Abstract
We report molecular modeling and functional confirmation of Ole and HT binding to HIV-1 integrase. Docking simulations identified two binding regions for Ole within the integrase active site. Region I encompasses the conserved D64-D116-E152 motif, while region II involves the flexible loop region formed by amino acid residues 140–149. HT, on the other hand, binds to region II. Both Ole and HT exhibit favorable interactions with important amino acid residues through strong H-bonding and van der Waals contacts, predicting integrase inhibition. To test and confirm modeling predictions, we examined the effect of Ole and HT on HIV-1 integrase activities including 3′-processing, strand transfer and disintegration. Ole and HT exhibit dose-dependent inhibition on all three activities, with EC50s in the nM range. These studies demonstrate that molecular modeling of target–ligand interaction coupled with structural-activity analysis should facilitate the design and identification of innovative integrase inhibitors and other therapeutics.
Keywords: HIV-1, AIDS, natural product, small molecule HIV-1 inhibitors, HIV-1 integrase inhibitor, Olive Leaf Extract (OLE), Oleuropein (Ole), Hydroxytyrosol (HT), structure-function, molecular modeling
INTRODUCTION
HIV-1 integrase is one of three viral enzymes required for viral replication, along with RT and protease [1–3]. Integration of HIV-1 cDNA into the host chromosome is essential for stable maintenance of the viral genome, efficient viral gene expression and productive infection. Thus, viral integrase is a critical target for anti-HIV therapy [4, 5]. The first step leading to viral DNA integration is the binding of viral integrase to HIV long terminal repeat (LTR) sequences. This is followed by three sequential reactions: 1) 3′-processing, the removal of two nucleotides, GT, from the 3′-end of HIV-LTR, 2) strand transfer, a concerted cleavage-ligation reaction, in which the integrase makes a staggered cut in the target DNA and ligates the recessed 3′ ends of the viral DNA to the 5′ ends of the target DNA, and 3) gap repair, removal of the two unpaired nucleotides at the 5′ end of the viral DNA and repair of the gap between the viral and the target DNA. In the presence of a DNA substrate that mimics the product of viral integration, integrase can catalyze the reversal of strand transfer, known as “disintegration” [6], in which viral DNA is released and the target DNA is sealed. In this article, we report our findings on molecular modeling of Ole and HT interactions with HIV-1 integrase, and confirmation of model predictions by analysis of functional activities.
MATERIAL AND METHODS
Oleuropein (Ole) and Hydroxytyrosol (HT)
Ole and HT used in this study were prepared and purified from olive leaf extract as described in the preceding article.
Target Cells, HIV-1, Anti-HIV and cytotoxicity assays
Target cells MT2, H9, HIV-1IIIB chronically infected H9 (H9/HIV-1IIIB) and HIV-1/IIIB virus, were obtained through the AIDS Research and Reference Reagent Program, NIAID, NIH. MT-2 cells were from D. Richman [7, 8]. H9 cells and HIV-1IIIB virus stocks were from R. Gallo [9, 10]. The cell lines were cultured in RPMI medium 1640 containing 100U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, and 10% heat-inactivated fetal calf serum. Viral stocks were prepared and standardized as described previously [11]. Anti-HIV activity was measured by the microtiter syncytial formation and HIV-1 p24 production assays [11]. Cytotoxicity was determined by the MTT assay [11].
Molecular Modeling
A combination of molecular docking, molecular dynamics (MD) simulation and free energy calculations [12, 13] were performed to probe the interactions of Ole or HT with viral targets. Docking was performed with AutoDock version 3.0.5 [14]. Relaxation of docking structure obtained was carried out by the program Discovery from Insight II (Accelrys Inc., San Diego, CA, U.S.A.), using 500 steps of Steepest Descent followed by Conjugate Gradient until the root mean square (RMS) of the energy gradient reaches 0.01 kcal/molÅ.
HIV-1 Integrase
HIV-1 integrase was expressed in E. coli from pIN (F185H/C280S) and purified according to previously reported method [15]. This recombinant clone makes the integrase protein more soluble and stable without affecting in vitro activity. HIV-1 integrase protein (F185H/C280S) was used as a standard for purification and assay. The integrase clone pIN (F185H/C280S) and the standard integrase protein were obtained through the NIH AIDS Research and Reference Reagent Program, NIAID, NIH from Dr. Robert Craigie [16].
