Abstract
The circulating enzyme, α2-antiplasmin cleaving enzyme (APCE), has very similar sequence homology and proteolytic specificity as fibroblast activation protein (FAP), a membrane-bound proteinase. FAP is expressed on activated fibroblasts associated with rapid tissue growth as in embryogenesis, wound healing, and epithelial-derived malignancies, but not in normal tissues. Its presence on stroma suggests that FAP functions to remodel extracellular matrix (ECM) during neoplastic growth. Precise biologic substrates have not been defined for FAP, although like APCE, it cleaves α2-antiplasmin to a derivative more easily crosslinked to fibrin. While FAP has been shown to cleave gelatin, evidence for cleavage of native collagen, the major ECM component, remains indistinct. We examined the potential proteolytic effects of FAP or APCE alone and in concert with selected matrix metalloproteinases (MMPs) on collagens I, III and IV. SDS-PAGE analyses demonstrated that neither FAP nor APCE cleaves collagen I. Following collagen I cleavage by MMP-1, however, FAP or APCE digested collagen I into smaller peptides. These peptides were analogous to, yet different from, those produced by MMP-9 following MMP-1 cleavage. Amino-terminal sequencing and mass spectrometry analyses of digestion mixtures identified several peptide fragments within the sequences of the two collagen chains. The proteolytic synergy of APCE in the cleavage of collagen I and III was not observed with collagen IV. We conclude that FAP works in synchrony with other proteinases to cleave partially degraded or denatured collagen I and III as ECM is excavated, and that derivative peptides might function to regulate malignant cell growth and motility.
Keywords: tumor-stromal cell interactions, fibroblast activation protein, α2-antiplasmin cleaving enzyme, collagen, extracellular matrix, cancer
Introduction
We recently reported the isolation of a plasma prolyl-specific serine proteinase, α2-antiplasmin cleaving enzyme (APCE), which cleaves a 12-residue methionine amino-terminal peptide from circulating precursive α2-antiplasmin (Met-α2AP) [1; 2]. Once this peptide is removed, the derivative form, which has asparagine as its amino-terminus (Asn-α2AP), can be cross-linked by activated fibrin-stabilizing factor (FXIIIa) to fibrin monomers during clot polymerization ~13x faster than precursive Met-α2AP. We noted that APCE shared strong amino acid sequence homology to the previously described fibroblast activation protein (FAP), which is a homodimeric 170 kDa type II integral membrane glycoprotein with both di- and endo-prolylpeptidase activities [3; 4]. Recently the crystallographic structure of the extracellular portion of FAP was reported [5]. It shows the enzyme to have an active-site within a tube-like structure that courses through each monomeric half with apertures of 14 and 24 diameters at the ends of each tube. Although previously stated that FAP or its derivatives do not circulate in blood [6], we concluded that plasma APCE, which lacks the cytoplasmic tail and transmembrane sequence of FAP, may indeed result from FAP [7], possibly as a consequence of unidentified cell membrane “sheddase” activity as occurs with certain other membrane peptidases [8; 9], or that it might be synthesized and secreted as a non-membrane-bound variant of FAP. To date Met-α2AP is the only definitive physiologic substrate identified for native APCE, and it is also cleaved by recombinant FAP [1; 7].
FAP attracted attention because of its significant production by activated stromal fibroblasts of epithelial-derived and sarcomatous malignancies [10–18]. While not detectably produced by normal, quiescent fibroblasts or other types of normal cells, FAP is present in developing embryonic stromal tissues [19; 20] and on activated fibroblasts during wound healing [10; 17]. Immunohistochemical patterns of FAP within and around edges of certain malignancies and its apparent effects on malignant cell growth [10–18; 21–23] have provided convincing data that the heightened expression of FAP may be important in the growth of various cancers. In the past few years, several reports indicate that besides malignancy-associated stroma, both neoplastic parenchymal cells and endothelial cells of budding capillaries may be positive for FAP [24–27]. Details about the role of FAP as it is transiently expressed in fetal growth remain largely unknown and are confounded by the apparent normal development of FAP-null mice embryos [19; 20]. Several reports suggest that FAP functions in the remodeling of extracellular matrix (ECM), and it has been reported that native collagen [4], denatured type I collagen [14; 17; 28] and gelatin [4; 14–17; 28–30] are FAP substrates. Importantly, however, while FAP appears to cleave gelatin, clearly defined effects on native collagen as a substrate have not been reported. Inadequate quantities of pure FAP for use in enzyme-substrate solutions; unknown effects on FAP activity when attached to immunoaffinity beads; an overall lack of described digestion conditions; and the relative slowness of proteolysis by this large complex dimeric enzyme have complicated interpretations about whether native collagen types are indeed cleaved by FAP. Consequently, FAP’s effect on collagen remains cloudy, as evidenced by results that appear to conflict [4; 14]. Since clinical trials targeting FAP are proposed or underway to determine if neoplastic invasion of ECM can be slowed or halted [31–33], and given that neither gelatin or denatured collagen is a component of ECM, the actual effect of this enzyme on its alleged ECM substrate, i.e., collagen, is important to clarify. The difficulty of producing FAP uncontaminated by other trace membrane proteinases could complicate efforts to obtain definitive collagen cleavage data; however, new purification methods developed in our laboratory for the production of significant amounts of pure human plasma-derived APCE as well as human rFAP prompted us to examine the potential proteolytic effects of each enzyme toward purified types I, III and IV collagens as substrates. In addition, the individual effect of APCE or rFAP in combination with selected metalloproteinases (MMPs) that also participate in ECM remodeling [34] was explored. We now report that APCE or rFAP acts in concert with MMP-1 to yield similar cleavage results that are specific for each collagen examined and unique from the effects of either MMP-2 (gelatinase A) or MMP-9 (gelatinase B).
Materials and Methods
Purification of APCE and expression and purification of recombinant FAP
APCE was isolated from citrated human plasma (Sylvan Goldman Blood Institute, Oklahoma City, OK) by sequential chromatographic steps as previously described [7]. A soluble form of recombinant human FAP beginning at residue 35 was expressed by Pichia pastoris and purified from culture media as previously described [7].
Collagen digestion
Rat tail collagen type I, human placenta collagen type III and EHS mouse tumor collagen type IV were purchased from BD Biosciences (Bedford, MA). MMP-1, MMP-2 and MMP-9 were from Sigma (St. Louis, MO). Type I collagen was gelled in 100 μl aliquots at a concentration of 2 mg/ml by raising the pH to 7.5 using 1 N NaOH, according to manufacturer’s instructions, followed by incubation at 37°C for 30 minutes. After gel formation, enzymes (APCE, rFAP, MMP-1, MMP-2, MMP-9, or selected combinations of these; see Figure legends for concentrations) dissolved in 12.5 mM sodium phosphate were layered on the collagen gel and incubated at 37°C for 24 hours. Following incubation, enzyme-containing buffer was removed and any remaining gel was solubilized by the addition of 0.5 N acetic acid. Buffer and gel solution were combined, lyophilized, reduced and analyzed on 4–12% SDS-PAGE gels (Invitrogen, Carlsbad, CA). According to the manufacturer, collagen types III and IV could not be induced to reform a gel, and therefore they were lyophilized to remove acid buffer, resolubilized in 25 mM sodium phosphate buffer, pH 7.5, at a concentration of 2 mg/ml, and divided into 100 μl aliquots prior to the addition of MMP-1, MMP-9, APCE or rFAP enzyme dilutions in 12.5 mM sodium phosphate, pH 7.5 (concentrations in Figure legends). After incubation at 37°C for 24 hours, collagen-enzyme mixtures were lyophilized, reduced and analyzed on 4–12% SDS-PAGE gels.
