Abstract
The primitive chordate Ciona intestinalis has emerged as a significant model system for the study of heart development. The Ciona embryo employs a conserved heart gene network in the context of extremely low cell numbers and reduced genetic redundancy. Here, I review recent studies on the molecular genetics of Ciona cardiogenesis as well as classic work on heart anatomy and physiology. I also discuss the potential of employing Ciona to decipher a comprehensive chordate gene network and to determine how this network controls heart morphogenesis.
Keywords: Heart development, Organogenesis, Gene regulation, Chordate evolution
1. Introduction
Ciona intestinalis is a member of the tunicates, a group of sessile marine invertebrates. Despite appearances (Fig. 1A) the tunicates are members of our own phylum, the Chordata. Indeed recent analysis indicate that the tunicates are the sister group to the vertebrates (Fig. 1B)[1]. Their close evolutionary relationship to the vertebrates is most evident during embryonic stages. Three chordate characters develop within the tunicate tadpole larvae, a dorsal hollow nerve cord, notochord and post-anal tail (Fig. 1C). Upon attachment to a substrate, the tail and associated chordate structures are resorbed while rudiments within the larval head differentiate into the adult organs, including the heart, body wall musculature, and gut.
Fig. 1.
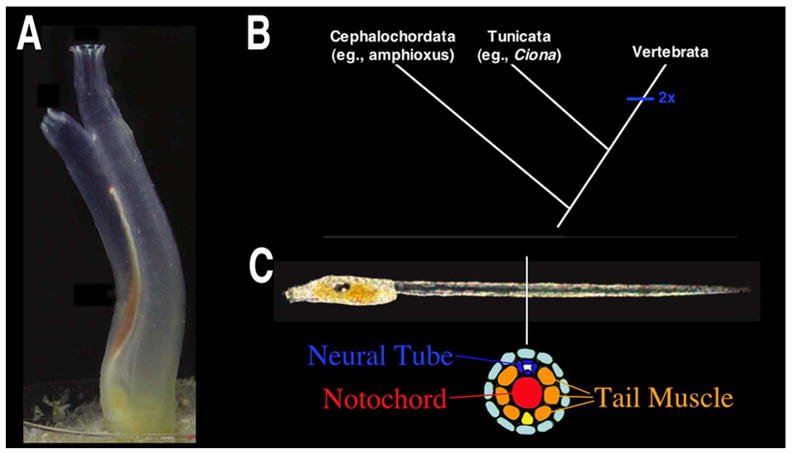
(A) Adult Ciona intestinalis. B. Current chordate phylogeny. C. Ciona intestinalis larvae with cross-sectional diagram of tail tissues. (A and C modified from [56])
The cellular simplicity of tunicate embryos has long made them an attractive model system for studying fundamental properties of animal development. Ciona embryos are a typical example, developing with extraordinarily low cell numbers. Gastrulation is initiated at the 110-cell stage, and the fully formed tadpole larva contains less then 3000 cells. This is in marked contrast to even the simplest vertebrate embryos, in which gastrulation takes place in the context of tens to hundreds of thousands of cells and larval/fetal stages consist of millions of cells. Additionally, Ciona cell lineages are invariant and an exhaustive cell fate map has been compiled [2]. Despite this extreme simplicity, embryonic development in Ciona is strikingly similar to that of vertebrate embryos. This similarity is particularly evident in comparing gastrulation and neurulation [3] and includes early steps of heart development (see below).
Ciona is also simple on a genetic level. Two Ciona genomes (Ciona intestinalis and savignyi) have recently been sequenced and assembled. The tunicates (Ciona included) diverged prior to vertebrate-specific gene duplications [4]. The resultant lack of genetic redundancy simplifies testing of gene function. Additionally, Ciona regulatory DNA is highly compact [5]. Availability of the fully sequenced Ciona savignyi genome allows for phylogenetic comparisons that can greatly assist in the identification of conserved, potential regulatory non-coding DNA [6]. Furthermore, putative regulatory regions can be quickly characterized due to the ease of generating transgenic embryos through electroporation. Despite this highly compact genome, genetic pathways underlying fundamental aspects of Ciona/vertebrate development, including cardiogenesis, are surprisingly well-conserved [7, 8].
