the anterior and posterior SCN slices are plotted against local time. Those from different mice are indicated with different colors. Ordinate, days in culture.
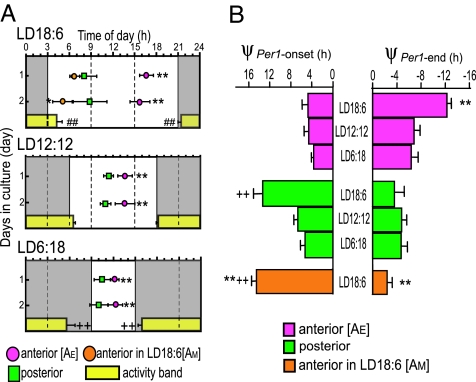
The phase relations were examined between the first day peaks and photoperiods as well as behavioral rhythms of mice from which the SCNs were sampled (Fig. 3A). The two peaks in the anterior SCN from LD18:6 were tentatively termed as AE and AM, where AE was closer to the light off and AM to the light on. Neither the anterior nor the posterior peaks from different photoperiods showed a consistent phase relation with the light on or light off. On the other hand, stable phase relations were observed between the anterior peaks (AE) and activity onset (ψPer1-onset), and between the posterior peaks and end of activity (ψPer1-end) in three photoperiods examined (Fig. 3B). The ψPer1-onset was kept at ≈4.3 h (4.70 ± 1.13 h in LD18:6; 4.59 ± 0.86 h in LD12:12; 3.63 ± 0.45 h in LD6:18) for the anterior perks and the ψPer1-end was kept at approximately −4.4 h (−3.63 ± 1.59 h in LD18:6; −4.80 ± 0.80 h in LD12:12; −4.70 ± 1.12 h in LD6:18) for the posterior peak. The correlations between the Per1 peak and four phase markers (activity onset, end of activity, light on, and light off) were calculated. The anterior peak (AE) was correlated most strongly with the activity onset (r = 0.904), and the posterior peak with the end of activity (r = 0.713).
Fig. 3.
Correlation between behavior rhythms and Per1-luc rhythms in anterior and posterior SCN. (A) Phase relationships between the anterior and posterior peaks of the first two days in different photoperiods. Symbols: peak phase (mean ± SD in hours; n = 8 in LD18:6, n = 9 in LD12:12, and n = 8 in LD6:18). Gray areas, subjective night. ∗ and ∗∗, P < 0.05 and 0.01, respectively, vs. posterior peak. Horizontal yellow bars, activity band (mean ± SD). + and ++, P < 0.05 and 0.01, respectively, vs. ψon or ψend in LD12:12. ##, P < 0.01 ψon or ψend in LD18:6 vs. those in LD6:18. (B) Phase differences (ψ) between the peak of Per1-luc expression rhythm and activity onset (ψPer1-onset, Left), and between the peak and end of activity (ψPer1-end, Right) in three photoperiods. Phase angle differences between the activity and bioluminescence rhythms were measured for each animal and averaged (mean ± SD). ∗∗, P < 0.01 vs. anterior SCN in LD12:12; + and ++, P < 0.05 and 0.01, respectively, vs. posterior SCN in LD12:12.
The posterior peak phase-led the anterior (AE) peak significantly. The phase difference was largest in LD18:6 (8.66 ± 1.78 h), intermediate in LD12:12 (2.08 ± 0.98h), and smallest in LD6:18 (1.55 ± 0.87 h) on day 1.
Per1-luc Expression Rhythms in the SCN Cells.
