Abstract
Epigenetic chromatin modification is a major regulator of eukaryotic gene expression, and aberrant epigenetic silencing of gene expression contributes to tumorigenesis. Histone modifications include acetylation, phosphorylation, and methylation, resulting in a combination of histone marks known collectively as the histone code. The chromatin marks at a given promoter determine, in part, whether specific promoters are in an open/active conformation or closed/repressed conformation. Dimethyl-lysine 4 histone H3 (H3K4me2) is a transcription-activating chromatin mark at gene promoters, and demethylation of this mark by the lysine-specific demethylase 1 (LSD1), a homologue of polyamine oxidases, may broadly repress gene expression. We now report that novel biguanide and bisguanidine polyamine analogues are potent inhibitors of LSD1. These analogues inhibit LSD1 in human colon carcinoma cells and affect a reexpression of multiple, aberrantly silenced genes important in the development of colon cancer, including members of the secreted frizzle-related proteins (SFRPs) and the GATA family of transcription factors. Furthermore, we demonstrate by chromatin immunoprecipitation analysis that the reexpression is concurrent with increased H3K4me2 and acetyl-H3K9 marks, decreased H3K9me1 and H3K9me2 repressive marks. We thus define important new agents for reversing aberrant repression of gene transcription.
Keywords: chromatin, histone, lysine demethylase, methylation
Epigenetic chromatin modification is a major regulator of eukaryotic gene expression (1, 2). In cancer, DNA promoter hypermethylation in combination with other chromatin modifications, including decreased activating marks and increased repressive marks on histone proteins 3 and 4, have been associated with the silencing of tumor suppressor genes (3). Histone modifications include acetylation, phosphorylation, and methylation, resulting in a combination of histone marks that is known collectively as the histone code (1). The combination of chromatin marks at a given promoter determines, in part, whether specific promoters are in an open/active conformation or closed/repressed conformation (1, 4). Histone acetylation is frequently associated with active genes and is a result of the dynamic interaction between activities of histone acetyltransferases and histone deacetylases (5). Histone methylation can be associated with either active or repressive signals and has also recently been discovered to be a dynamic process regulated not only by the addition of methyl groups by histone methyltransferases, but also by removal of methylation catalyzed by lysine-specific demethylase 1 (LSD1) and JmjC domain demethylases (6–10).
A key positive chromatin mark found associated with promoters of active genes is histone 3 dimethyl-lysine 4 (H3K4me2) (11, 12). LSD1, also known as BHC110 (6, 13), catalyzes the demethylation of H3K4me1 and H3K4me2 and is associated with transcriptional repression. Therefore, LSD1 has the potential to be a major regulator of gene expression through the modulation of chromatin structure, and its inhibition has considerable importance in the study of the biology underlying chromatin regulation of gene transcription. A better understanding of the role of LSD1 in the regulation of gene expression should aid in the discovery of strategies for reexpressing inappropriately silenced genes as a rational approach for the treatment of cellular pathologies, including cancer.
LSD1 shares considerable homology with FAD-dependent polyamine oxidases, including spermine oxidase (SMO/PAOh1) (6, 14). As guanidines have been shown to inhibit both SMO/PAOh1 and other polyamine oxidases (15, 16), we sought to determine whether unique biguanide and bisguanidine polyamine analogues were effective inhibitors of LSD1 and whether cellular inhibition of LSD1 could lead to the reexpression of aberrantly repressed genes important in cancer.
Results and Discussion
A small library of bisguanidine (Fig. 1A, 1a–1g) and biguanide (Fig. 1B, 2a–2f) polyamine analogues (17) was tested for the ability to inhibit recombinant LSD1 in vitro. Nine compounds were found to inhibit demethylase activity by >50% at 1 μM (Fig. 1C). The two most potent inhibitors, 1c (1,11-bis{N2,N3-dimethyl-N1-guanidino}-4,8-diazaundecane) and 2d (1,15-bis{N5-[3,3-(diphenyl)propyl]-N1-biguanido}-4,12-diazapentadecane), were chosen for further study. In inhibition studies with purified LSD1 protein, both compounds exhibited noncompetitive inhibition kinetics at concentrations <2.5 μM (Fig. 1 D and E), suggesting that, although the polyamine compounds could be considered analogues of the natural methyl lysine substrate of LSD1, they do not appear to compete with H3K4me2 at the active site. It is, however, possible that the same kinetics may not apply in the context of the nucleosome in situ.