Recent efforts have leveraged Ciona’s simplicity and tractability to initiate construction of comprehensive gene regulatory networks. Almost all of the transcription and signaling factors identified in the genome have been systematically assayed for their developmental expression patterns and many of them have also been functionally tested by morpholino knock down assays leading to the generation of detailed gene networks for early embryos [9, 10]. Techniques have been developed to permit generation and rearing of stable transgenic lines [11]. Ascidian stock centers for the creation and maintenance of stable transgenic lines are being developed in California (Smith Lab), Japan (Sasakura Lab) and France (Joly Lab).
2. Heart anatomy and physiology in the tunicates
The tunicate heart is a simple, single compartment organ with some interesting physiological properties. Almost all studies of the mature heart were conducted more than 25 years ago [12, 13]. Thus, updated studies employing modern techniques are overdue.
2.1. Heart anatomy
The tunicate heart is located ventrally (Fig. 2A–B), just posterior to the pharynx and anterior to the stomach. This simple organ consists of a valve-less, myocardial tube encased within a pericardial coelom (Fig. 2C). In Ciona and many other tunicates, the myocardial tube is V shaped, fitting into a roughly triangular pericardial sac. The pericardium and myocardium are derived from a single continuous epithelial tube invaginated along the dorsal side. Although this folded sheet of epithelial cells is only one cell layer thick it includes a number of distinct cell types (Fig. 2D–G). The myocardial cells are epithelial but have striated myofilaments concentrated along the luminal surface while the nuclei are found in the opposite surface (facing the pericardium). These cells are therefore described as myoepithelial. The myofilaments do not form fibers but instead a compact but loosely organized field [14]. Excitation-contraction coupling appears to rely on peripheral couplings or diads, as in vertebrate cardiac muscle [14]. The junctions between the Ciona myoepithelial cells bear some resemblance to the intercalated discs of the vertebrate myocardium [14, 15]. There is no discernible endocardium in the Ciona heart. Instead, the lumen of the heart is lined with the basal lamina of the myoepithelial cells. Along the junction of the pericardial and myocardial tubes there is a dense extracellular matrix, termed the raphe (Fig. 2D). Opposing the raphe is a line of smaller non-contractile cells, termed the undifferentiated line (Fig. 2D, F). At either end of the heart is a ring of smaller cells that have been characterized as a growth zone (Fig. 2E) [12]. The Ciona heart grows and regenerates throughout the lifespan of the organism, continually shedding degenerating myocardial cells. Generation of novel heart cells appears to occur at the terminal growth zones and possibly along the undifferentiated line [12].
Fig. 2.
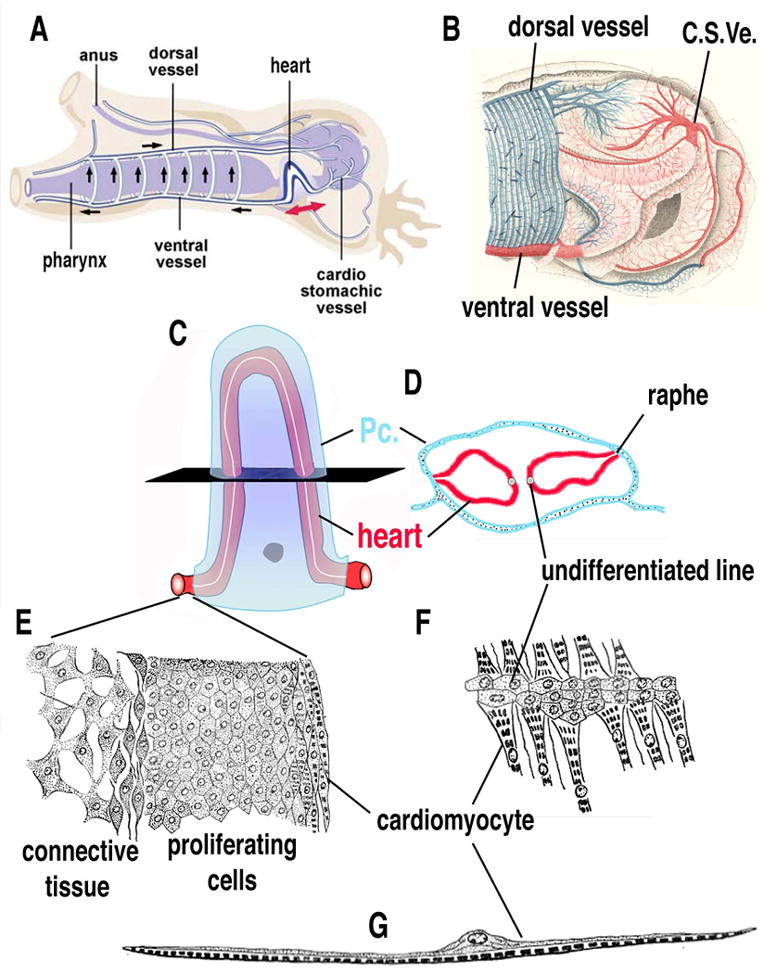
(A) Diagram of Ciona heart anatomy and blood circulation, anterior to the left (modified from [34]. (B) Close up illustration of Ciona heart and vasculature, proximal vessels in red, distal vessels in blue, anterior to the left (modified from [57]). (C–D) Cartoon (C) and cross-sectional diagram (D) of Ciona heart, myocardial tube (red), pericardial coelom (blue), undifferentiated line indicated in white. (E) Illustration of growth zone found at the ends of the heart tube. (F) Illustration of undifferentiated line. (G) Illustration of cardiac myoepithelial cell. Pc.- Pericardium, C. S. Ve. – Cardiac-stomachic vessel. (C–G) modified from [12].