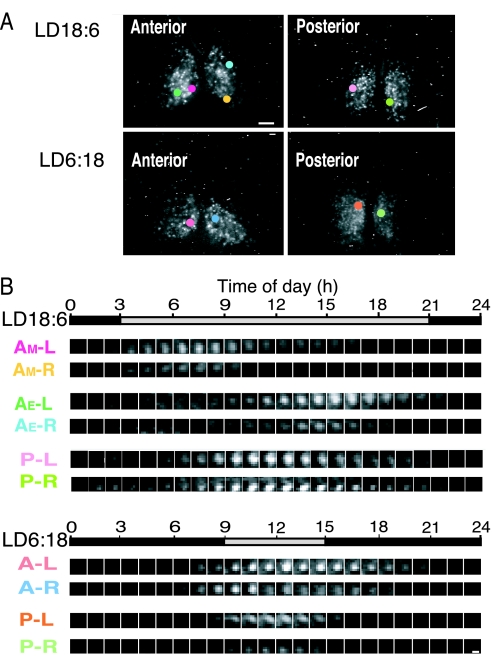
To clarify the nature of two peaks in the anterior SCN slices from mice in LD18:6 and to identify the localizations of anterior and posterior peaks in the SCN, Per1-luc expression was measured in individual SCN cells for more than three circadian cycles by using bioluminescent cell imaging (Fig. 4 and 5A). Regardless of the anterior or posterior SCN, almost all cells that exhibited bioluminescence at detectable levels showed circadian rhythms in Per1-luc expression with a single peak. The intensity of bioluminescence was slightly and nonsystematically different between the right and left SCN, but no evidence was detected to suggest the lateralization of the circadian peak.
Fig. 4.
CCD images of Per1-luc rhythms in single SCN cells. (A) Representative CCD images from the anterior and posterior SCN in different photoperiods. Single SCN cells indicated by colored circles correspond to cells with the number in the same color in Fig. 4B. (Scale bar: 100 μm.) (B) Serial demonstration of bioluminescence in a single cell at 1-h intervals on day 1. In LD18:6 (Upper), bioluminescence is strong either at the early or late subjective day in the anterior SCN, whereas it is strong at the middle of subjective day in the posterior. In LD6:18 (Lower), individual rhythms are essentially the same between the anterior and posterior SCN with respect to circadian phase. (Scale bar, 10 μm.) A, anterior; P, posterior; L, left; R, right. AM and AE, cells with morning and evening peaks for the anterior SCN in LD18:6, respectively. Gray and black horizontal bars, the subjective day and night, respectively.
Fig. 5.
Spatial and temporal distributions of circadian rhythms in Per1-luc from single SCN cells in different photoperiods. (A) Forty-eight-hour profiles of bioluminescence of individual cells in the anterior and posterior SCN slices obtained from a mouse in LD18:6 (Top), LD12:12 (Middle) and LD6:18 (Bottom). (B) Temporal distribution of individual circadian peaks (on day 1) from the same SCNs indicated in A. Distribution peaks were expressed in each graph with the mean and SD of individual circadian peaks. See SI Fig. 8 for the peak distribution of the total cells from 3 SCNs. (C) Spatial distribution of cells with the morning (AM, orange) and evening peaks (AE, pink) in the anterior SCN under LD18:6 shown in A. (Scale bar: 100 μm.)
Circadian peaks in individual SCN cells detected on the first culture day were plotted against the local time (Fig. 5B). In LD18:6, the temporal distribution was bimodal in the anterior SCN, whereas it was unimodal in the posterior. Bimodal patterns were detected in the peak distribution in all three SCN slices examined. The distribution of the early group peaked at 7.77 ± 1.67 h, whereas the late group at 14.54 ± 1.42 h, respectively (SI Fig. 8). The early group corresponded to AM, and the late one corresponded to AE in the SCN slices. The number of AM and AE cells were almost equal in three slices examined, and of 292 cells in total, 142 belonged to the AM cells and 142 to AE cells. Eight cells (2.7%) between AM and AE at 1100 hours could not be classified. The distribution peak in the posterior SCN cells was located at 12.35 ± 2.83 h (n = 152). By contrast, the distribution of circadian peaks in individual cells was unimodal in LD12:12 and LD6:18 in both the anterior and posterior SCN, with peaks at approximately the middle of subjective day (SI Fig. 8). When the range of distribution was defined as SD, the distributions in the anterior SCN were significantly wider than those in the respective posteriors (P < 0.01). The range of distribution in the posterior SCN was significantly narrower in LD12:12 and LD6:18 than in LD18:6 (P < 0.01 and P < 0.05, respectively).
Fig. 5C illustrates spatial distribution of individual cells in the anterior SCN from LD18:6, which belong to either the AM or AE cells. The AM and AE cells were widely distributed in the anterior SCN and intermixed.