Integrase Substrates
Oligonucleotide substrates were synthesized and purified as described previously [17]. Three types of substrates were synthesized with sequences that correspond to the U3 and U5 ends of HIV-LTR: (i) the 21-nucleotide minus strand of U3 end HIV-LTR 5′-GAGTGAATTAGCCCTTCCAGT-3′, and the U5 HIV-LTR, 5′-GTGTGGAAAATCTCTAGCAGT-3′, as well as their complementary strands, for assaying the 3′-processing reaction; (ii) the 19-mer U3-GT, and U5-GT (i.e., U3 and U5 minus the 3′-end dinucleotide GT) for assaying heterologous integration (strand transfer); and (iii) a 38-mer dumbbell substrate with the sequence 5′-TGCTAGTTCTAGCAGGCCCTTGGGCCGGCGCTTGCGCC-3′, for assaying the dis-integration.
Radiolabeling and Preparation of the Substrates
The integrase substrates were 5′-end radiolabeled as reported previously [17]. Briefly, 1μg of the oligonucleotide was 5′-end labeled with 100 μCi of [α-32P]ATP (3000 Ci/mmol; 1 Ci = 37 GBq; A-mersham) by 20 units of polynucleotide T4 kinase in a final volume of 40 μl of kinase buffer (Boehringer Mannheim) at 37°C for 60 min. The reaction was stopped by EDTA (25 mM) and heat inactivation. Unincorporated label was removed by two passages through a Sephadex G-25 spin column (Boehringer Mannheim). The purified labeled oligonucleotide was then annealed with an equimolar amount of unlabeled complementary strand in 10 mM Tris HCl, pH 7.5/1 mM EDTA/100 mM NaCl at 95°C for 5 min followed by slow cooling. The dumbbell substrate was self-annealed under the same conditions.
Integrase Assays
Integrase assays were carried out in 20 mM Hepes, pH 7.5/10 mM MgCl2 or MnCl2 /10 mM dithiothreitol/0.05% Nonidet P-40 (integrase buffer) with 40 pmol of HIV-1 integrase and 20 ng of 5′-end radiolabeled substrates specific for 3′-processing (21-mer U3), strand-transfer (19-mer U3-GT)) or disintegration (38-mer dumbbell) in the presence or absence of Ole or HT in a final volume of 10 μl at 37°C for 60 min. For 3′-processing and disintegration, the reactions were stopped by the addition of 10 μl of 90% formamide/ 0.025% bromophenol blue/0.025% xylene cyanol/89 mM Tris/89 mM boric acid/2 mM EDTA, pH 8.0. Samples were heated at 75°C for 3 min, load at 10μl/well onto 18 % polyacrylamide denaturing (7.5 M urea) gels in TBE buffer and electrophoresed at 200 V constant voltage at room temperature for 2 h. The results were visualized by autoradiography of wet gels. For strand-transfer (integration), pUC18 plasmid DNA (50 ng) was used as the target for the integration of viral DNA into heterologous plasmid DNA. The reaction was stopped by 0.1% SDS and integration products monitored on 1% agarose gels at 10 ul/well in TBE buffer at 100 V constant voltage at room temperature for 45 min. The results were visualized by autoradiography of dried gels.
RESULTS
Ole and HT Bind to the Catalytic Site of HIV-1 Integrase
To examine the molecular interactions of Ole and HT with HIV integrase, we performed a series of docking simulations. In these studies, we focused on the catalytic core domain (CCD) of the viral enzyme, because the linkages between the CCD and both the N- and C-terminal domains (NTD and CTD) are flexible and have been not precisely determined. We used the crystal structure 1QS4 (pdb ID) as the starting structure [18], because it is the only structure has an inhibitor (5CITEP) bound in the active site. To keep the binding pocket available, we removed the ligand (5CITEP) from the CCD. Before docking, the missing residues in the loop region and the point mutation at position 185 were restored with AMBER software. The K185 mutation was converted to the native F185.