Determination of APCE and MMP-1 cleavage sites in type I collagen
Type I collagen was gelled as described and incubated with MMP-1 alone; MMP-1 and APCE; or MMP-1, APCE and EDTA at 37°C for 24 hours. An aliquot of each sample was reduced and separated on a 4–12% SDS-PAGE gel after which bands were transferred to PVDF membranes for amino-terminal sequencing of selected fragments. The remaining sample was brought to 70% acetonitrile, which precipitated large proteins, but left peptides in the supernatant. The supernatant was lyophilized and dissolved in 5% acetic acid in preparation for analysis by liquid chromatography/mass spectrometry (LC/MS). A Paradigm HPLC system (Michrom BioResources, Auburn, CA) equipped with a Haisil C18 column (1.0 mm X 150 mm, Higgins Analytical Inc., Mountain View, CA) was utilized for the liquid chromatography. The column was equilibrated with 2% acetonitrile/water containing 0.1% TFA. Upon sample injection, a linear gradient was performed to 50% acetonitrile over 40 min, followed by an increase to 70% acetonitrile over 10 min. Chromatographic separations were monitored at 215 nm wavelength and peptides from selected peaks were subjected to mass analysis on an electrospray mass spectrometer (QSTAR, Applied Biosystems, Foster City, CA) equipped with an ion spray source operated in the positive ion mode. Amino-terminal sequencing was used to characterize the peptides identified by mass. After identification, peptides were located within the published sequence of collagen α1(I) Rattus norvegicus (gi: 2894106) and collagen α2(I) Rattus norvegicus (gi: 16758080) based on mass obtained from MS analysis in conjunction with amino-terminal sequence.
Two peptides, ~115 kDa and ~96 kDa, recovered from the blot of MMP-1 digestion of collagen I appeared to have blocked amino-termini and could not be sequenced by Edman degradation. Since previous reports indicated that amino-terminal Gln of both collagen α1(I) and collagen α2(I) are blocked by cyclization [35; 36], a duplicate sample of the MMP-1 digestion was reduced and subjected to SDS-PAGE, after which the ~115 kDa and ~96 kDa bands were excised, reduced, alkylated and digested with trypsin [37]. Each trypsin digest was analyzed by liquid chromatography-mass spectrometry-mass spectrometry (LC/MS/MS) to obtain molecular weights and MS/MS fragment ion spectra so that the MASCOT MS/MS ion search engine could be used to query the NCBI comprehensive non-identical protein database (http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=MIS).
Results
Cleavage of collagen I
Collagen type I contains two α1(I) chains and one α2(I) chain in a triple-helical “coiled-coil” structure that is resistant to digestion by most proteinases [38; 39]. The chains are crosslinked at each end via acid-soluble bonds between short non-helical stretches referred to as telopeptides [39]. Acid-solubilized collagen I will reform fibrils at neutral pH; however, collagen I obtained by proteinase extraction does not, since the telopeptides are removed by proteolytic cleavage, and therefore crosslinking necessary for triple helix formation cannot occur. As indicated earlier, other groups have reported that FAP cleaves collagen, but the degree of FAP purity was not clearly defined nor were cleavage products or specific cleavage sites shown or identified; in some other reports it was unclear whether intact collagen was used or whether the substrate was actually gelatin [4; 14; 17]. Hence, to clarify and establish whether native collagen is a biologic substrate of FAP, we used acid-extracted collagen I so that when gelled, cross-linking could take place between telopeptides, thus providing collagen in a natural triple-helical structure and crosslinked as it would be in a physiologic ECM milieu. We examined both recombinant FAP and APCE for ability to cleave intact collagen I. As shown in Figure 1a, incubating APCE or rFAP with intact collagen I for 24 hours at 37°C did not yield cleavage products, which prompted the question of whether initial partial proteolysis by another protease might trigger further digestion by APCE or rFAP. Since MMP-1 is a frequent active component of disrupted ECM, and since it is known to make a single cleavage in collagen I at Gly775-Ile776 of α1(I) or Gly775-Leu776 of α2(I) chain to yield two fragments comprising one-fourth and three-fourths of the collagen I molecule [40; 41], MMP-1 was examined for proteolytic synergy with APCE or rFAP with collagen I as substrate. As expected, MMP-1 did cleave intact collagen I to yield the two dominant fragments and a few other very minor fragments as evidenced by the additional faint ~70 kDa bands on SDS-PAGE. Neither MMP-2 nor MMP-9 cleaved intact collagen I. We reasoned that intact collagen I was not digested by either APCE or rFAP prolyl endoproteinase since its triple-helical structure must protect amino acid sequences that contain possible cleavage sites. After limited hydrolysis of type I collagen by MMP-1, however, our results indicate that APCE or rFAP proteolytic sites become exposed and accessible. As evident in Figure 1b, SDS-gel analyses of collagen I digests by APCE or rFAP with MMP-1 show extensive proteolytic degradation to much smaller peptide species (<15 kDa) than produced by MMP-1 alone, and not at all by APCE or rFAP alone.
Figure 1. Digestion of collagen I by APCE, rFAP, MMP-1, MMP-2 and MMP-9.
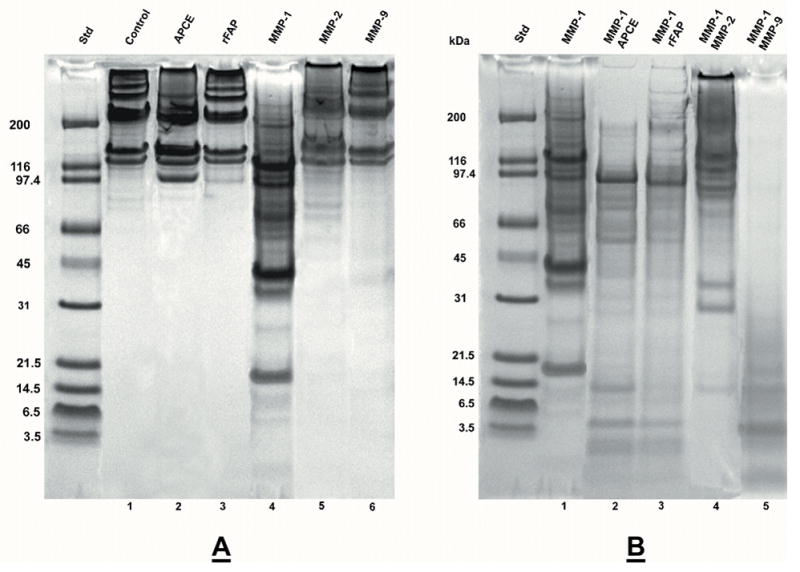
Rat tail collagen type I was gelled at 2 mg/ml and incubated at 37 °C for 24 hours with enzymes in 12.5 mM Na PO4 buffer, pH 7.5. Panel A. Lane: 1) Control-buffer; 2) 16 μg APCE; 3) 16 μg rFAP; 4) 1.5 μg MMP-1; 5) 1 μg MMP-2; 6) 1 μg MMP-9. Panel B. Lane: 1) 1.5 μg MMP-1; 2) 1.5 μg MMP-1, 16 μg APCE; 3) 1.5 μg MMP-1, 16 μg rFAP; 4) 1.5 μg MMP-1, 2.5 μg MMP-2; 5) 1.5 MMP-1, 3.7 μg MMP-9. Digests were reduced and analyzed by 4–12% SDS-PAGE. Where present, the ~97 kDa band is either APCE or rFAP.
Given that perturbations of ECM are also characterized by increases of MMP-2 and MMP-9 [42], the effect of either in place of APCE or rFAP was also analyzed. Digestion mixtures containing MMP-1 and MMP-2 extended collagen I fragmentation by MMP-1 alone only modestly. In contrast, those containing MMP-1 and MMP-9 produced a terminal digest composed dominantly of peptides <30 kDa, whereas the MMP-1 plus MMP-2 terminal digest still contained protein bands >90 kDa. Figures 2a and 2b demonstrate APCE and rFAP concentration-dependent cleavages of collagen I in the presence of a constant amount of MMP-1 and clearly show that a decrease in higher molecular weight bands and an increase in lower molecular weight bands occur with increasing concentrations of APCE or rFAP.
Figure 2. Concentration dependent digestion of collagen I by MMP-1 and APCE or rFAP.