Fig. 1.
Inhibition of LSD1 by polyamine analogues. (A) Bisguanidines (1a–1g). (B) Biguanides (2a–2f). (C) Three micrograms of purified LSD1 protein were incubated with 5 μM H3K4me2 (1–21 aa) as substrate in the presence of 1 μM of the indicated analogue. The results represent the mean of three determinations ± SD. (D and E) The effects of increasing concentrations of 1c (D) or 2d (E) on LSD1 activity in the presence of increasing substrate concentrations. Double reciprocal plots indicate inhibition of LSD1 by 1c and 2d to be noncompetitive.
To determine whether compounds 1c and 2d had cellular activity, the effects on global H3K4 methylation were examined after exposure of HCT116 human colon carcinoma cells to increasing concentrations of each compound for 48 h. This exposure produced significant increases in both H3K4me1 and H3K4me2, without affecting global H3K9me2 levels (Fig. 2 A and B). By contrast, compounds 1d and 2b, which are poor inhibitors of purified LSD1, were significantly less effective at increasing global levels of H3K4me2 in treated HCT116 cells [supporting information (SI) Fig. 7]. Levels of H3K4me3, which is not a substrate for LSD1 (6), were not affected.
Fig. 2.
Inhibition of LSD1 by polyamine analogues increases global H3K4me1 and H3K4me2, resulting in reexpression of aberrantly silenced genes in treated human colon cancer cells. (A) HCT116 cells were exposed to increasing concentrations of the indicated compound for 48 h, and 30 μg of nuclear protein per lane were analyzed for expression of H3K4me1, H3K4me2, H3K9me2, and PCNA as a loading control. (B) Histograms represent the mean protein expression levels of three determinations relative to PCNA ± SD, as determined by quantitative immunoblotting using infrared detection and analysis. (C) HCT116 cells were treated for 48 h with the indicated compounds, and total RNA was extracted for RT-PCR analysis of SFRP1, SFRP4, SFRP5, and GATA5 expression. GAPDH is included as an internal control. The results shown are from a single experiment repeated at least three times with similar results. (D) RKO cells were exposed to increasing concentrations of the indicated compound for 48 h, and 30 μg of nuclear protein per lane were analyzed for expression of H3K4me2 and PCNA. The histogram represents the mean protein amount of three determinations relative to PCNA ± SD. (E) RKO cells were treated for 48 h with the indicated compounds and total RNA was extracted for RT-PCR analysis of SFRP4 and SFRP5 expression. GAPDH is included as an internal control. The results shown are from a single experiment repeated at least three times with similar results.
Promoter region H3K4me2 is associated with expressed genes (11, 12), and although this mark can occur outside the promoter region, in vertebrates it is predominantly found proximal to active genes (18, 19). In cancer cells, this mark is depleted in the promoters of several epigenetically silenced, and aberrantly DNA hypermethylated genes important in tumorigenesis (20). Multiple such suppressed genes are present in HCT116 cells, as well as in many primary human colon carcinomas (3, 20–23). Therefore, we examined whether such genes could be reexpressed after treatment with 1c or 2d. We examined six genes: four members of the secreted frizzle-related protein family, SFRP1, SFRP2, SFRP4, and SFRP5, that are each important in the normal regulation of the WNT signaling pathway (24), and two GATA family transcription factors, GATA4 and GATA5 (25). Of these, SFRP1, SFRP4, SFRP5, and GATA5 were reexpressed after 48 h treatment with either compound (Fig. 2C). In HCT116 cells, compound 2d exhibited greater potency than 1c, inducing reexpression at 1 μM. However, for SFRP4, SFRP5, and GATA5, 10 μM 2d actually produced less expression than did lower doses, suggesting that the growth inhibition observed in HCT116 cells at higher concentrations of 2d may lead to lower reexpression of specific genes (SI Fig. 8). The effects of 1c and 2d were not, however, limited to HCT116 cells, as similar results with respect to reexpression of SFRP4 and SFRP5, and global H3K4me2 levels were observed in RKO colon cancer cells (Fig. 2 D and E).