Discussion
Continuous monitoring of Per1 expression activity in the cultured SCN cells revealed that at least three different oscillating cell populations exist in the mouse SCN: two in the anterior and one in the posterior SCN. They appear to form functional cell networks with humoral or neural communications and respond differentially to photoperiod. Changes in the 24-h profiles of bioluminescence rhythm in the SCN slice under different photoperiods are mostly the reflection of changes in the phase relation, not the waveform, of individual oscillating cells (Fig. 5), as recently suggested by electrophysiology (20) and simulation studies (21).
The one of two oscillating cell populations (AE cells) in the anterior SCN was correlated most strongly with the activity onset, whereas the cell population in the posterior SCN was correlated most strongly with the end of activity (Fig. 3B). The length of an activity band, which reflects the phase relation between the activity onset and end of activity, depended on photoperiod (Fig. 1), which increased (decompressed) in a shorter photoperiod (LD6:18) and decreased (compressed) in a longer photoperiod (LD18:6). In parallel with compression and decompression of an activity band, the phase relation between the anterior (AE) and posterior oscillations was changed in different photoperiods. The posterior oscillation phase-led the anterior in three photoperiods examined (Fig. 3A). Therefore, the cell populations in the anterior and posterior SCN are likely to correspond to the E and M oscillators, respectively. The findings, in part, support a model advanced recently by Johnston (19). In their model, however, the circadian peaks in the SCN are correlated with photoperiod, whereas our results indicate strong correlations between the circadian oscillations in the SCN and behavioral rhythms. In addition, our results demonstrated the third oscillation in the anterior SCN. The circadian system in the SCN for photoperiodic responses seems to be more complex.
The AM cell population in the anterior SCN became evident in a longer photoperiod (LD18:6), especially in the first several days of culture. Eventually, their oscillation peaks merged with the peaks of AE oscillation, suggesting that they are not direct responses to light but expressions of endogenous oscillation, and there are coupling mechanisms between them. The numbers of cells categorized to the AE or AM population were nearly the same, and both groups of cells seem to spatially distribute rather diffusely and are intermixed (Fig. 5C). The role of AM oscillation was unidentified, but it seems to be related to photoperiodic time measurement. The AE oscillation in the anterior SCN and the oscillation in the posterior SCN are closely related with behavioral rhythms, but it is not known whether they are directly phase-locked by photoperiod or through other mechanisms. In LD18:6, the circadian peak of the posterior cell population was located in between those of the AE and AM cell populations. In shorter photoperiods, the AM cell population seemed to disappear, but the ranges of peak distribution were still wider in the anterior SCN than in the posterior (Fig. 5B). The AM oscillation may have some roles in determining the phases of the AE oscillation.
We reported a dissociation of circadian rhythms in AVP and VIP secretion during the prolonged culture of rat SCN (9, 22) and suggested that AVP and VIP secretion was regulated by separate circadian oscillators in the SCN. Using GFPs as a reporter, Quitero et al. (8) reported differentially phased three Per1 expressing cell groups and suggested their role in photoperiodic responses. Jagota et al. showed two distinct peaks of electrophysiological activity in the horizontally sectioned SCN culture in hamsters (10) but not in rats and mice (11), which responded differentially to photoperiods. In addition, the circadian rhythms in Per1 and Per2 expression were reported to change in different photoperiods (12–19, 23). The duration of gene expression was extended in long photoperiods, whereas the rhythm amplitudes were increased in short photoperiods, suggesting that the mutual coupling of individual cell oscillations was altered by photoperiods.
The E and M oscillators have been proposed as the mechanism of rhythm splitting in hamsters under prolonged constant light (LL) (1). Previously, the circadian rhythm in clock gene expression was demonstrated to be antiphasic in the right and left SCNs from hamsters exhibited behavioral splitting in LL (24). More recently, two antiphase-oscillating subregions were reported in each side of the SCN of split hamsters (25). However, it is not known whether the E and M oscillators for the photoperiodic responses are identical to those for behavioral splitting, because a unilateral SCN has a redundancy for behavioral splitting as demonstrated by SCN transplants (26) and unilateral SCN lesion (27).