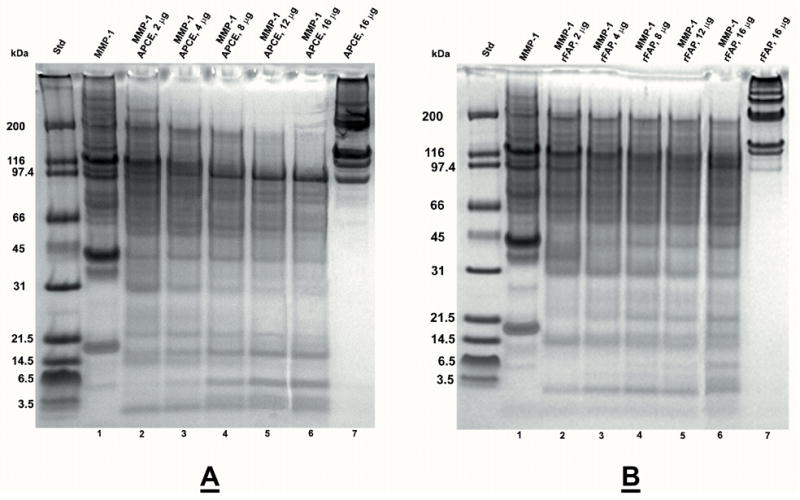
Rat tail collagen type I was gelled at 2 mg/ml and incubated at 37 °C for 24 hours with enzymes in 12.5 mM Na PO4 buffer, pH 7.5. Panel A. Lane: 1) 1.5 μg MMP-1; 2) 1.5 μg MMP-1, 2 μg APCE; 3) 1.5 μg MMP-1, 4 μg APCE; 4) 1.5 μg MMP-1, 8 μg APCE; 5) 1.5 μg MMP-1, 12 μg APCE; 6) 1.5 μg MMP-1, 16 μg APCE; 7) 16 μg APCE. Panel B. Lane: 1) 1.5 μg MMP-1; 2) 1.5 μg MMP-1, 2 μg rFAP; 3) 1.5 μg MMP-1, 4 μg rFAP; 4) 1.5 μg MMP-1, 8 μg rFAP; 5) 1.5 μg MMP-1, 12 μg rFAP; 6) 1.5 μg MMP-1, 16 μg rFAP; 7) 16 μg rFAP. Digests were reduced and analyzed by 4–12% SDS-PAGE. Where present, the ~97 kDa band is either APCE or rFAP.
As a member of the MMP family, MMP-1 is a zinc-dependent endoproteinase. It has two domains that act in concert: a hemopexin domain for unwinding the triple-helical structure of collagen and a catalytic domain for cleavage. Chung et al. [43] demonstrated that the unwinding and catalytic activities of MMP-1 are clearly distinct, yet both are required for collagen cleavage by MMP-1. Importantly, MMP-1 promotes only a local unwinding of collagen and the overall collagen triple helix is not denatured (41). It has not been reported whether zinc is required for the unwinding function, which prompted us to examine whether MMP-1 could unwind collagen in the presence of EDTA to expose sites otherwise not accessible for cleavage by rFAP or APCE. As expected (41) and shown in Figure 3, EDTA did prevent the cleavage of collagen I by MMP-1. Even at a significantly higher concentration of MMP-1 (1.5 μg), but with EDTA still in the digestion mixture, no new bands were noted in analyses of the reduced digestion mixture on SDS-PAGE as evidenced by the gel pattern being identical to the control sample in Figure 1a. Interestingly, despite the lack of MMP-1 proteolytic function with EDTA present, Figure 3 shows that APCE still cleaved collagen I in the presence of MMP-1 and EDTA. Although digestion occurred slower and less completely than in the absence of EDTA, three distinct lower molecular weight bands (<20 kDa) are clearly evident. These data indicate that 1) while zinc is necessary for MMP-1 catalytic activity, it is not required for the hemopexin domain of MMP-1 to unwind collagen; and 2) the extent of unwinding, which is less denaturing than the conversion of collagen to gelatin, appears to be localized and to not affect the overall triple-helical structure of collagen [41], yet it does allow access to APCE cleavage sites.
Figure 3. Digestion of collagen I by MMP-1 and APCE with or without EDTA.
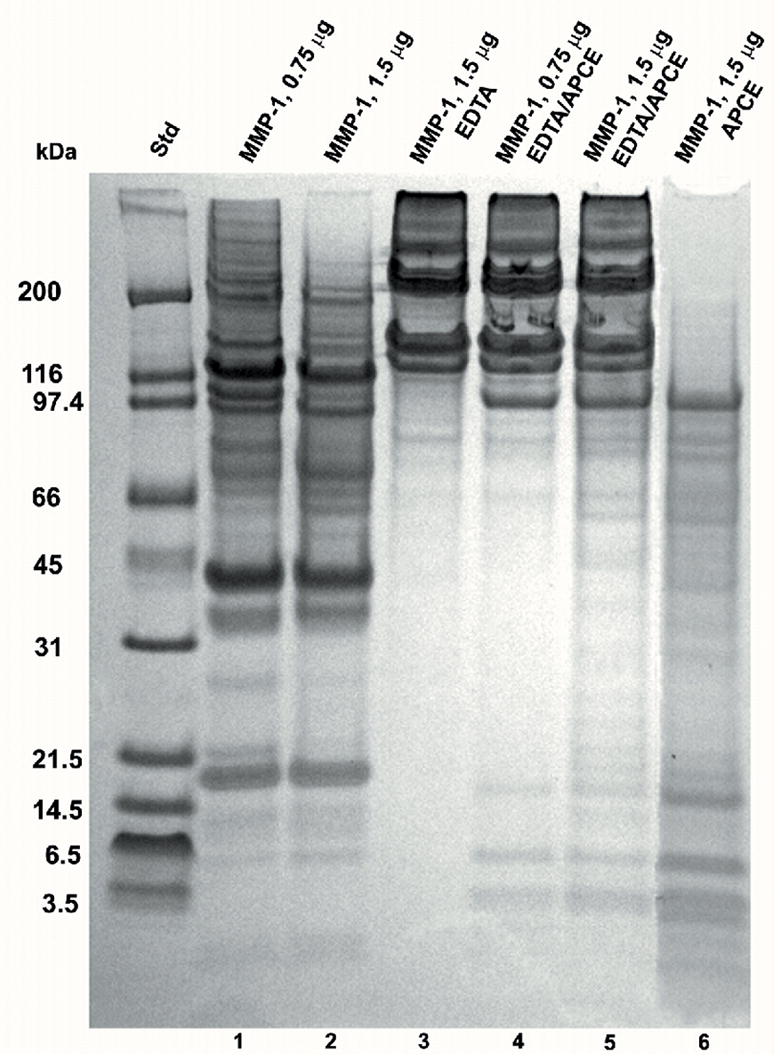
Rat tail collagen type I was gelled at 2 mg/ml and incubated at 37 °C for 24 hours with enzymes in 12.5 mM Na PO4 buffer, pH 7.5. Lane: 1) 0.75 μg MMP-1; 2) 1.5 μg MMP-1; 3) 1.5 μg MMP-1, 20 Mm EDTA; 4) 0.75 μg MMP-1, 20 mM EDTA, 15 μg APCE; 5) 1.5 μg MMP-1, 20 mM EDTA, 15 μg APCE; 6) 1.5 μg MMP-1, 15 μg APCE. Digests were reduced and analyzed by 4–12% SDS-PAGE. Where present, the ~97 kDa band is either APCE or rFAP.