Figures 1A and 1B represent the predicted bound conformations of Ole and HT within the active site of HIV-1 integrase. Two unique binding regions have been identified within the integrase active site [2, 4], referred to as regions I and II (Figure 1B). Region I, near the active site center, encompasses the conserved DDE motif, D64-D116-E152 in HIV-1 integrase. These residues are highly conserved in all integrases, retrotransposases and other DNA-processing enzymes (polynucleotide transferases). Mutation of any of these acidic amino acids abolishes integrase activities and viral replication. D64 and D116 are involved in the formation of coordination complex with divalent metal (Mg2+ or Mn2+). A second metal (Mg2+ or Mn2+) can be coordinated between D116 and E152 once HIV-1 integrase binds its DNA substrates (20, 21). Metal ion coordination with viral integrase and the phosphodiester backbone of the DNA substrates are likely to occur during 3′-processing and strand-transfer reactions. Region II is close to the active catalytic loop (amino acid residues 139–147), and involves the flexible loop formed by amino acid residues 140–149. This loop region has been identified as the DNA binding site which is important for integrase action [19].
Figure 1. Molecular docking of Ole and HT with HIV-1 integrase.
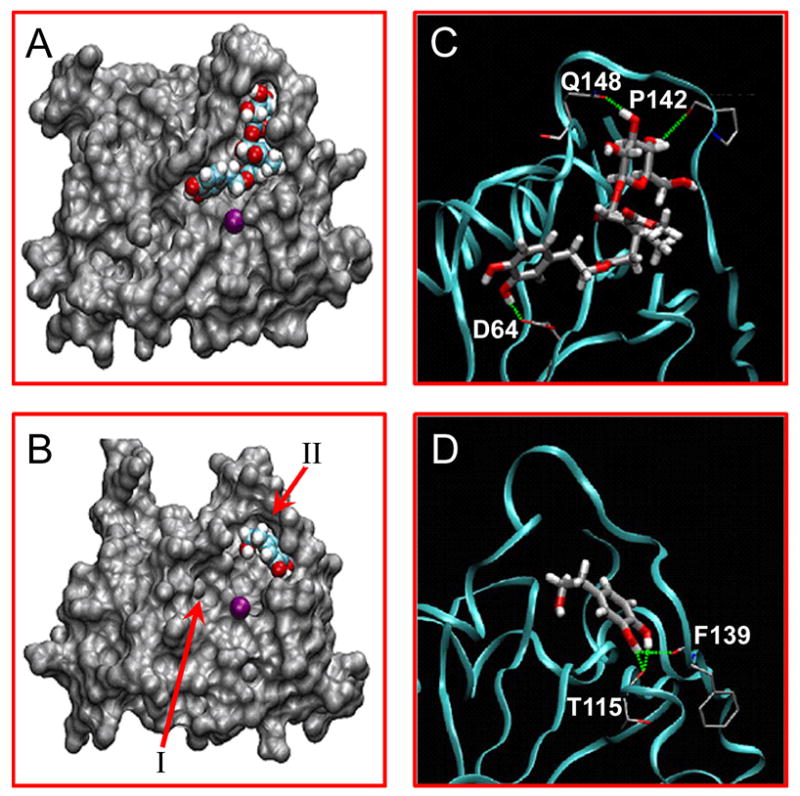
The predicted binding structures of Ole (A) and HT (B) inside the HIV-1 integrase catalytic site. Integrase is shown as a surface model, while Ole and HT are shown as van der Waals models and the purple sphere represents Mg2+. Hydrogen bonds formed by Ole (C) and HT (D) with integrase are indicated as green dotted lines, and the integrase backbone is represented by the cyan ribbon.
As seen in Figure 1A, Ole binds to both regions I and II. The β-glycopyranose moiety of Ole interacts with residues in region II whereas the dihydroxyphenol ring occupies region I. Figures 1C and 1D are ribbon representation of the HIV-integrase CCD showing major strong H-bonding sites with Ole and HT. The flexible loop widens the active site region and allows the sugar ring of Ole to dock with strong H-bond with P142 and Q148 as well as form weak interactions with S147. The dihydroxyphenol moiety of Ole binds to region I with a strong H-bond interaction with D64 and a weak H bond-network interactions with K156 and K159. This suggests that Ole would be a strong integrase binding inhibitor because it interacts with residues in both regions I and II. On the other hand, the dihydroxyphenol ring of HT binds to region II with strong H bond interaction with F139 and nearby T115, and weak interactions with E138 and Q 148. Since the dihydroxyphenol ring is capable of binding both regions I and II, HT maintains the ability to bind the integrase active site even if mutations occur. Thus the likelihood of resistance development should be less than inhibitors that bind to a single site. Thus, interaction modeling suggests that Ole and HT bind to both regions I and II, so they would be expected to be effective against metal coordination as well as substrate binding. Modeling results therefore predict that Ole and HT would inhibit all of the three HIV-1 integrase activities.