To quantify the level of gene reexpression achieved with 1c and 2d treatment compared with other agents affecting chromatin structure, HCT116 cells were treated with 1 μM of the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (DAC) (22), 300 nM of the class I/II histone deacetylase inhibitor trichostatin A (TSA) (26), or the less effective LSD1-inhibiting analogues, compounds 1d and 2b (Fig. 3). DAC and TSA concentrations were chosen based on maximal inhibitor effect without cytotoxicity. After 48-h treatment with each individual agent, total RNA was extracted, and expression of SFRP4 and SFRP5 was determined by quantitative real-time PCR (qPCR) relative to expression achieved by DAC treatment. Treatment with 1c or 2d resulted in substantial reexpression of both genes (≈20–35% that achieved by DAC treatment). This is in contrast to a lack of measurable expression after treatment with TSA, 1d, or 2b. These results demonstrate that both 1c and 2d, although not as potent as DAC, are effective at producing highly significant reexpression of epigenetically silenced genes. Furthermore, the inability of 1d and 2b treatment to result in gene reexpression is consistent with the hypothesis that the reexpression of silenced genes by 1c and 2d is a result of their potent LSD1 inhibition.
Fig. 3.
Relative reexpression of SFRP4 and SFRP5 induced by polyamine analogue inhibitors of LSD1. HCT116 cells were treated with 5 μM 1c, 2d, 1d, or 2b; 1 μM DAC; or 300 nM TSA for 48 h. (A) qPCR analysis of SFRP4 and SFRP5 expression. Results are presented relative to expression induced by DAC and represent the mean of three independent experiments, each performed in triplicate ± SD. (B) Representative qPCR products were analyzed on GelStar-stained 2% agarose gel.
To examine whether inhibitor-induced gene reexpression was accompanied by changes in regulatory chromatin marks at the specific gene promoters, chromatin immunoprecipitation (ChIP) analysis was used. ChIP analysis of analogue-treated HCT116 cells revealed that gene reexpression was accompanied by increased H3K4me1 and H3K4me2 at the promoters of all reexpressed genes (Fig. 4 A–H). Consistent with results observed above for global methylation, no change in promoter H3K4me3 levels was detected (SI Fig. 9). The increased H3K4 methylation was accompanied by increased acetyl-H3K9, a chromatin mark associated with active transcription, and concurrent with decreases in the repressive marks H3K9me1 and H3K9me2 (Fig. 4 A–H and SI Fig. 10). H3K9me3 levels and H3K27 methylation status remained unchanged (SI Fig. 9), similar to findings observed in the reexpression of silenced genes in cells treated with the DNA demethylating agent, DAC (20). It is important to note that the inhibition of demethylase activity by 1c and 2d appears to be selective for LSD1 at the promoter sites examined here, and thus may not affect the activity of the JmjC domain-containing histone demethylases (7, 8, 27), because no increase in H3K9 methylation (mono-, di-, or tri-) was observed and H3K9me1 and H3K9me2 levels actually decreased in the promoters of the reexpressed genes. However, this is not direct proof of selective inhibition of LSD1 and further study will be necessary to probe the selectivity of the analogues among the growing family of lysine demethylases (6–10, 28).
Fig. 4.
Inhibition of LSD1 by polyamine analogues increases activating H3K4me2 and acetyl H3K9 marks and decreases repressive H3K9me1 and H3K9me2 marks at the promoters of reexpressed genes. HCT116 cells were treated with 5 μM of the indicated compound for 48 h. (A–D) ChIP analysis was used to determine the occupancy of the indicated promoters by multiple activating and repressive marks. (E–H) The quantified results are the means of three independent experiments with an SD as indicated.