Collagen I cleavage sites for MMP-1 and APCE
To determine cleavage sites for MMP-1 and APCE, digestion mixtures from collagen I cleavage by MMP-1; MMP-1 and APCE; and MMP-1, APCE and EDTA were precipitated by acetonitrile and the remaining soluble peptides were then separated by liquid chromatography. Figure 4 is an overlay of the three chromatograms. As evident, different peptides are produced by the three treatments. Selected peaks were analyzed by MS followed by amino-terminal sequencing to identify peptides. Those resulting from APCE cleavage were identified by mass and amino-terminal sequence as indicated by black underlining in the sequence of the α1(I) chain in Figure 5a and in the sequence of the α2(I) chain in Figure 5b. In addition, prior to precipitation, an aliquot of each cleavage mixture was reduced and electrophoresed on SDS-PAGE gels, after which each band was transferred for amino acid sequencing by Edman degradation to allow placement in the sequence of either collagen chain. The carboxy-terminus of each peptide was estimated from the molecular weight based on SDS-PAGE and the known cleavage preference for APCE, i.e., ...-Gly-Pro-X-... [44]. When both MMP-1 and APCE were present in the incubation mixture, the peptides from the supernatant are indicated by red underlining in the sequence of collagen α1(I) chain and collagen α2(I) chain as shown in Figure 5a and b, respectively. Green underlining signifies collagen I peptides identified from the digestion mixture containing MMP-1/APCE/EDTA where the catalytic activity of MMP-1 was eliminated by EDTA. These peptides were produced by APCE cleavage without prior cleavage by MMP-1, since the latter’s proteolytic activity was absent due to the presence of EDTA. Collagen I cleavage sites for MMP-1 were identified by amino-terminal sequencing of bands blotted to PVDF and indicated in the figures as black triangles. Two of the bands from the blot, ~115 kDa and ~96 kDa, did not yield sequences by Edman degradation, likely because of cyclization of amino-terminal glutamine [35; 36]. It was established that these bands were from the amino-terminal region of the collagen α1(I) and collagen α2(I) chains by tryptic digestion and subsequent LC/MS/MS analyses of duplicate samples. Figures 5a and 5b show that APCE, in the presence of MMP-1, generated more cleavages than MMP-1 by itself. These cleavages occurred at sites distant to the MMP-1 site in both chains, indicating that initial cleavage by MMP-1 must relax the triple-helix sufficiently for APCE/FAP to recognize and cleave multiple sites within the partially digested-monomer to yield smaller fragments. In addition, identical APCE cleavages by MMP-1 plus APCE, or MMP-1 plus APCE and EDTA, were identified in the amino-terminal region of both collagen chains; whether EDTA was present or not, the same ~8 kDa peptide from the α1(I) chain and the same ~16 kDa peptide from the α2(I) chain were produced.
Figure 4. Liquid chromatography of peptides produced from collagen I digestion.
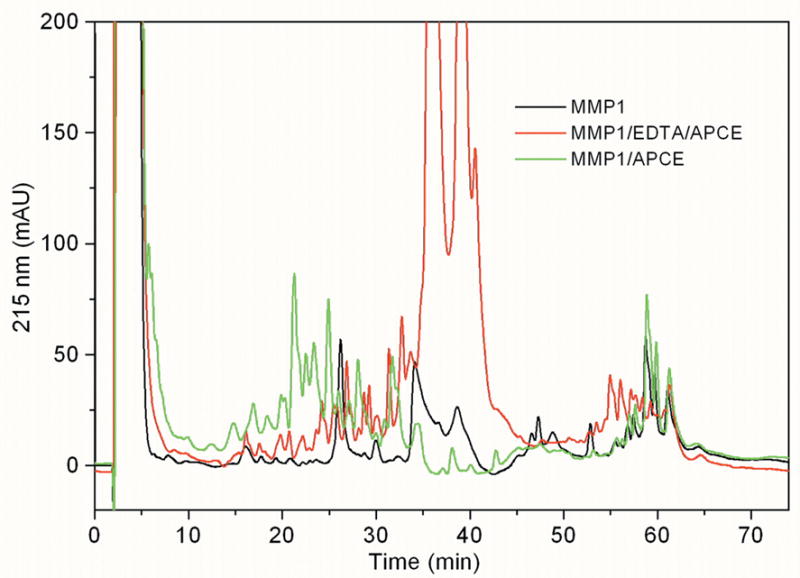
Rat tail collagen type I, gelled at 2 mg/ml was incubated at 37 °C for 24 hours with 1.5 μg MMP-1 (black), 1.5 μg MMP-1, 15 μg APCE (green) or 1.5 μg MMP-1, 20 mM EDTA, 15 μg APCE (red). Samples were precipitated with 70% acetonitrile and the supernatant analyzed by reverse phase chromatography.
Figure 5. Peptides identified from digestion of collagen I.
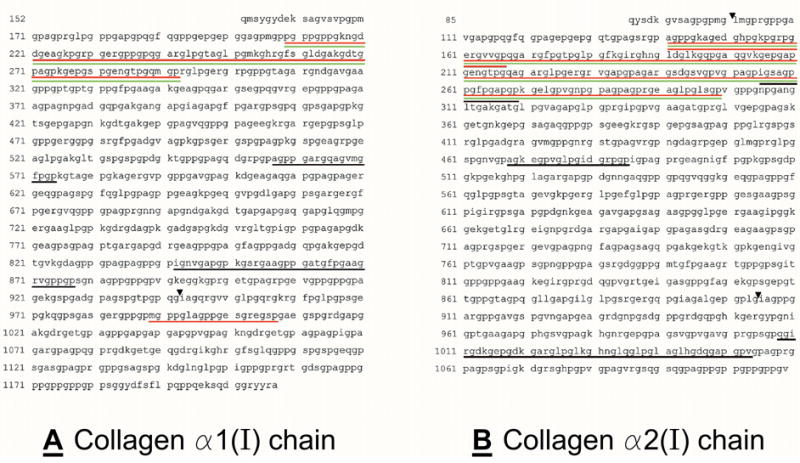
Rat tail collagen type I was gelled at 2 mg/ml and incubated at 37 °C for 24 hours with 1.5 μg MMP-1; 1.5 μg MMP-1, 15 μg APCE or 1.5 μg MMP-1, 20 mM EDTA, 15 μg APCE. Panel A, collagen α1(I) chain and Panel B, collagen α2(I) chain. Peptides resulting from APCE cleavage in the presence of MMP-1: red underlining indicates peptides identified by amino-terminal sequencing; black indicates peptides identified by amino-terminal sequence and mass spectrometry analysis. Peptides identified by amino-terminal sequencing and mass spectrometry analysis resulting from APCE cleavage in the presence of MMP-1/EDTA are indicated by green underlining. MMP-1 cleavage sites identified are indicated by ▾.
Cleavage of collagen III and collagen IV
Type III collagen is found along with type I collagen in different ratios in almost all tissues [39] and appears to co-assemble with collagen type I in heterotypical fibrils in skin and tendon [38]. Collagen III is a fibrillar collagen like type I and likewise has a triple helical structure resistant to digestion by a number of proteolytic enzymes. Besides type I collagen [42], MMP-1 also cleaves type III, thereby suggesting that APCE or rFAP in conjunction with MMP-1 might also proteolytically degrade type III collagen into smaller fragments than MMP-1 alone. As seen in Figure 6a, neither APCE nor rFAP cleaved collagen type III, but following incubation of collagen III with MMP-1 and APCE or rFAP, significant proteolysis occurred that caused further fragmentation and production of smaller peptides (< 30 kDa) than those produced by MMP-1 alone. Hence, the pattern of collagen type III cleavage by MMP-1, with or without APCE or rFAP, is analogous to that of collagen type I.
Figure 6. Digestion of collagen III and collagen IV. Panel A.