The heart is attached at either end to large vessels that branch out to form smaller vessels throughout the body of the animal (Fig. 2B). The extent to which these vessels are lined by endothelia is unclear and tunicates are generally considered to have an open circulatory system. More detailed study is required to determine the extent and nature of Ciona vasculature and its evolutionary relationship to vertebrate blood vessels.
2.2. Heart physiology
The tunicate heart has been described as driving blood circulation through peristalsis. The striated myofibrils along the wall of the heart run parallel to each other and at a 60–70 degree angle to the long axis of the heart [12]. Thus these fibers form a “tight spiral around the heart” and rhythmic contractions proceed in a gradual wringing motion (see Supplemental Movie 1). This may be critical in preventing backflow, in a manner similar to the recently described spiral wringing of the early vertebrate heart tube [16]. The recent analysis of the embryonic zebrafish heart suggests that circulation is achieved through a dynamic suction pump mechanism rather than peristalsis [17]. A similar mechanism may apply to the Ciona heart.
Regular contractile waves can originate at either end of the heart, driving periodic reversals in blood flow. This fascinating, although not unique [13], feature of the tunicate heart has been the subject of numerous studies. The core mechanism for this reversibility is the presence of two myogenic pacemakers, one at either end of the heart [18]. Alternating predominance of one or the other appears to determine the direction of peristalsis. However, regulation of reversibility may also relate to external stimuli provided by accumulating pressure or possibly nervous input (observations on innervation of the tunicate heart remain inconclusive). There is no evidence for any specialized conduction system in the Ciona heart. Tight junctions between adjacent myocardial cells are likely to direct electrical coupling [19].
3. Ciona heart development: Lineage and morphogenesis
3.1. Relationship between the emerging heart lineage and the germ line
In Ciona and other tunicates the heart lineage is closely associated with the germ-plasm (also called pole plasm)[8]. The putative Ciona ‘germ plasm’ is segregated to the posterior/vegetal pole of the fertilized egg. Blastomeres containing this germ plasm are transcriptionally inactive [20]. At the 32-cell stage, the pair of blastomeres containing the heart lineage (B6.3) divides asymmetrically to produce two large, germ plasm free, daughter cells (B7.5) and two small germ plasm containing cells (B7.6) (Fig. 3A). The B7.5 cells constitute the pre-cardiac mesoderm [21], while progeny of the B7.6 cells go on to express Vasa and form the germ line [22]. We have speculated that the presence of germ plasm in the emergent heart lineage up to the 32-cell stage prevents these cells from responding to early fate determination factors. Intriguingly, the chordate heart specification factor, Mesp (see below), was first isolated in a screen for mRNAs localized in mouse primordial germ cells [23]. Whether germ plasm has any role in Ciona or vertebrate heart development has not been determined.
Fig. 3.