Ole and HT Inhibit 3′-Processing Activity of HIV-1 Integrase
Modeling predictions that Ole and HT may affect HIV-integrase activity were tested in all of the three activities, namely 3′-processing, strand transfer (integration) and disintegration. Results of HIV-1 integrase inhibitory activities of Ole and HT are summarized in Table I with their anti-HIV activities.
Figure 2A shows the 3′-processing reaction and results. 5′ [32P] labeled 21-mer double-stranded oligonucleotide that mimics the U3 HIV-1 LTR was used as a substrate. 3′-processing by HIV-1 integrase, removes the dinucleotide GT from the 3′ end of the labeled minus strand of the 21 -mer substrate and yields a 3′ recessed product (U3-GT) with 19 nucleotides in length. Ole or HT inhibits the 3′-processing activity of HIV-1 integrase. Figure 2A demonstrates dose-dependent inhibition of 3′-processing by Ole and HT as detected by 7.5 M urea denaturating polyacrylamide gel electrophoresis. Inhibition of the formation of the 19 -mer product from the 21 -mer substrate increases with the increase of Ole or HT concentration from 25 to 100 nM. The degree of inhibition depends on the concentrations of the HIV-1 integrase, the substrate and the inhibitor. Under our assay conditions, EC50s of 46 and 54 nM were obtained for Ole and HT respectively, and total inhibition was observed at 100 nM. Substrate U5 HIV-LTR showed similar results.
Figure 2. The effect of Ole and HT on HIV-1 integrase 3′-processing, strand transfer and disintegration activities.
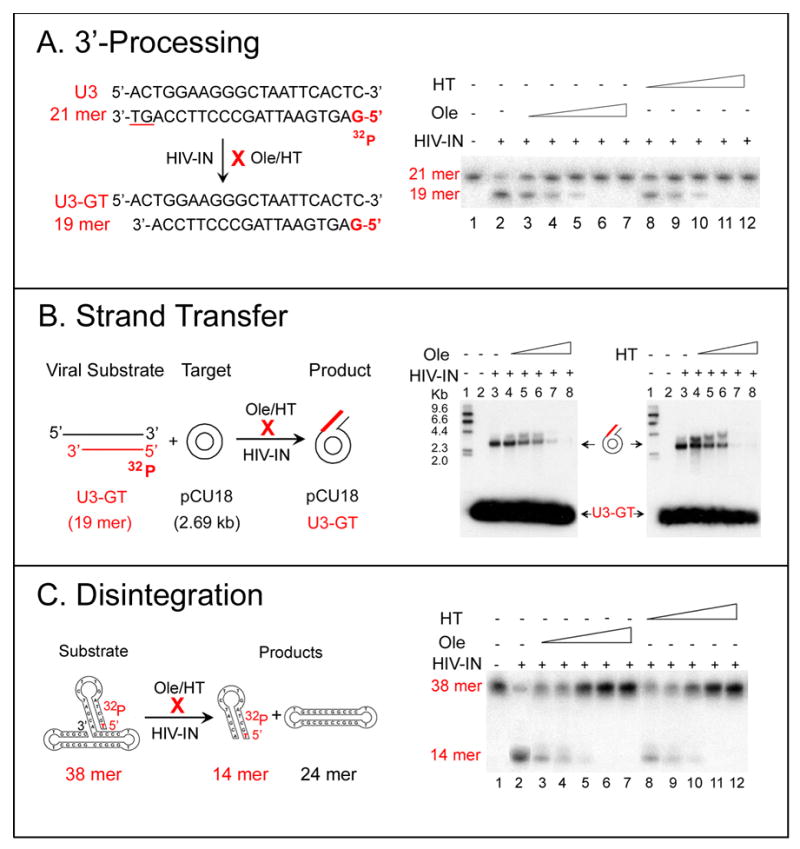
A. The effect of Ole and HT on HIV-1 integrase 3′-processing activity
Left: Schematic representation of the 3′-processing activity of HIV-1 integrase. A 5′-32P-labeled 21-mer of HIV-1 LTR U3 double-stranded (ds) DNA was used as the substrate. Specific cleavage of the dinucleotide GT from the 3′ end of the substrate results in the formation of 19-mer 3′-recessed U3-GT.