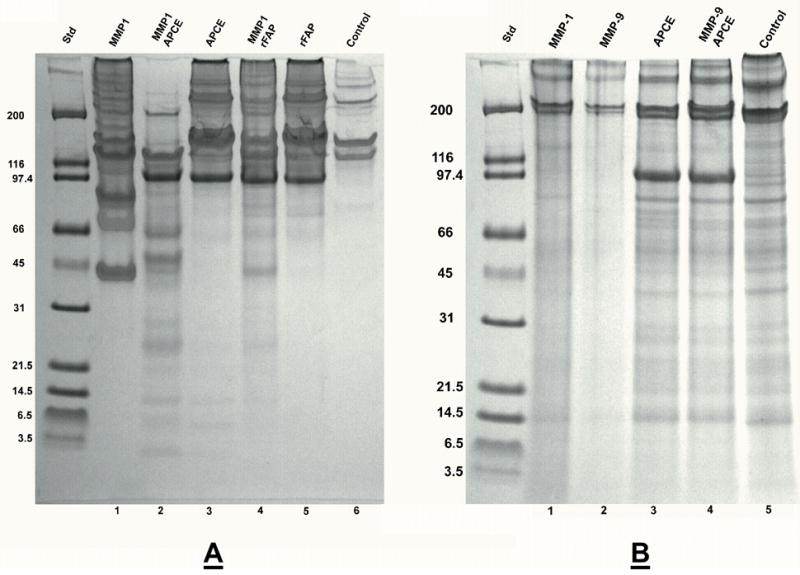
Human placenta collagen type III at 2 mg/ml was incubated at 37 °C for 24 hours with enzymes in 12.5 mM Na PO4 buffer, pH 7.5. Lane: 1) 1.5 μg MMP-1; 2) 1.5 μg MMP-1, 16 μg APCE; 3) 16 μg APCE; 4) 1.5 μg MMP-1, 16 μg rFAP; 5) 16 μg rFAP; 6) Control-buffer. Panel B. EHS mouse tumor collagen type IV at 2 mg/ml was incubated at 37 °C for 24 hours with enzymes in 12.5 mM Na PO4 buffer, pH 7.5. Lane: 1) 1.5 μg MMP-1; 2) 1 μg MMP-9; 3) 16 μg APCE; 4) 1 μg MMP-9, 16 μg APCE; 5) Control-buffer. Digests were reduced and analyzed by 4–12% reduced SDS-PAGE. Where present, the ~97 kDa band is either APCE or rFAP.
Unlike collagen types I and III, collagen type IV is a network-forming collagen and does not have the triple-helical structure associated with fibrillar collagens. It is reported to not be cleaved by MMP-1. Although reported that MMP-9 will cleave type IV collagen [42], most studies question this, unless Type IV collagen is denatured or mildly degraded [45]. As shown in Figure 6b, we found that collagen type IV was not significantly cleaved by MMP-9, and just as reported [45], SDS-PAGE analysis of reduced collagen type IV alone revealed a minor amount of cleavage even prior to enzyme treatment. When exposed to MMP-9, these smaller fragments apparently yielded peptides so small as to not be visible by SDS-PAGE. At most, only partial cleavage of intact collagen type IV (~200 kDa and larger bands) occurred as evident by the remaining intact bands and lack of smaller fragments. The addition of APCE to MMP-9 for digestion of collagen type IV was essentially indistinguishable from APCE alone, except for minor bands in the 70–90 kDa range that differed only slightly in mass and intensity by SDS-PAGE. An explanation for the lack of cleavage might be that MMP-9 possibly digested and destroyed the enzymatic activity of APCE; however, the 97 kDa APCE band remained intact (Figure 6b, lane 4) and the proteolytic capacity of APCE was retained throughout the incubation of the digestion mixture (data not shown). Hence, our results show that neither MMP-1 nor APCE alone cleaves native collagen type IV and that there is no adjunctive role for APCE with MMP-9, even if the latter caused minimal degradation.
Discussion
FAP is expressed by activated fibroblasts in the stroma of embryos, healing wounds and invasive epithelial-derived cancers, but not by normal, quiescent stromal tissue [12; 13]. Proteolytic activity associated with a malignancy is thought to digest ECM as extracellular space is expanded and remodeled to accommodate continued growth. In the process, derivative peptide fragments with growth promoting properties are produced, and abnormal ECM attachment sites become exposed for neoplastic cells as they divide, migrate and escape to distant locations [18; 46–53]. FAP is believed to digest ECM proteins as activated fibroblasts simultaneously multiply, migrate, synthesize and deposit collagen, fibronectin and other ECM constituents [4; 10; 13; 30; 54–57]. These processes appear to occur in synchrony with angiogenesis so that ECM adjacent to a neoplasm becomes remodeled as a malignancy expands. To date, the exact role of FAP in a neoplastic microenvironment remains unknown. No physiologic substrate for FAP within ECM or on cell membranes has been convincingly demonstrated, but despite this, the apparent relationship of the proteinase to epithelial cell cancer growth continues to spur efforts to develop inhibitors or immunological approaches with the hope of mitigating malignant expansion [31–33; 58–61]. Prior reports suggest that FAP cleaves collagen I, the principle protein component of ECM [4; 14; 17; 28]. Fragmentation patterns, however, were not substantiated by SDS-PAGE analyses or by identification of cleavage sites or new amino-terminal sequences. Recent studies from our laboratory strongly suggest that the human plasma prolyl serine proteinase, termed antiplasmin cleaving enzyme (APCE), could be a derivative of cell membrane FAP or possibly, an alternately spliced gene product [1; 7]. FAP is reported to not circulate [6], yet it is functionally indistinguishable from plasma-derived APCE despite the latter’s lacking the intra-membrane and cytoplasmic amino-terminal domains of FAP [7]. Our ability to prepare relatively large amounts of homogeneously pure APCE from human plasma, and recombinant FAP from a yeast expression system [1; 7], allowed us to examine a range of well-defined concentrations for either enzyme’s proteolytic activity towards purified collagens I, III and IV.
We found that neither APCE nor FAP cleaved collagen I in its native state. Consistent with previous reports [40; 41], we also observed that MMP-1 cleaved fibrillar collagen I to yield peptides in two dominant molecular weight ranges: two bands ~95 to 120 kDa and another pair ~ 40 kDa. These peptides represent the terminal digestion pattern of collagen I by MMP-1, which is in accord with prior reports that show MMP-1 cleaves collagen I into ¾ and ¼ length fragments of its intact 220 kDa fiber; however, neither MMP-2 nor MMP-9 alone digests non-denatured collagen I. If APCE or rFAP is also present in the collagen I and MMP-1 incubation mixture, then a complex series of small peptides are generated, with the majority below ~80 kDa, including three prominent low molecular weight peptides between ~3 and 16 kDa. When collagen I digestion by MMP-1 occurred with MMP-9 present, we observed a pattern of low molecular weight peptides less than ~30 kDa with the most prominent peptide being ~4.5 kDa and aligning with one of the fragments from MMP-1 plus rFAP or MMP-1 plus APCE collagen I digests. In contrast, the inclusion of MMP-2 in MMP-1-collagen I incubation mixtures generated a majority of bands > 80 kDa, another two distinct lower molecular weight peptides of ~30 to 35 kDa, and a single minor band at ~15 kDa. Clearly, APCE or rFAP behaved analogously to MMP-9 with respect to further digesting MMP-1-cleaved fibrillar collagen I. It is to be emphasized, however, that neither APCE nor rFAP alone digests collagen I unless the latter has undergone initial cleavage or mild denaturation, i.e., partial helical unwinding. Under the conditions of our experiments, the digestion of partially degraded collagen I by APCE or rFAP proceeded slowly, yielding a stable pattern at 24 hours that we arbitrarily defined as “terminal,” given the high APCE or rFAP concentrations relative to collagen. The stability of the terminal SDS-PAGE pattern for either enzyme and its difference from that given by MMP-1 plus MMP-9 raises speculations about the potential importance of specific peptides generated in the MMP-1 plus APCE or MMP-1 plus rFAP incubation mixture. In view of the growth promoting scenarios, e.g., embryogenesis, wound healing and epithelial-derived cancers associated with the expression of membrane FAP, which is otherwise absent or in extremely low abundance in normal quiescent tissue, some of these digestion products may possess unusual bioactivities necessary for tissue growth. In an expanding cancer, the generation of unique growth or motility promoting peptides, or ones which interfere with normal attachment or apoptosis of a cell, might produce effects different from those observed in more common tissue-destroying inflammatory disorders where MMP-1 or MMP-9 generation of collagen peptide fragments appears to prevail [62; 63].
Proteolysis by MMP-1 is abrogated by chelating agents that bind Zn++, yet MMP-1 retains the ability to initiate the unwinding of the triple-coiled helix of collagen I structure. Even with a very limited loss of conformation, APCE or FAP cleaved the mildly denatured collagen I. The three low molecular weight fragments generated from denatured collagen I in the absence of prior proteolysis by MMP-1 gave the same gel pattern as noted in the MMP-1 plus APCE or MMP-1 plus rFAP terminal patterns. This, combined with amino-terminal sequence as shown in Figure 5, indicates that these three peptides were derived from the amino-terminal regions of the two α chains of type I collagen, ~ 40 residues beyond the telopeptides. Hence, albeit hydrolysis proceeded slowly, the resulting peptides were not dependent on intermediate band formation as occurred with cleavage of collagen I by MMP-1 without EDTA.