Per1 and Cry1 have been hypothesized to represent the components of M oscillator, and Per2 and Cry2 have been hypothesized to represent the components of E oscillator (28). The hypothesis is based on differential responses to light in the respective mutant animals (14, 28). From the present studies, the photoperiodic response of behavioral rhythms in nocturnal rodents is explained without assuming differential roles of Per1 and Per2, although the possibility remains that a single SCN cell contains the coupled E and M oscillators, in which Per1 and Per2 compete for dominancy. Specific pacemaker cells may exist in the SCN, which drive other cells to constitute separate oscillations as reported recently in Drosophila in which specific pacemaker cell groups (29): one processes the light information to convey into circadian networks, whereas the other dominates in activity output (30). Alternatively, specific cell communication may enable coherent oscillations of different nature.
In conclusion, the present findings indicate that three distinct cellular oscillations in the SCN change their phases in response to the photoperiod, and at least two of them are strongly coupled to the onset and end of activity, separately, in mice.
Methods
Per1-luc Transgenic Mice.
We developed mPer1-luc transgenic mice with a firefly luciferase reporter on a C57BL/6J background. A 6.7-kb region upstream of the transcription-translation codon of the mPer1 gene was linked to a firefly luciferase cDNA (a generous gift of H. Tei, Mitsubishi Kagaku Institute of Life Sciences, Tokyo, Japan; ref. 31) to make the transgene. Per1-luc mice were born and reared in our animal quarters where environmental conditions were controlled (LD12:12; lights: 0600–1800 hours). Animals were cared for according to the Guidelines for the Care and Use of Laboratory Animals in Hokkaido University Graduate School of Medicine.
Measurement of Behavioral Rhythms.
Male Per1-luc mice of 5–8 weeks old (n = 18) were housed individually in a cage placed in a light-tight chamber. Spontaneous locomotor activity was monitored by thermal sensors (32). Photoperiod was changed by phase-delaying the light off and phase-advancing the light on by 3 h each for LD18:6, and by phase advancing the light off and phase-delaying the light on by 3 h each for LD6:18. The onset and end of nocturnal activity were determined by using behavioral rhythms of the last 5 days in each of three photoperiods (LD12:12, LD18:6, and LD6:18) by visual inspection (Clock Lab; Actimetrics, Wilmette, IL).
SCN Culturing and Bioluminescent Measurement.
After monitoring behavioral rhythms for 3 weeks in the respective photoperiod, mice (n = 30) were decapitated after cervical dislocation, and brains were rapidly removed between 1100 and 1500 hours. Using a microslicer (D.S.K: DTK-1000; Dosaka EM, Kyoto, Japan), two successive 300-μm-thick coronal brain slices containing the anterior and posterior regions of each SCN were cut for bioluminescence measurement with photomultiplier tubes (PMT). Bilateral SCNs with optic chiasms were trimmed to ≈3 × 3 mm square in ice cold Hanks's balanced salt solution and placed on a culture membrane (Millicell-CM; Millipore; pore size 0.4 μm) in a 35-mm Petri dish. SCN slices were cultured in air at 37°C with 130 μl of DMEM (Invitrogen) supplemented with 10 mM Hepes/2.74 mM NaHCO3/0.1 mM d-Luciferin K salt and serum-free growth supplements described in ref. 33. Bioluminescence from each SCN slice was measured by a microplate luminometer equipped with PMTs (Lumicycle; Actimetrics) for 1 min at 9-min intervals for 5 successive days.
For single-cell analysis, five successive 100-μm-thick coronal slices were prepared through the rostrocaudal extent of the SCN from nine mice. The second and fourth slices were chosen as anterior and posterior SCN slices, respectively. Slices were cultured at 37°C in a miniincubator (Phoenix; IMP, Tokyo, Japan) installed on the stage of an inverted microscope (DM IRB; Leica). Bioluminescence from SCN cells was measured with a high-sensitive cryogenic CCD camera (ORCA-II ER; Hamamatsu Photonics, Hamamatsu, Japan) mounted at the bottom port of the microscope with a camera adaptor of ×0.6. Bioluminescence was measured every hour with a 59-min exposure and a 1-min interval for data transfer for 3 consecutive days. CCD imaging of an SCN slice was performed by using a ×10 objective lens (N.A. 0.4; Leica) at 4 × 4 binning of the 1,344 (horizontal) × 1,024 (vertical) pixel array. The final pixel number and pixel size of an image for detection were 336 × 256 and 4.3 μm × 4.3 μm, respectively.