ChIP analysis also confirmed that LSD1 is present at the promoter of each gene examined, and treatment of cells with 1c or 2d has no effect on LSD1 promoter occupancy (Fig. 5). Interestingly, SFRP2 and GATA4, two genes that are not reexpressed after analogue exposure, demonstrate higher levels of LSD1 present at their promoter relative to those genes that are reexpressed and exhibit no change in H3K4me2 or H3K4me1 levels in response to treatment (Fig. 5 A and B). These results suggest that the lack of SFRP2 and GATA4 reexpression may be a result of an inability to sufficiently inhibit the elevated levels of promoter-associated LSD1 at these specific sites (Fig. 5C). Other events, in addition to levels of promoter LSD1 occupancy, may also contribute to the observed selective reexpression of silenced genes. Such events include recruitment of transcriptional activation complexes containing histone acetyltransferase activity and chromatin remodeling proteins, in combination with the recruitment of specific histone lysine methyltransferases to produce activating marks (5, 9, 29–32). These events combined with binding of the basal transcriptional machinery to the promoter may act in a selective manner to complement the inhibition of LSD1. Consequently, other factors responsible for the selective gene reexpression that results from LSD1 inhibition remain to be identified.
Fig. 5.
Promoter occupancy by LSD1. (A) HCT116 cells were treated with 5 μM of the indicated compounds for 48 h. ChIP analysis was performed to determine the occupancy of the indicated promoters by LSD1. (B) The quantified results are the mean of three independent experiments ± SD, relative to LSD1 detected at the SFRP1 promoter of untreated cells. (C) After HCT116 cells were treated with 5 μM of the indicated compounds for 48 h, ChIP was used to determine the levels of H3K4me2 and H3K4me1 at the promoters of SFRP2 and GATA4.
Promoter CpG island DNA hypermethylation collaborates with specific histone marks in the epigenetic inactivation of tumor suppressor genes (3), and this is true for all such genes examined in HCT116 cells (23). Therefore, we examined whether analogue inhibition of LSD1 was accompanied by changes in promoter DNA methylation. Methylation-sensitive PCR (MSP) suggested that analogue exposure produced a decrease in promoter methylation of SFRP4 and SFRP5, but not in the SFRP1 and GATA5 promoters (data not shown). However, the small changes in DNA methylation observed with bisulfite sequencing (SI Fig. 11) after treatment with 2d suggest that such demethylation plays a relatively minor role in reexpression and may be a consequence of reactivation rather than a cause. These results indicate that analogue-induced increases in H3K4 methylation alone are potent enough as activating marks to produce some reexpression of even heavily methylated genes.
The natural polyamines are known to associate with and alter the conformation of DNA and chromatin (33–35). Additionally, treatment of cells with specific polyamine analogues are known to alter polyamine metabolism and polyamine pools, and may thus have secondary effects on chromatin (36). Therefore, the effects of treatment with 1c or 2d on polyamine biosynthesis and polyamine pools were determined in HCT116 (SI Table 1). No changes in polyamine pools were observed after 48-h exposure to 5 μM of either compound and only treatment with 2d was observed to result in a modest decrease (≈40%) in activity of the rate-limiting step of polyamine biosynthesis, ornithine decarboxylase (37). These results are consistent with the hypothesis that the changes in chromatin marks and gene reexpression observed are not a result of changes in polyamine pools, but are due to inhibition of LSD1.
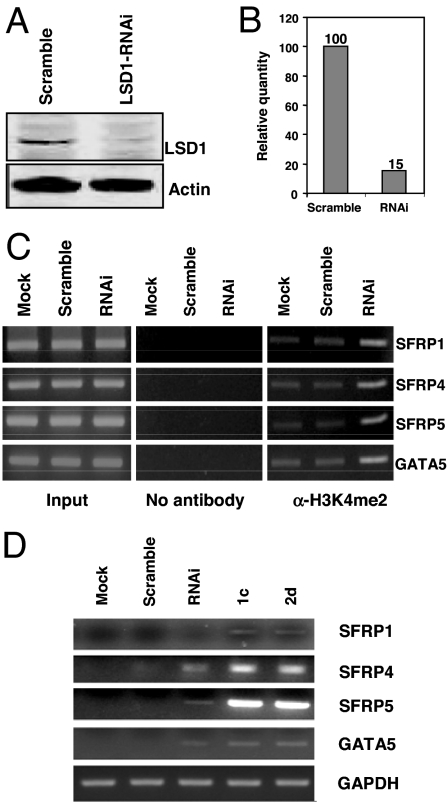
Finally, the effects of the inhibition of LSD1 by 1c and 2d were compared with an RNAi-mediated decrease in LSD1 expression (Fig. 6). Forty-eight-hour exposure of HCT116 cells to LSD1-targeting siRNA resulted in an 85% decrease in LSD1 protein (Fig. 6 A and B), which was accompanied by increased H3K4me2 at the promoters of SFRP1, SFRP4, SFRP5, and GATA5, and reexpression of each gene (Fig. 6 C and D). However, 48-h treatment with 5 μM of either 1c or 2d was more effective in inducing the reexpression of silenced genes than siRNA treatment, particularly in the case of SFRP4 and SFRP5. These data indicate that pharmacologic inhibition of LSD1 is more effective than RNAi with respect to reexpression of silenced genes, and may reflect inherent differences in chromatin structure resulting from inhibitor/LSD1 complexes versus siRNA-induced decreases in LSD1.