While mixtures of MMP-1 and FAP cleave collagen I to yield terminal digestion products between ~3 to 90 kDa, the combined enzyme effects on type III collagen resulted principally in a band of ~110 kDa, two ~50 to 60 kDa, and a series of faintly staining low molecular weight peptides that were similar in size, but not identical to those observed in the pattern for collagen I. With respect to collagen type IV, MMP-9 alone yielded essentially the same pattern as MMP-1, except for the digestion of the lower molecular weight cleavage products already present in the collagen solution; the addition of APCE caused no additional cleavage. Moreover, we observed little or no evidence of Type IV collagen digestion by APCE alone or APCE plus MMP-9 when the SDS-PAGE patterns were compared with buffer control. Hence, our results indicate that APCE, and likely rFAP, given its homology, do not act secondarily to MMP-9 in the digestion of type IV collagen. Although not examined in our studies, it is likely that the initial degradation of type IV collagen occurs in response to other enzymes such as elastase or plasmin, which then allows proteolytic attack by MMP-9 on the resulting fragments [64]. At this time, we speculate that APCE or rFAP may act similarly, primarily based on their obvious proteolysis of mildly denatured or partially digested type I collagen. Lastly, in accord with work by Park et al [4], we also found that fibronectin was not cleaved by rFAP or APCE.
Our direct demonstrations that FAP or APCE only digests partially degraded or denatured collagen I or III indicate that other enzyme(s) must play an initial role. Given its up-regulation in migrating, dividing fibroblasts, and its tissue concentrations, MMP-1 is probably the best prospect for cleaving collagen I to fragments that then serve as substrates for FAP [42]. While this process has gelatinase-like features, the cellular and chemical events involved suggest that FAP possesses several unique properties that separate it from the several MMPs with analogous activities. MMPs seem to be produced by essentially all cells and activated in almost all cellular reactive inflammatory processes, and are associated with normal and pathological conditions characterized by tissue destruction such as menstruation, liver, cardiac and lung fibroses, atherosclerosis, arthritis, congestive heart failure and neurodegenerative diseases [62]. This suggests that MMP expression and activation may be less restricted than FAP. In contrast, FAP’s expression appears unique to activated fibroblasts and possibly endothelial cells [27] as they migrate and multiply during tissue expansion as exemplified by embryogenesis, wound healing, and epithelial-derived neoplastic growth. FAP possesses unique properties in several respects. It only cleaves at a prolyl scissile bond, (...X1-Gly-Pro-X2...); its endopeptidase activity is 10-fold greater than its dipeptidase activity (unpublished data); and it yields a terminal cleavage pattern of partially digested collagen I unique from that of MMP-2 or MMP-9. Given these latter characteristics and FAP’s apparent paucity of physiologic substrates, except for Met-α2AP and partially degraded or denatured collagen I or III, questions surface about whether the enzyme might also be giving rise to specific peptide fragments important in growth regulation as reported for other collagens when digested by selected proteinases [65–67]. It is likewise possible that FAP may be more involved in other cleavage pathways essential for cell function, such as membrane adhesive proteins (cadherins or integrins) requisite for cell-cell attachment, or cell anchoring to ECM as cells migrate and divide; or FAP may activate pro-paracrine proteins that govern cell migration within ECM.
We conclude that recognition and digestion of partially degraded or denatured collagen I by FAP imputes to it a significant role in tissue growth and malignant expansion with respect to ECM preparation and remodeling, indeed suggesting a “tilling the soil” function as posed by Cheng [68]. Further studies of the structure/function relationships of FAP and its role in cell growth—especially stromal cells—will eventually determine its potential usefulness as a target for novel cancer therapies or as a cellular marker for diagnosing and monitoring cancer.
Acknowledgments
We thank Ms. C. S. Lee and Mr. J-G Chun for excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Lee KN, Jackson KW, Christiansen VJ, Chung KH, McKee PA. A novel plasma proteinase potentiates alpha2-antiplasmin inhibition of fibrin digestion. Blood. 2004;103:3783–3788. doi: 10.1182/blood-2003-12-4240. [DOI] [PubMed] [Google Scholar]
- 2.Lee KN, Jackson KW, Christiansen VJ, Chung KH, McKee PA. alpha(2)-Antiplasmin: potential therapeutic roles in fibrin survival and removal. Curr Med Chem Cardiovasc Hematol Agents. 2004;2:303–310. doi: 10.2174/1568016043356228. [DOI] [PubMed] [Google Scholar]
- 3.Niedermeyer J, Scanlan MJ, Garin-Chesa P, Daiber C, Fiebig HH, Old LJ, Rettig WJ, Schnapp A. Mouse fibroblast activation protein: molecular cloning, alternative splicing and expression in the reactive stroma of epithelial cancers. Int JCancer. 1997;71:383–389. doi: 10.1002/(sici)1097-0215(19970502)71:3<383::aid-ijc14>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 4.Park JE, Lenter MC, Zimmermann RN, Garin-Chesa P, Old LJ, Rettig WJ. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J Biol Chem. 1999;274:36505–36512. doi: 10.1074/jbc.274.51.36505. [DOI] [PubMed] [Google Scholar]
- 5.Aertgeerts K, Levin I, Shi L, Snell GP, Jennings A, Prasad GS, Zhang Y, Kraus ML, Salakian S, Sridhar V, Wijnands R, Tennant MG. Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha. J Biol Chem. 2005;280:19441–19444. doi: 10.1074/jbc.C500092200. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt A, Muller D, Mersmann M, Wuest T, Gerlach E, Garin-Chesa P, Rettig WJ, Pfizenmaier K, Moosmayer D. Generation of human high-affinity antibodies specific for the fibroblast activation protein by guided selection. Eur J Biochem. 2001;268:1730–1738. [PubMed] [Google Scholar]
- 7.Lee KN, Jackson KW, Christiansen VJ, Lee CS, Chun JG, McKee PA. Antiplasmin-cleaving enzyme is a soluble form of fibroblast activation protein. Blood. 2006;107:1397–1404. doi: 10.1182/blood-2005-08-3452. [DOI] [PubMed] [Google Scholar]
- 8.Gorrell MD. Dipeptidyl peptidase IV and related enzymes in cell biology and liver disorders. Clin Sci(Lond) 2005;108:277–292. doi: 10.1042/CS20040302. [DOI] [PubMed] [Google Scholar]
- 9.Hooper NM, Karran EH, Turner AJ. Membrane protein secretases. Biochem J. 1997;321( Pt 2):265–279. doi: 10.1042/bj3210265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rettig WJ, Garin-Chesa P, Beresford HR, Oettgen HF, Melamed MR, Old LJ. Cell-surface glycoproteins of human sarcomas: differential expression in normal and malignant tissues and cultured cells. Proc Natl Acad Sci USA. 1988;85:3110–3114. doi: 10.1073/pnas.85.9.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rettig WJ, Garin-Chesa P, Healey JH, Su SL, Ozer HL, Schwab M, Albino AP, Old LJ. Regulation and heteromeric structure of the fibroblast activation protein in normal and transformed cells of mesenchymal and neuroectodermal origin. Cancer Res. 1993;53:3327–3335. [PubMed] [Google Scholar]