Double-Labeling Immunohistochemistry.
SCN slices were immunohistochemically labeled by using anti-mouse AVP monoclonal antibody (generous gift of H. Gainer, National Institutes of Health, Bethesda, MD; ref. 34) and anti-rabbit VIP polyclonal antibody (Peptide Institute, Osaka, Japan).
Data and Statistical Analyses.
Circadian rhythms in Per1-luc measured by Lumicycle were smoothed by a five-point moving average method, and the peak phases were calculated after detrending bioluminescent levels by using first-order regression for each cycle. To analyze circadian rhythms in single cells, individual cells were identified by inspection, and the region of interest (ROI) was defined to cover most of the cell bodies. Bioluminescence of a given ROI was expressed with an averaged intensity per pixel by Aquacosmos (Hamamatsu Photonics). The circadian peak of bioluminescent rhythms was defined as the midpoint of two succeeding troughs in each cycle. For the analyses of phase relation between circadian rhythms in Per1-luc activity and behavioral rhythms, 5-day records of behavioral rhythms immediately before the brain sampling were used.
Statistical significance of differences in parameters among three photoperiods was evaluated with one-way ANOVA and a post hoc Tukey–Kramer test. Unpaired Student's t test was used for group comparisons. Paired Student's t test was used for comparison of parameters of behavioral rhythms in different photoperiods. Differences in the peak distribution among photoperiods were evaluated by F test after a Bartlett test. Linear regression was obtained between the Per1 peak of SCN slice and one of four phase markers; activity onset, activity end, light on, and light off.
Supplementary Material
Acknowledgments
We thank Dr. H. Tei for kindly donating the Per1-luc plasmid vector and Dr. H. Gainer for providing anti-mouse AVP monoclonal antibody. We are grateful to M. P. Butler for advice on the manuscript.
Abbreviations
- AVP
arginine vasopressin
- E
evening
- M
morning
- LD
light–dark cycle
- SCN
suprachiasmatic nucleus
- VIP
vasoactive intestinal peptide.
Note:
A related report by van der Leest et al. (35) has been published during the revision of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at wMww.pnas.org/cgi/content/full/0607713104/DC1.
References
- 1.Pittendrigh CS, Daan S. J Comp Physiol A. 1976;106:333–355. [Google Scholar]
- 2.Klein DG, Moore RY, Reppert SM. Suprachiasmatic Nucleus: The Mind's Clock. New York: Oxford Univ Press; 1991. [Google Scholar]
- 3.Reppert SM, Weaver DR. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 5.Antle MC, Silver R. Trends Neurosci. 2005;28:145–151. doi: 10.1016/j.tins.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Welsh DK, Logothetis DE, Meister M, Reppert SM. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 7.Honma S, Shirakawa T, Katsuno Y, Namihira M, Honma K. Neurosci Lett. 1998;250:157–160. doi: 10.1016/s0304-3940(98)00464-9. [DOI] [PubMed] [Google Scholar]
- 8.Quintero JE, Kuhlman SJ, McMahon DG. J Neurosci. 2003;23:8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shinohara K, Honma S, Katsuno Y, Abe H, Honma K. Proc Natl Acad Sci USA. 1995;92:7396–7400. doi: 10.1073/pnas.92.16.7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jagota A, de la Iglesia HO, Schwartz WJ. Nat Neurosci. 2000;3:372–376. doi: 10.1038/73943. [DOI] [PubMed] [Google Scholar]
- 11.Burgoon PW, Lindberg PT, Gillette MU. J Comp Physiol A. 2004;190:167–171. doi: 10.1007/s00359-003-0486-z. [DOI] [PubMed] [Google Scholar]