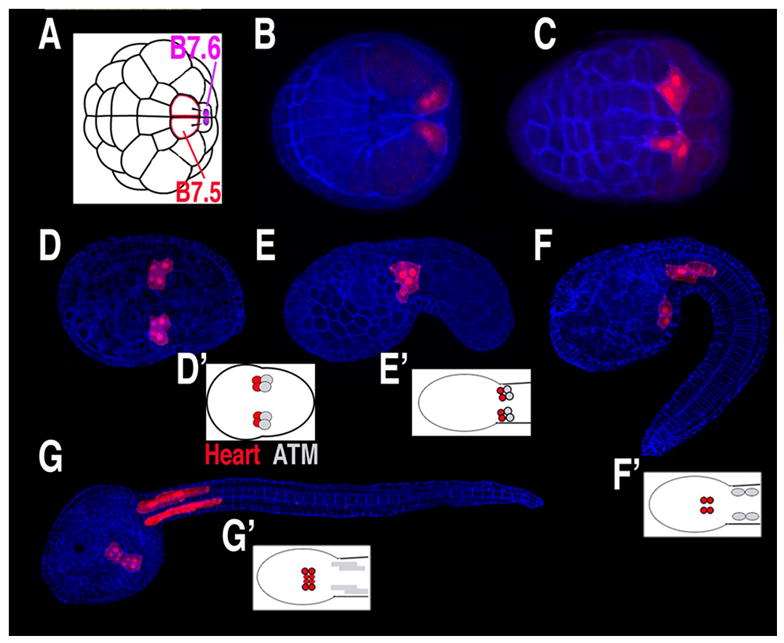
(A–G) Diagrams and confocal micrographs of Ciona embryos, stained with phalloidin (blue) and with B7.5 lineage cells expressing fluorescent proteins under the control of the Mesp enhancer (red), anterior to the left. (A) Diagram of 64-cell stage embryo, position of putative germ plasm in purple. (B) Dorsal view of gastrula stage embryo. (C–D) Early and late neurula stage embryos, ventral view. (E–F) Early and late tailbud stage embryos, lateral view. (G) Larva, ventra-lateral view. (D’–G’) Diagrams of B7.5 lineage fates from a ventral perspective, heart lineage in red, anterior tail muscle (ATM) in grey. (D and G modified from [24]).
3.2. Migration of the heart lineage during embryogenesis
The Ciona heart lineage can be traced back to a single pair of cells in the 64-cell embryo, the B7.5 cells. During gastrulation, the B7.5 cells involute at the posterior margin of the blastopore and migrate anteriorly (Fig. 3B,C). They also undergo two rounds of mitosis, resulting in bilateral clusters of 4 cells positioned at the border of the future head and tail regions (Fig. 3D–E)[24]. As the tail extends, the anterior two cells in each cluster separate from their posterior sisters. These anterior cells (termed trunk ventral cells, or TVCs) constitute the heart lineage. As the tail elongates, the heart precursor cells migrate rostrally and ventrally along the endoderm rudiment (Fig. 3F). The two bilateral clusters of migrating cells then fuse at the ventral midline and initiate a series of asymmetric cell divisions (Fig. 3G). Meanwhile, the posterior sister cells remain at the juncture of the head and tail and differentiate into anterior tail muscle cells (ATMs). The migration of the Ciona heart precursor cells and subsequent fusion on the ventral midline bears a striking resemblance to the behavior of vertebrate cardioblasts. Thus, research in Ciona has the potential to elucidate conserved mechanisms underlying the initiation of migratory behavior, response to guidance cues, and fusion at the midline.
3.3. Post-larval morphogenesis of the heart tube
In hatched Ciona larvae, dividing heart progenitors form a flat plate of cells. Larvae can survive for a number of days without undergoing metamorphosis but they do not develop beating heart tissue. However, larvae generally settle and metamorphose within 12 hours of hatching and the resulting juveniles will develop a beating heart 2–3 days after metamorphosis. The molecular basis for the arrest and subsequent re-initiation of heart development has not been characterized.
In early juveniles, the heart rudiment consists of a seemingly unstructured mass of cells (B. D. unpublished observations). In diagrams made from sectioned juveniles, researchers have described the formation of two distinct hollow tubes that fuse into a single tube [25]. An invagination along the dorsal surface of this tube gives rise to the myocardium while the outer surface differentiates as pericardium. Further studies are required to confirm these classic observations of post-larval cardiogenesis. If these observations prove accurate, they indicate that Ciona heart morphogenesis may exhibit some ancestral chordate characteristics. In Xenopus and the basal chordate amphioxus, the myocardium is also derived from a dorsal invagination [26, 27]. Whether this process is actually homologous among the chordates must await more detailed molecular characterizations.