- 12.Rettig WJ, Su SL, Fortunato SR, Scanlan MJ, Raj BK, Garin-Chesa P, Healey JH, Old LJ. Fibroblast activation protein: purification, epitope mapping and induction by growth factors. Int J Cancer. 1994;58:385–392. doi: 10.1002/ijc.2910580314. [DOI] [PubMed] [Google Scholar]
- 13.Garin-Chesa P, Old LJ, Rettig WJ. Cell surface glycoprotein of reactive stromal fibroblasts as a potential antibody target in human epithelial cancers. Proc Natl Acad Sci USA. 1990;87:7235–7239. doi: 10.1073/pnas.87.18.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pineiro-Sanchez ML, Goldstein LA, Dodt J, Howard L, Yeh Y, Chen WT. Identification of the 170-kDa melanoma membrane-bound gelatinase (seprase) as a serine integral membrane protease. J Biol Chem. 1997;272:7595–7601. doi: 10.1074/jbc.272.12.7595. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein LA, Ghersi G, Pineiro-Sanchez ML, Salamone M, Yeh Y, Flessate D, Chen WT. Molecular cloning of seprase: a serine integral membrane protease from human melanoma. Biochim Biophys Acta. 1997;1361:11–19. doi: 10.1016/s0925-4439(97)00032-x. [DOI] [PubMed] [Google Scholar]
- 16.Nakahara H, Nomizu M, Akiyama SK, Yamada Y, Yeh Y, Chen WT. A mechanism for regulation of melanoma invasion. Ligation of alpha6beta1 integrin by laminin G peptides. J Biol Chem. 1996;271:27221–27224. doi: 10.1074/jbc.271.44.27221. [DOI] [PubMed] [Google Scholar]
- 17.Ghersi G, Dong H, Goldstein LA, Yeh Y, Hakkinen L, Larjava HS, Chen WT. Regulation of fibroblast migration on collagenous matrix by a cell surface peptidase complex. J Biol Chem. 2002;277:29231–29241. doi: 10.1074/jbc.M202770200. [DOI] [PubMed] [Google Scholar]
- 18.Mori Y, Kono K, Matsumoto Y, Fujii H, Yamane T, Mitsumata M, Chen WT. The expression of a type II transmembrane serine protease (Seprase) in human gastric carcinoma. Oncology. 2004;67:411–419. doi: 10.1159/000082926. [DOI] [PubMed] [Google Scholar]
- 19.Niedermeyer J, Kriz M, Hilberg F, Garin-Chesa P, Bamberger U, Lenter MC, Park J, Viertel B, Puschner H, Mauz M, Rettig WJ, Schnapp A. Targeted disruption of mouse fibroblast activation protein. Mol Cell Biol. 2000;20:1089–1094. doi: 10.1128/mcb.20.3.1089-1094.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niedermeyer J, Garin-Chesa P, Kriz M, Hilberg F, Mueller E, Bamberger U, Rettig WJ, Schnapp A. Expression of the fibroblast activation protein during mouse embryo development. Int J Dev Biol. 2001;45:445–447. [PubMed] [Google Scholar]
- 21.Huang Y, Wang S, Kelly T. Seprase promotes rapid tumor growth and increased microvessel density in a mouse model of human breast cancer. Cancer Res. 2004;64:2712–2716. doi: 10.1158/0008-5472.can-03-3184. [DOI] [PubMed] [Google Scholar]
- 22.Kelly T, Yan Y, Osborne RL, Athota AB, Rozypal TL, Colclasure JC, Chu WS. Proteolysis of extracellular matrix by invadopodia facilitates human breast cancer cell invasion and is mediated by matrix metalloproteinases. Clin Exp Metastasis. 1998;16:501–512. doi: 10.1023/a:1006538200886. [DOI] [PubMed] [Google Scholar]
- 23.Kelly T, Kechelava S, Rozypal TL, West KW, Korourian S. Seprase, a membrane-bound protease, is overexpressed by invasive ductal carcinoma cells of human breast cancers. Mod Pathol. 1998;11:855–863. [PubMed] [Google Scholar]
- 24.Okada K, Chen WT, Iwasa S, Jin X, Yamane T, Ooi A, Mitsumata M. Seprase, a membrane-type serine protease, has different expression patterns in intestinal- and diffuse-type gastric cancer. Oncology. 2003;65:363–370. doi: 10.1159/000074650. [DOI] [PubMed] [Google Scholar]
- 25.Iwasa S, Jin X, Okada K, Mitsumata M, Ooi A. Increased expression of seprase, a membrane-type serine protease, is associated with lymph node metastasis in human colorectal cancer. Cancer Lett. 2003;199:91–98. doi: 10.1016/s0304-3835(03)00315-x. [DOI] [PubMed] [Google Scholar]
- 26.Jin X, Iwasa S, Okada K, Mitsumata M, Ooi A. Expression patterns of seprase, a membrane serine protease, in cervical carcinoma and cervical intraepithelial neoplasm. Anticancer Res. 2003;23:3195–3198. [PubMed] [Google Scholar]
- 27.Aimes RT, Zijlstra A, Hooper JD, Ogbourne SM, Sit ML, Fuchs S, Gotley DC, Quigley JP, Antalis TM. Endothelial cell serine proteases expressed during vascular morphogenesis and angiogenesis. Thromb Haemost. 2003;89:561–572. [PubMed] [Google Scholar]
- 28.Aoyama A, Chen WT. A 170-kDa membrane-bound protease is associated with the expression of invasiveness by human malignant melanoma cells. Proc Natl Acad Sci USA. 1990;87:8296–8300. doi: 10.1073/pnas.87.21.8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monsky WL, Lin CY, Aoyama A, Kelly T, Akiyama SK, Mueller SC, Chen WT. A potential marker protease of invasiveness, seprase, is localized on invadopodia of human malignant melanoma cells. Cancer Res. 1994;54:5702–5710. [PubMed] [Google Scholar]
- 30.Kelly T. Evaluation of seprase activity. Clin Exp Metastasis. 1999;17:57–62. doi: 10.1023/a:1026430206274. [DOI] [PubMed] [Google Scholar]
- 31.Scott AM, Wiseman G, Welt S, Adjei A, Lee FT, Hopkins W, Divgi CR, Hanson LH, Mitchell P, Gansen DN, Larson SM, Ingle JN, Hoffman EW, Tanswell P, Ritter G, Cohen LS, Bette P, Arvay L, Amelsberg A, Vlock D, Rettig WJ, Old LJ. A Phase I dose-escalation study of sibrotuzumab in patients with advanced or metastatic fibroblast activation protein-positive cancer. Clin Cancer Res. 2003;9:1639–1647. [PubMed] [Google Scholar]
- 32.Cheng JD, Valianou M, Canutescu AA, Jaffe EK, Lee HO, Wang H, Lai JH, Bachovchin WW, Weiner LM. Abrogation of fibroblast activation protein enzymatic activity attenuates tumor growth. Mol Cancer Ther. 2005;4:351–360. doi: 10.1158/1535-7163.MCT-04-0269. [DOI] [PubMed] [Google Scholar]
- 33.Adams S, Miller GT, Jesson MI, Watanabe T, Jones B, Wallner BP. PT-100, a small molecule dipeptidyl peptidase inhibitor, has potent antitumor effects and augments antibody-mediated cytotoxicity via a novel immune mechanism. Cancer Res. 2004;64:5471–5480. doi: 10.1158/0008-5472.CAN-04-0447. [DOI] [PubMed] [Google Scholar]
- 34.Lauer-Fields JL, Juska D, Fields GB. Matrix metalloproteinases and collagen catabolism. Biopolymers. 2002;66:19–32. doi: 10.1002/bip.10201. [DOI] [PubMed] [Google Scholar]
- 35.Bornstein P. Comparative sequence studies of rat skin and tendon collagen. II. The absence of a short sequence at the amino terminus of the skin alpha-1 chain. Biochemistry. 1969;8:63–71. doi: 10.1021/bi00829a010. [DOI] [PubMed] [Google Scholar]
- 36.Kang AH, Bornstein P, Piez KA. The amino acid sequence of peptides from the cross-linking region of rat skin collagen. Biochemistry. 1967;6:788–795. doi: 10.1021/bi00855a019. [DOI] [PubMed] [Google Scholar]
- 37.Jeno P, Mini T, Moes S, Hintermann E, Horst M. Internal sequences from proteins digested in polyacrylamide gels. Anal Biochem. 1995;224:75–82. doi: 10.1006/abio.1995.1010. [DOI] [PubMed] [Google Scholar]