Fig. 6.
Knockdown of LSD1 by siRNA leads to specific gene reexpression. (A) HCT116 cells were transfected with scrambled or LSD1-targeted siRNA oligonucleotides for 48 h. Proteins isolated from transfected cells were subjected to quantitative immunoblotting with an antibody to LSD1. (B) The histogram represents the relative LSD1 protein quantity in scrambled and LSD1 siRNA transfectants. (C) ChIP analysis was used to determine the levels of H3K4me2 in the promoters of indicated genes. (D) HCT116 cells were treated for 48 h with 5 μM 1c or 2d or transfected with scrambled or LSD1 siRNA oligonucleotides for 48 h. RNA was extracted for RT-PCR analysis of expression of the indicated genes. GAPDH is included as an internal control.
Abnormal epigenetic silencing of tumor suppressor genes is associated with the development and progression of multiple human cancers (3, 38). The recognition of the important role of epigenetic regulation in cancer has led to active efforts to develop drugs that can be used to restore the genes of interest to a transcriptionally active state. Particular focus has been on combinations of histone deacetylase inhibitors and DAC, which are currently in clinical trial. The few studies demonstrating inhibition of LSD1, primarily by siRNA, have focused on changes in global and promoter specific H3K4me2 and the increased expression, not reactivation, of LSD1 target genes (6, 9, 13). Lee et al. (13), who reported an increase in H3K4me2 in P19 EC cells treated with the nonspecific monoamine oxidase inhibitor, tranylcypromine, examined only one other chromatin mark, the activating acetyl-H3K9 at the Oct4 promoter. However, instead of observing the expected increase in acetyl-H3K9, they reported a decrease in this activating mark. The present study is thus the first report to present data regarding changes in other chromatin marks and activation of silenced genes as a consequence of LSD1 inhibition.
The results presented here reveal that two promising lead compounds, 1c and 2d, are potent inhibitors of LSD1, and that this inhibition results in increased active chromatin marks and decreased repressive marks in the promoter regions of specific genes. Recent data indicate that the ratios between repressive and active chromatin marks may regulate gene transcription states (19), and this may be an important factor in the degree of gene reactivation and selectivity observed here. It is also of note that emerging data suggest LSD1 activity is involved in regulating ligand-dependent transcription of both androgen- and estrogen-dependent genes (39–41). Therefore, further studies may be necessary to determine the optimal strategies for improving selective reexpression of target genes.
The fact that the polyamine analogue LSD1 inhibitors as single agents are capable of leading to reexpression of several important aberrantly silenced genes suggests the usefulness of such a strategy in a clinical setting. The combination of histone deacetylase inhibitors and DAC has demonstrated significant clinical responses in specific leukemias, presumably in part through the reexpression of epigenetically silenced genes (42). Therefore, the potential of using LSD1 inhibitors, both alone and in combination with other chromatin-modifying agents, presents itself as an intriguing possibility that merits further study.
Materials and Methods
Compounds, Peptides, Histones, and Culture Conditions.
Biguanide and bisguanidine polyamine analogues were synthesized as reported previously (17). Stock solutions (10 mM in double distilled H2O) of each compound were diluted with medium to the desired concentrations for specific experiments. Synthetic H3K4me2 peptides were purchased from Upstate Biotechnology (Charlottesville, VA). Bulk histones were purchased from Sigma (St. Louis, MO). HCT116 colorectal carcinoma cells were maintained in McCoy's 5A medium and RKO cells were maintained in MEM medium, both supplemented with 10% FBS (Gemini Bio-Products, Woodland, CA) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA), and grown at 37°C in 5% CO2 atmosphere.