- 12.Messager S, Ross AW, Barrett P, Morgan PJ. Proc Natl Acd Sci USA. 1999;96:9938–9943. doi: 10.1073/pnas.96.17.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nuesslein-Hildesheim B, O'Brien JA, Ebling FJP, Maywood ES, Hastings MH. Eur J Neurosci. 2000;12:2856–2864. doi: 10.1046/j.1460-9568.2000.00173.x. [DOI] [PubMed] [Google Scholar]
- 14.Steinlechner S, Jacobmeier B, Scherbarth F, Dernbach H, Kruse F, Albrecht U. J Biol Rhythms. 2002;17:202–209. doi: 10.1177/074873040201700303. [DOI] [PubMed] [Google Scholar]
- 15.Carr A-J, Johnston JD, Semikhodskii AG, Nolan T, Cagampang FR, Stirland JA, Loudon AS. Curr Biol. 2003;13:1543–1548. doi: 10.1016/s0960-9822(03)00619-5. [DOI] [PubMed] [Google Scholar]
- 16.Sumova A, Jac M, Sladek M, Sauman I, Illnerova H. J Biol Rhythms. 2003;18:134–144. doi: 10.1177/0748730403251801. [DOI] [PubMed] [Google Scholar]
- 17.Johnston JD, Ebling FJP, Hazlerigg DG. Eur J Neurosci. 2005;21:2967–2974. doi: 10.1111/j.1460-9568.2005.04148.x. [DOI] [PubMed] [Google Scholar]
- 18.Hazlerigg DG, Ebling FJP, Johnston JD. Curr Biol. 2005;15:449–450. doi: 10.1016/j.cub.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Johnston JD. J Neuroendocrinol. 2005;17:459–465. doi: 10.1111/j.1365-2826.2005.01326.x. [DOI] [PubMed] [Google Scholar]
- 20.Schaap J, Albus H, van der Leest HT, Eilers PHC, Détári L, Meijer JH. Proc Natl Acad Sci USA. 2003;100:15994–15999. doi: 10.1073/pnas.2436298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohling J, Wolters L, Meijer JH. J Biol Rhythms. 2006;21:301–313. doi: 10.1177/0748730406290317. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura W, Honma S, Shirakawa T, Oguchi H, Honma K. Eur J Neurosci. 2001;14:666–674. doi: 10.1046/j.0953-816x.2001.01684.x. [DOI] [PubMed] [Google Scholar]
- 23.de la Iglesia HO, Meyer J, Schwartz WJ. Mol Brain Res. 2004;127:121–127. doi: 10.1016/j.molbrainres.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 24.de la Iglesia H, Meyer J, Carpino A, Jr, Schwartz WJ. Science. 2000;290:799–801. doi: 10.1126/science.290.5492.799. [DOI] [PubMed] [Google Scholar]
- 25.Yan L, Foley NC, Bobula JM, Kriegsfeld LJ, Silver R. J Neurosci. 2005;25:9017–9026. doi: 10.1523/JNEUROSCI.2538-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis FC, Viswanathan N. J Biol Rhythms. 1996;11:291–301. doi: 10.1177/074873049601100402. [DOI] [PubMed] [Google Scholar]
- 27.Davis FC, Gorski RA. J Comp Physiol A. 1984;154:221–232. [Google Scholar]
- 28.Daan S, Albrecht U, van der Horst GT, Illnerova H, Roenneberg T, Wehr TA, Schwartz WJ. J Biol Rhythms. 2001;16:105–116. doi: 10.1177/074873001129001809. [DOI] [PubMed] [Google Scholar]
- 29.Stoleru D, Peng Y, Nawathean P, Rosbash M. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- 30.Stoleru D, Nawathean P, de la Paz Fernandez M, Menet JS, Ceriani MF, Rosbash M. Cell. 2007;129:207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 31.Hida A, Koike N, Hirose M, Hattori M, Sakaki Y, Tei H. Genomics. 2000;65:224–233. doi: 10.1006/geno.2000.6166. [DOI] [PubMed] [Google Scholar]
- 32.Honma K, Honma S, Shirakawa T, Hiroshige T. Am J Physiol. 1987;252:256–261. doi: 10.1152/ajpregu.1987.252.2.R256. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura W, Honma S, Shirakawa T, Honma K. Nat Neurosci. 2002;5:399–400. doi: 10.1038/nn843. [DOI] [PubMed] [Google Scholar]
- 34.Whitnall MH, Key S, Ben-Barak Y, Ozato K, Gainer H. J Neurosci. 1985;5:98–109. doi: 10.1523/JNEUROSCI.05-01-00098.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Leest H, Houben T, Michel S, Deboer T, Albus H, Vansteensel M, Block G, Meijer J. Curr Biol. 2007;17:468–473. doi: 10.1016/j.cub.2007.01.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.