Right: Inhibition of HIV-1 integrase 3′-processing activity by Ole and HT. Inhibition was monitored by the formation of labeled 19-mer product. Lane 1, the 21-mer substrate, 5′-32P-labeled U3. Lane 2, cleavage of the 3′ GT of the 21-mer substrate by HIV-1 integrase results in the formation of the 19-mer 3′-recessed U3-GT. Lanes 3-7 or 8-12, in the presence of 25, 50, 75, 100, and 200 nM Ole or HT.
B. The effect of Ole and HT on HIV-1 integrase strand transfer activity
Left: Schematic representation of strand transfer (integration). Pre-cleaved 5′-32P-labeled U3-GT 19-mer was used as the viral substrate and, unlabeled pUC18 DNA (2.69 kb) was used as the heterologous target substrate.
Right: Inhibition of strand-transfer activity of HIV-1 integrase by Ole and HT. Integration was monitored by the conversion of the unlabeled plasmid into labeled DNA. Lanes 1, 5′-32P-labeled size marker, HindIlI fragments of λ phage DNA. Lanes 2, target substrate, pUC18; because it is unlabeled, it is not seen in the autoradiogram. Lanes 3, the product of integration (ST) by HIV-1 integrase. The integration of the 5′32P-labeled U3-GT into pUC18 results in the appearance of labeled band at 2.69 kb, corresponding to the size of pUC18. Lanes 4–8, in the presence of 25, 50, 75, 100 and 200 nM Ole or HT.
C. The effect of Ole and HT on HIV-1 integrase disintegration activity
Left: Schematic representation of the disintegration activity of HIV-1 integrase. The 5′ 32P-labeled 38-mer dumbbell was used as the substrate and shown with the predicted secondary structure. Disintegration yields a 32P-labeled 14-mer consisting of the viral sequences in the hairpin stem and a 24-mer unlabeled target sequence that has been repaired.
Right: Inhibition of disintegration activity of HIV-1 integrase by Ole and HT. Lane 1, the 5′ 32P-labeled 38-mer dumbbell substrate. Lane 2, treatment with HIV-1 integrase results in the formation of the 5′ 32P-labeled14-mer disintegration product. Lanes 3, 4, 5, 6, 7 disintegration assays in the presence of 25, 50, 75, 100, and 200 nM Ole. Lanes 8, 9,10,11,12, in the presence of 25, 50, 75, 100, and 200 nM HT.
Ole and HT Inhibit Strand-Transfer Activity of HIV-1 Integrase
The effect of Ole and HT on the strand-transfer activity of HIV-1 integrase was tested by a quantitative heterologous integration assay. Figure 2B shows the design and results of this assay: U3-GT, a 5′ [32P] labeled and 3′-recessed 19-mer was used as the viral substrate. To focus on strand transfer, a supercoiled pUC18 plasmid DNA of 2.69 kb was used as the heterologous target. Incubation of the 5′ [32P] labeled viral substrate with unlabeled target in the presence of HIV-1 integrase results in the integration of the labeled 19-mer viral substrate into the 2.69 kb target plasmid. Integration was monitored by the conversion of unlabeled plasmid to labeled DNA in agarose gel electrophoresis as seen in the figure. Under these conditions, any inhibition detected must be specific for strand transfer and not for 3′ processing. Ole and HT demonstrated dose-dependent inhibition of the strand-transfer activity of HIV-1 integrase with EC50s of 56 and 43 nM respectively.