Expression, Purification, and Demethylase Assay of Recombinant Proteins.
Full-length human LSD1 cDNA was subcloned into the pET15b bacterial expression vector (Novagen, Madison, WI) in-frame with an N-terminal 6× HIS-tag and transformed into the BL21(DE3) strain of Escherichia coli. Selection, expression, and purification of recombinant LSD1 protein were performed as described previously (SI Fig. 12) (6). Briefly, expression of LSD1-HIS protein was induced by 1 mM isopropyl β-d-thiogalactoside for 4 h at 37°C. The HIS-tagged protein was purified by using Ni-nitrilotriacetic acid affinity purification resin and column as recommended by the manufacturer (Qiagen, Valencia, CA). Bound protein was eluted by imidazole, and the eluate was dialyzed in PBS at 4°C. Enzymatic activity of LSD1 was examined by using luminol-dependent chemiluminescence to measure the production of H2O2, as described previously (43). In brief, LSD1 activity was assayed in 50 mM Tris (pH 8.5), 50 mM KCl, 5 mM MgCl, 5 nmol of luminol, and 20 μg/ml horseradish peroxidase with the indicated concentrations of H3K4me2 (1–21 aa) peptide as substrate. The integral values were calibrated against standards containing known concentrations of H2O2, and the activities expressed as picomoles of H2O2 per milligram of protein per minute. In addition, 5 μg of purified bulk histones were incubated with or without 5 μg of purified LSD1 in 50 mM Tris (pH 8.5), 50 mM KCl, 5 mM MgCl, 0.5% BSA, and 5% glycerol for 3 h at 37°C. This reaction mixture was analyzed by Western blotting with antibodies (Upstate Biotechnology) that specifically recognize the dimethyl group of H3K4.
Western Blotting.
Cytoplasmic and nuclear fractions were prepared for Western blot analysis by using the NE-PER Nuclear and Cytoplasmic Extraction kit (Pierce, Rockford, IL). Primary antibodies against H3K4me2, H3K9me2, and LSD1 were from Upstate Biotechnology. The PCNA monoclonal antibody was purchased from Oncogene Research Products (Cambridge, MA). Dye-conjugated secondary antibodies were used to quantify Western blot results with the Odyssey Infrared Detection system and software (LI-COR Biosciences, Lincoln, NE).
RNA Isolation, RT-PCR, and qPCR.
RNA for RT-PCR and qPCR was extracted by using TRIzol reagent (Invitrogen) and used for RT-PCR as described previously (22). First-strand cDNA was synthesized by using M-MLV reverse transcriptase with an oligo(dT) primer (Invitrogen). PCR was performed by using the following primers: SFRP1, sense, TCT GAG GCC ATC ATT GAA CA; SFRP1, antisense, GAA GTG GTG GCT GAG GTT GT; SFRP2, sense, AAG CCT GCA AAA ATA AAA ATG ATG; SFRP2, antisense, TGT AAA TGG TCT TGC TCT TGG TCT; SFRP4, sense, TCT ATG ACC GTG GCG TGT GC; SFRP4, antisense, ACC GAT CGG GGC TTA GGC GTT TAC; SFRP5, sense, ACC GCG CCT CCA GTG ACC A; SFRP5, antisense, TCT CCT TGA TGC GCA TTT TGA CCA; GATA4, sense, GGC CGC CCG ACA CCC CAA TCT; GATA4, antisense, ATA GTG ACC CGT CCC ATC TCG; GATA5, sense, CCT GCG GCC TCT ACC ACA A; GATA5, antisense, GGC GCG GCG GGA CGA GGA C. A total of 35 cycles of amplification was performed for each of the RT-PCR experiments. GAPDH was amplified as an internal control. Amplified products were analyzed on 2% agarose gels with GelStar staining (Cambrex, Walkersville, MD).
qPCR of SFRP4 and SFRP5 was performed as published previously (44) by using the iQ SYBR Green Supermix kit (Bio-Rad, Hercules, CA). The same forward and reverse primers as were used for RT-PCR were used for qPCR in a MyiQ single color real-time PCR machine (Bio-Rad) with GAPDH as an internal control. Amplification conditions for SFRP4 consisted of a 15-min denaturation step followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s. Identical conditions for SFRP5 were used except the annealing temperature was 57°C. To quantify relative expression, the comparative cycle threshold (Ct) method was used, normalizing the Ct values for the gene of interest to the Ct values of GAPDH relative to untreated control.