- 38.Linsenmayer TF. Collagen. In: Hay ED, editor. Cell Biology of Extracellular Matrix. Plenum Press; New York, N.Y: 1991. pp. 7–44. [Google Scholar]
- 39.Kuhn K. The Classical Collagens: Type I, II, and III. In: Mayne R, Burgeson RE, editors. Structure and Function of Collagen Types. Academic Press; New York: 1987. pp. 1–42. [Google Scholar]
- 40.Fields GB. A model for interstitial collagen catabolism by mammalian collagenases. J Theor Biol. 1991;153:585–602. doi: 10.1016/s0022-5193(05)80157-2. [DOI] [PubMed] [Google Scholar]
- 41.Gross J, Harper E, Harris ED, McCroskery PA, Highberger JH, Corbett C, Kang AH. Animal collagenases: specificity of action, and structures of the substrate cleavage site. Biochem Biophys Res Commun. 1974;61:605–612. doi: 10.1016/0006-291x(74)91000-6. [DOI] [PubMed] [Google Scholar]
- 42.Chakraborti S, Mandal M, Das S, Mandal A, Chakraborti T. Regulation of matrix metalloproteinases: an overview. Mol Cell Biochem. 2003;253:269–285. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
- 43.Chung L, Dinakarpandian D, Yoshida N, Lauer-Fields JL, Fields GB, Visse R, Nagase H. Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J. 2004;23:3020–3030. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson KW, Christiansen VJ, Lee KN, McKee PA. Determination of antiplasmin cleaving enzyme substrate specificity and inhibitor development by peptide library analyses. FASEB Journal. 2005;19:A864. [Google Scholar]
- 45.Mackay AR, Hartzler JL, Pelina MD, Thorgeirsson UP. Studies on the ability of 65-kDa and 92-kDa tumor cell gelatinases to degrade type IV collagen. J Biol Chem. 1990;265:21929–21934. [PubMed] [Google Scholar]
- 46.Stetler-Stevenson WG. Matrix metalloproteinases in angiogenesis: a moving target for therapeutic intervention. J Clin Invest. 1999;103:1237–1241. doi: 10.1172/JCI6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Werb Z. ECM and cell surface proteolysis: regulating cellular ecology. Cell. 1997;91:439–442. doi: 10.1016/s0092-8674(00)80429-8. [DOI] [PubMed] [Google Scholar]
- 48.Erickson AC, Barcellos-Hoff MH. The not-so innocent bystander: the microenvironment as a therapeutic target in cancer. Expert Opin Ther Targets. 2003;7:71–88. doi: 10.1517/14728222.7.1.71. [DOI] [PubMed] [Google Scholar]
- 49.Coussens LM, Werb Z. Matrix metalloproteinases and the development of cancer. Chem Biol. 1996;3:895–904. doi: 10.1016/s1074-5521(96)90178-7. [DOI] [PubMed] [Google Scholar]
- 50.Rosenthal EL, McCrory A, Talbert M, Carroll W, Magnuson JS, Peters GE. Expression of proteolytic enzymes in head and neck cancer-associated fibroblasts. Arch Otolaryngol Head Neck Surg. 2004;130:943–947. doi: 10.1001/archotol.130.8.943. [DOI] [PubMed] [Google Scholar]
- 51.Chambers AF, Matrisian LM. Changing views of the role of matrix metalloproteinases in metastasis. J Natl Cancer Inst. 1997;89:1260–1270. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- 52.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 53.Nanus DM. Of peptides and peptidases: the role of cell surface peptidases in cancer. Clin Cancer Res. 2003;9:6307–6309. [PubMed] [Google Scholar]
- 54.Wuest T, Moosmayer D, Pfizenmaier K. Construction of a bispecific single chain antibody for recruitment of cytotoxic T cells to the tumour stroma associated antigen fibroblast activation protein. J Biotechnol. 2001;92:159–168. doi: 10.1016/s0168-1656(01)00355-8. [DOI] [PubMed] [Google Scholar]
- 55.Levy MT, McCaughan GW, Abbott CA, Park JE, Cunningham AM, Muller E, Rettig WJ, Gorrell MD. Fibroblast activation protein: a cell surface dipeptidyl peptidase and gelatinase expressed by stellate cells at the tissue remodelling interface in human cirrhosis. Hepatology. 1999;29:1768–1778. doi: 10.1002/hep.510290631. [DOI] [PubMed] [Google Scholar]
- 56.Chen WT. Proteases associated with invadopodia, and their role in degradation of extracellular matrix. Enzyme Protein. 1996;49:59–71. doi: 10.1159/000468616. [DOI] [PubMed] [Google Scholar]
- 57.Martin P. Wound healing--aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 58.Cheng JD, Cohen SJ, Stzempkowski-Brun B, Magalong K, Christiansen VJ, McKee PA, Alpaugh RK, Lee H, Weiner LM, Meropol NJ. Phase II pharmacodynamic study of the fibroblast activation protein inhibitor Val-boro-Pro in patients with metastatic colorectal cancer. J Clin Oncology. 2006;24:3594. [Google Scholar]
- 59.Mersmann M, Schmidt A, Rippmann JF, Wuest T, Brocks B, Rettig WJ, Garin-Chesa P, Pfizenmaier K, Moosmayer D. Human antibody derivatives against the fibroblast activation protein for tumor stroma targeting of carcinomas. Int J Cancer. 2001;92:240–248. doi: 10.1002/1097-0215(200102)9999:9999<::aid-ijc1170>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 60.Fassnacht M, Lee J, Milazzo C, Boczkowski D, Su Z, Nair S, Gilboa E. Induction of CD4(+) and CD8(+) T-cell responses to the human stromal antigen, fibroblast activation protein: implication for cancer immunotherapy. Clin Cancer Res. 2005;11:5566–5571. doi: 10.1158/1078-0432.CCR-05-0699. [DOI] [PubMed] [Google Scholar]
- 61.Lee J, Fassnacht M, Nair S, Boczkowski D, Gilboa E. Tumor immunotherapy targeting fibroblast activation protein, a product expressed in tumor-associated fibroblasts. Cancer Res. 2005;65:11156–11163. doi: 10.1158/0008-5472.CAN-05-2805. [DOI] [PubMed] [Google Scholar]
- 62.Mandal M, Mandal A, Das S, Chakraborti T, Sajal C. Clinical implications of matrix metalloproteinases. Mol Cell Biochem. 2003;252:305–329. doi: 10.1023/a:1025526424637. [DOI] [PubMed] [Google Scholar]
- 63.Corbel M, Boichot E, Lagente V. Role of gelatinases MMP-2 and MMP-9 in tissue remodeling following acute lung injury. Braz J Med Biol Res. 2000;33:749–754. doi: 10.1590/s0100-879x2000000700004. [DOI] [PubMed] [Google Scholar]
- 64.Mainardi CL, Dixit SN, Kang AH. Degradation of type IV (basement membrane) collagen by a proteinase isolated from human polymorphonuclear leukocyte granules. J Biol Chem. 1980;255:5435–5441. [PubMed] [Google Scholar]
- 65.O'Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS, Flynn E, Birkhead JR, Olsen BR, Folkman J. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 66.Maeshima Y, Colorado PC, Torre A, Holthaus KA, Grunkemeyer JA, Ericksen MB, Hopfer H, Xiao Y, Stillman IE, Kalluri R. Distinct antitumor properties of a type IV collagen domain derived from basement membrane. J Biol Chem. 2000;275:21340–21348. doi: 10.1074/jbc.M001956200. [DOI] [PubMed] [Google Scholar]
- 67.Kamphaus GD, Colorado PC, Panka DJ, Hopfer H, Ramchandran R, Torre A, Maeshima Y, Mier JW, Sukhatme VP, Kalluri R. Canstatin, a novel matrix-derived inhibitor of angiogenesis and tumor growth. J Biol Chem. 2000;275:1209–1215. doi: 10.1074/jbc.275.2.1209. [DOI] [PubMed] [Google Scholar]
- 68.Cheng JD, Weiner LM. Tumors and their microenvironments: tilling the soil. Commentary re: A. M. Scott et al., A Phase I dose-escalation study of sibrotuzumab in patients with advanced or metastatic fibroblast activation protein-positive cancer. Clin Cancer Res. 2003;9:1590–1595. [PubMed] [Google Scholar]