Ole and HT are Effective Against Disintegration Activity of HIV-1 Integrase
Figure 2C shows a schematic representation of disintegration reaction and assay results. Disintegration is the reverse of integration and involves concerted strand-cleavage and ligation reactions [20]. Strand cleavage takes place precisely at the junction between the viral and the target sequences and is coupled with the rejoining of the cleaved target sequences. The disintegration substrate is a dumbbell shaped 38-mer oligonucleotide that mimics the recombination intermediate of HIV-1 integration. It contains 5′ [32P] labeled virus-specific U5-LTR sequence of 14-mer in the stem of the hairpin loop and arbitrary target DNA sequences of 24-mer in the base of the dumbbell [21, 22]. The folded structure of the annealed substrate shown is based on reports obtained from hairpin formation by similar sequences [23, 24]. Disintegration of the 5′ [32P] labeled dumbbell by HIV-1 integrase is expected to give two products, a 5′ [32P] labeled 14-mer hairpin loop viral sequence and an unlabeled 24-mer closed circular target DNA. In the presence of HIV-integrase, the production of labeled 14-mer hairpin loop was detected by 7.5 M urea denaturating polyacrylamide gel electrophoresis and autoradiography of the gel, whereas the 24-mer product, the target DNA, was not seen in the autoradiography because it is unlabeled. This product can be detected by UV shadowing of the gel or by the use of 3′-labeled substrate. In the presence of Ole or HT, dose-dependent inhibition on the formation of the labeled 14-mer disintegration product was observed with EC50s of 28 or 18 nM respectively.
DISCUSSION
Several classes of HIV integrase inhibitors have been reported [4, 23], but none is clinically available yet. Lack of structural information for the intact protein, issues regarding different active site conformations dependent on crystal structure, and uncertain oligo-meric character of the enzyme protein have impeded the discovery of a clinically useful HIV-integrase inhibitor [4, 23]. Thus, molecular modeling becomes a key component in both the design of new integrase inhibitors and the identification of important protein–ligand interactions. There are 14 crystal structures of HIV-integrase available from the Protein Data Bank (PDB); however, only one has an inhibitor bound in the active site: 1QS4 [18]. We believe that this crystal structure contains the inhibitor would be the most relevant active site conformation on which to conduct the docking simulations with our anti-HIV small molecules, Ole and HT, despite previously reported crystal-packing effects associated with this structure [24].
The docking results reported here show good correlation with experimental data and provide a valuable tool for both evaluating compounds and designing more potent inhibitors. Ole and HT exhibit dose dependent inhibition in all of the three activities of HIV-1 integrase: 3′-processing, strand transfer and disintegration with EC50s all in the nM range. These compounds also showed good antiviral efficacy both in cell-to-cell transmission of HIV-1 as assayed by syncytial formation and in HIV-1 replication as assayed by p24 production. However, they are not toxic in the effective dose ranges and even at the concentration of 1,000 times EC50 (Table 1).
Table 1.
Anti-HIV Activities and Inhibition on HIV-1 Integrase Activity
| Compound | Anti-HIV Activitya | Inhibitory Activityb EC50 (nM) | ||||
|---|---|---|---|---|---|---|
| EC50 (nM) | IC50 (nM) | HIV-1 Integrase Activities | ||||
| Syncytium | P24 | Cytotoxicity | 3′-Proc | ST | Dis-In | |
| Ole | 55±5 | 73±8 | >10,000 | 46±6 | 56±5 | 28±3 |
| HT | 61±6 | 68±8 | >10,000 | 54±5 | 43±5 | 18±2 |
EC50, effective concentration at 50% inhibition; IC50, cytotoxicity concentration at 50% inhibition.
3′-Proc, 3′-processing; ST, strand transfer; Dis-In, disintegration. Values are means ±SD of triplicates in three independent determinations
To our knowledge, Ole and HT are the first group of small molecules capable of multiple actions against the AIDS virus, inhibiting both viral entry and integration. To act both outside and inside of the cellular environments represents a great advantage of this novel class of drugs. The structure-function information described here should facilitate the design of innovative multi-functional HIV-1 inhibitors.