ChIP.
All reagents and antibodies for ChIP were purchased from Upstate Biotechnology, and ChIP was performed as reported previously (20). Briefly, 2 × 106 cells were used for each ChIP assay. Control and treated cells were exposed to 1% formaldehyde to cross-link proteins. The cell pellets were resuspended in SDS lysis buffer with protease inhibitors, and chromatin samples were sonicated on ice three times for 10 s each, to give average lengths of 1- to 1.5-kb sheared genomic DNA. Lysates were then centrifuged for 10 min, diluted 10-fold, and precleared with protein A-agarose beads diluted in ChIP dilution buffer. Antibodies against H3K4me2, acetyl-H3K9, mono-, di-, or trimethylated H3K9 or H3K27, and LSD1 were used as indicated for immunoprecipitation of protein–DNA complexes. After overnight agitation at 4°C, immune complexes were collected and washed with protein A-agarose beads, low salt immune complex wash buffer, high salt immune complex wash buffer, and finally TE buffer. The immune complexes were eluted with 1% SDS, 0.1 M NaHCO3. The cross-links were reversed overnight at 65°C, and DNA was recovered by phenol extraction, ethanol precipitation, and resuspension in 50 μl of sterile water. PCR primer sets used for amplification of precipitated fragments were as published previously (22). Sheared genomic DNA was used as a positive control (input). Chromatin eluted from immunoprecipitations lacking antibodies was used as a “no antibody” control. PCR products were run on 2% agarose gels and quantified by using Kodak (Rochester, NY) Digital Science 1D Image Analysis software as reported previously (20, 44, 45). The concordance of this method with quantitative ChIP was confirmed for changes in H3K4me2 and acetyl H3K9 at the promoters of SFRP4 and SFRP5 by using qPCR (SI Fig. 10). qPCR was performed with the same primers used for standard ChIP on the MyiQ single color real-time PCR machine (Bio-Rad) by using 35 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s for both sets of primers. Results were quantified as described above for qPCR except that input DNA was used for normalization.
Ornithine Decarboxylase and Polyamine Pool Analysis.
Ornithine decarboxylase enzyme activity was measured as we have published previously (46). Intracellular polyamine pools were determined by the methods of Kabra et al. (47). Protein concentrations were determined by using the Bio-Rad Protein Assay (Bio-Rad).
Methylation-Specific PCR and Bisulfite Sequencing.
Genomic DNA from HCT116 cells was bisulfite modified and amplified by using primers specific for either the methylated or unmethylated DNA under the conditions described previously (48). The methylation-sensitive PCR primer sets and conditions for SFRP1, SFRP4, SFRP5, and GATA5 were described previously (23).
Bisulfite sequencing of the promoters of the SFRP4 and SFRP5 genes was performed as reported previously (23) by using the published primer sequences and conditions.
RNAi.
The previously published (39) siRNA oligonucleotide duplexes targeting LSD1 (5′-CACAAGGAAAGCTAGAAGA-3′), and the scrambled control (5′-CTTGCTATGAGAACAAATT-3′), were purchased from Dharmacon (Lafayette, CO). Transient transfections were performed in HCT116 cells with LipofectAMINE 2000 reagent (Invitrogen) as recommended by the manufacturer. After 48 h of exposure to 200 pmol per 1 × 105 cells, cells were harvested and lysates analyzed for LSD1 expression, gene expression, and changes in H3K4me2 levels.
Acknowledgments
We thank Drs. James Herman, Ken Kinzler, and Bert Vogelstein for helpful comments on the manuscript; Kelly McGarvey and Dr. Joyce Ohm (both of Johns Hopkins University School of Medicine) for supplying PCR primers and advice; and Amy Hacker for her technical expertise. This work was supported by National Institutes of Health Grants CA43318, CA51085, CA98454, and ES11858.
Abbreviations
- LSD1
lysine-specific demethylase 1
- H3K4
lysine residue 4 of histone protein 3
- H3K9
lysine residue 9 of histone protein 3
- SFRP
secreted frizzle-related protein
- DAC
5-aza-2′-deoxycytidine
- TSA
trichostatin A
- qPCR
quantitative real-time PCR
- ChIP
chromatin immunoprecipitation.
Footnotes
Conflict of interest statement: With respect to competing financial interests, a patent application has been filed by P.M.W. and R.A.C. to cover the compounds reported in this manuscript. As of January 2007, P.M.W. and R.A.C. serve as consultants to Cellgate, Inc., which has an option to license these compounds.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700720104/DC1.
References
- 1.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Baylin SB, Ohm JE. Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 4.Lachner M, O'Sullivan RJ, Jenuwein T. J Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 5.Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 8.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 11.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, et al. Proc Natl Acad Sci USA. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 13.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Chem Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Devereux W, Woster PM, Stewart TM, Hacker A, Casero RA., Jr Cancer Res. 2001;61:5370–5373. [PubMed] [Google Scholar]
- 15.Federico R, Leone L, Botta M, Binda C, Angelini R, Venturini G, Ascenzi P. J Enzyme Inhib. 2001;16:147–155. doi: 10.1080/14756360109162364. [DOI] [PubMed] [Google Scholar]
- 16.Bianchi M, Polticelli F, Ascenzi P, Botta M, Federico R, Mariottini P, Cona A. FEBS J. 2006;273:1115–1123. doi: 10.1111/j.1742-4658.2006.05137.x. [DOI] [PubMed] [Google Scholar]
- 17.Bi X, Cacchi CJ, Rattendi D, Woster PA. Bioorg Med Chem Lett. 2006;16:3229–3232. doi: 10.1016/j.bmcl.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 20.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 21.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fahrner JA, Eguchi S, Herman JG, Baylin SB. Cancer Res. 2002;62:7213–7218. [PubMed] [Google Scholar]
- 23.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 24.Rattner A, Hsieh JC, Smallwood PM, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. Proc Natl Acad Sci USA. 1997;94:2859–2863. doi: 10.1073/pnas.94.7.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akiyama Y, Watkins N, Suzuki H, Jair KW, van Engeland M, Esteller M, Sakai H, Ren CY, Yuasa Y, Herman JG, et al. Mol Cell Biol. 2003;23:8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 27.Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 28.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Heng Qi H, Whetstine JR, Bonni A, Roberts TM, Shi Y. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Dillon SC, Zhang X, Trievel RC, Cheng X. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y, Whetstine JR. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Li B, Carey M, Workman JL. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Basu HS, Schwietert HC, Feuerstein BG, Marton LJ. Biochem J. 1990;269:329–334. doi: 10.1042/bj2690329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feuerstein BG, Williams LD, Basu HS, Marton LJ. J Cell Biochem. 1991;46:37–47. doi: 10.1002/jcb.240460107. [DOI] [PubMed] [Google Scholar]
- 35.Hobbs CA, Paul BA, Gilmour SK. Exp Cell Res. 2003;290:427–436. doi: 10.1016/s0014-4827(03)00352-5. [DOI] [PubMed] [Google Scholar]
- 36.Casero RA, Jr, Woster PM. J Med Chem. 2001;44:1–26. doi: 10.1021/jm000084m. [DOI] [PubMed] [Google Scholar]
- 37.Pegg AE. J Biol Chem. 2006;281:14529–14532. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- 38.Jones PA, Baylin SB. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 40.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, et al. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, et al. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, Grever M, Galm O, Dauses T, Karp JE, et al. Cancer Res. 2006;66:6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Murray-Stewart T, Devereux W, Hacker A, Frydman B, Woster PM, Casero RA., Jr Biochem Biophys Res Commun. 2003;304:605–611. doi: 10.1016/s0006-291x(03)00636-3. [DOI] [PubMed] [Google Scholar]
- 44.Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casero RA, Jr, Go B, Theiss HW, Smith J, Baylin SB, Luk GD. Cancer Res. 1987;47:3964–3967. [PubMed] [Google Scholar]
- 47.Kabra PM, Lee HK, Lubich WP, Marton LJ. J Chromatogr. 1986;380:19–32. doi: 10.1016/s0378-4347(00)83621-x. [DOI] [PubMed] [Google Scholar]
- 48.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]