Acknowledgments
This work was supported in part by Public Health Service grant R01 AT0343 to SLH. We are grateful to Drs. Morgan Jackson and Helen C. Lin for their support and encouragement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown PO. In: Retroviruses. Coffin J, Hughes S, Varmus H, editors. Cold Spring Harbor Press; Cold Spring Harbor: 1998. pp. 161–203. [Google Scholar]
- 2.Chiu TK, Davies DR. Structure and function of HIV-1 integrase. Curr Top Med Chem. 2004;4:965–977. doi: 10.2174/1568026043388547. [DOI] [PubMed] [Google Scholar]
- 3.Zhu K, Dobard C, Chow SA. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J Virol. 2004;78:5045–5055. doi: 10.1128/JVI.78.10.5045-5055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pommier Y, Johnson AA, Marchand C. Integrase inhibitors to treat HIV/AIDS. Nat Rev Drug Discov. 2005;4:236–248. doi: 10.1038/nrd1660. [DOI] [PubMed] [Google Scholar]
- 5.Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci U S A. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow SA, Vincent KA, Ellison V, Brown PO. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 7.Haertle T, Carrera CJ, Wasson DB, Sowers LC, Richman DD, Carson DA. Metabolism and anti-human immunodeficiency virus-1 activity of 2-halo-2′,3′-dideoxyadenosine derivatives. J Biol Chem. 1988;263:5870–5875. [PubMed] [Google Scholar]
- 8.Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 9.Mann DL, O'Brien SJ, Gilbert DA, Reid Y, Popovic M, Read-Connole E, Gallo RC, Gazdar AF. Origin of the HIV-susceptible human CD4+ cell line H9. AIDS Res Hum Retroviruses. 1989;5:253–255. doi: 10.1089/aid.1989.5.253. [DOI] [PubMed] [Google Scholar]
- 10.Popovic M, Sarngadharan MG, Read E, Gallo RC. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 11.Lee-Huang S, Maiorov V, Huang PL, Ng A, Lee HC, Chang YT, Kallenbach N, Huang PL, Chen HC. Structural and functional modeling of human lysozyme reveals a unique nonapeptide, HL9, with anti-HIV activity. Biochemistry. 2005;44:4648–4655. doi: 10.1021/bi0477081. [DOI] [PubMed] [Google Scholar]
- 12.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, -merz KM, Jr, Onufriev A, Sim-merling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 14.Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins TM, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multi-merization. J Biol Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Craigie R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J Biol Chem. 2005;280:29334–29339. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee-Huang S, Huang PL, Bourinbaiar AS, Chen HC, Kung HF. Inhibition of the integrase of human immunodeficiency virus (HIV) type 1 by anti-HIV plant proteins MAP30 and GAP31. Proc Natl Acad Sci U S A. 1995;92:8818–8822. doi: 10.1073/pnas.92.19.8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldgur Y, Craigie R, Cohen GH, Fujiwara T, Yoshinaga T, Fujishita T, Sugimoto H, Endo T, Murai H, Davies DR. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc Natl Acad Sci U S A. 1999;96:13040–13043. doi: 10.1073/pnas.96.23.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchand C, Johnson AA, Karki RG, Pais GC, Zhang X, Cowansage K, Patel TA, Nicklaus MC, Burke TR, Jr, Pommier Y. Metal-dependent inhibition of HIV-1 integrase by beta-diketo acids and resistance of the soluble double-mutant (F185K/C280S) Mol Pharmacol. 2003;64:600–609. doi: 10.1124/mol.64.3.600. [DOI] [PubMed] [Google Scholar]
- 20.Vincent KA, Ellison V, Chow SA, Brown PO. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blom-mers MJ, Walters JA, Haasnoot CA, Aelen JM, van der Marel GA, van Boom JH, Hilbers CW. Effects of base sequence on the loop folding in DNA hairpins. Biochemistry. 1989;28:7491–7498. doi: 10.1021/bi00444a049. [DOI] [PubMed] [Google Scholar]
- 22.Chow SA, Brown PO. Substrate features important for recognition and catalysis by human immunodeficiency virus type 1 integrase identified by using novel DNA substrates. J Virol. 1994;68:3896–3907. doi: 10.1128/jvi.68.6.3896-3907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Mawsawi LQ, Fikkert V, Dayam R, Witvrouw M, Burke TR, Jr, Borchers CH, Neamati N. Discovery of a small-molecule HIV-1 integrase inhibitor-binding site. Proc Natl Acad Sci U S A. 2006;103:10080–10085. doi: 10.1073/pnas.0511254103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sotriffer CA, Ni H, McCammon JA. Active site binding modes of HIV-1 integrase inhibitors. J Med Chem. 2000;43:4109–4117. doi: 10.1021/jm000194t. [DOI] [PubMed] [Google Scholar]