In older juveniles (approximately 2 weeks after metamorphosis) the Ciona heart tube begins to fold into its characteristic V-shape (BD, personal observations). This shape change may be caused by the growth of the heart tube in length while the two ends remain at a relatively fixed distance [28]. It is possible that the bending is also influenced by the spiral orientation of the cardiac myofibers. Whether this event bears any homology to cardiac looping in vertebrates remains purely speculative. A detailed characterization of the mechanistic and genetic basis for heart bending in Ciona is required to resolve this issue.
4. Questions of homology and evolution
A key question in assessing the value of Ciona as a model for vertebrate cardiogenesis is the evolutionary relationship between the single compartment Ciona heart and the more complex multi-chambered heart of vertebrates. Despite differences in myocardial cell structure, similarities in structural and regulatory gene expression argue for myocardial cell homology [29–31] (also see below). Further characterization of regulatory gene function will clarify the extent of this homology while detailed characterization of structural gene regulation and function will reveal the evolutionary mechanisms driving structural divergence. Similarities in cardioblast migratory behavior and heart tube morphogenesis argue for homology on the tissue/organ level. Further characterization of post-larval morphogenesis of the Ciona heart tube will clarify the extent of this homology.
It has recently become apparent that amniote (avian, reptilian and mammalian) hearts are constructed from at least two distinct fields [32]. The left ventricle and atrial chambers arise from a primary field. The right ventricle, outflow tract and portions of the atrial chambers are derived from a secondary heart field (originating in the neighboring pharyngeal mesoderm). The ancestral chordate heart has been proposed to be equivalent to the primary heart field with the resulting left ventricle representing the ancestral single chamber. Studies of primary heart field patterning indicate that the default fate of this lineage is ventricular and that the atrial fate is the result of secondary patterning events, including a critical role for RA signaling [33]. Thus the Ciona heart is probably homologous to the vertebrate left ventricular field (Fig. 4). According to this assumption, during the transition from invertebrate to vertebrate chordates, RA was employed to re-pattern this primary field into two distinct chambers. This re-patterning may have occurred through two distinct mechanisms. One possibility is that the ancestral primary field expanded through increased cell division. The anterior portion of this enlarged field maintained the ancestral ventricular character while the posterior portion was modified to form the atrial chamber. Alternatively, the primary field may have been expanded by recruitment of neighboring mesoderm (as appears to have occurred during evolution of the right ventricle). This recruited posterior portion may have maintained a distinct “atrial” character relating to its historical function. Intriguingly, in Ciona the RA metabolic gene RALDH2 is expressed specifically in the posterior sister lineage to the embryonic heart field (the anterior tail muscle lineage) [34]. This provides preliminary support for the hypothesis that a newly recruited atrial lineage carried a distinct genetic signature that was subsequently exploited for patterning of two distinct chambers (Fig. 4). Further characterization of the role of RA in Ciona heart development and the relationship between the Ciona heart lineage and the posterior sister lineage will provide a test of this highly speculative scenario.
Fig. 4.
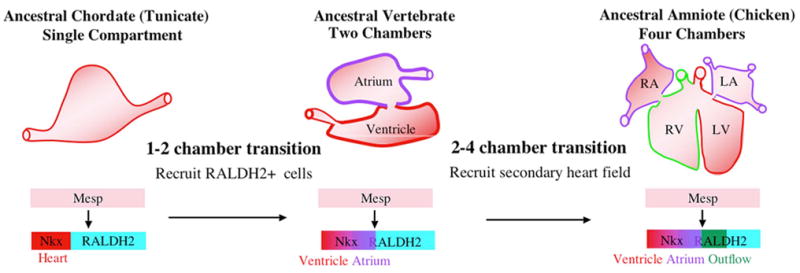
Model for chordate heart evolution. RA - Right atrium, LA – Left atrium, RV – Right ventricle, LV – Left ventricle.
Recent research on Ciona cardiogenesis has provided further support for the importance of recruitment in chordate heart evolution [30]. Normally, only half of the B7.5 lineage cells form heart. We have demonstrated that this decision is mediated by FGF signaling (see below). Targeted expression of a constitutively active form of the FGF transcriptional mediator Ets1/2 causes all B7.5 lineage cells to form heart (Fig. 5). Surprisingly, this expansion in the number of heart precursor cells can generate a two-compartment heart. A simple two-compartment heart resulting from expanded cardioblast recruitment may represent an evolutionary intermediate between the ancestral chordate single compartment heart and the basal vertebrate two-chambered heart.
Fig. 5.
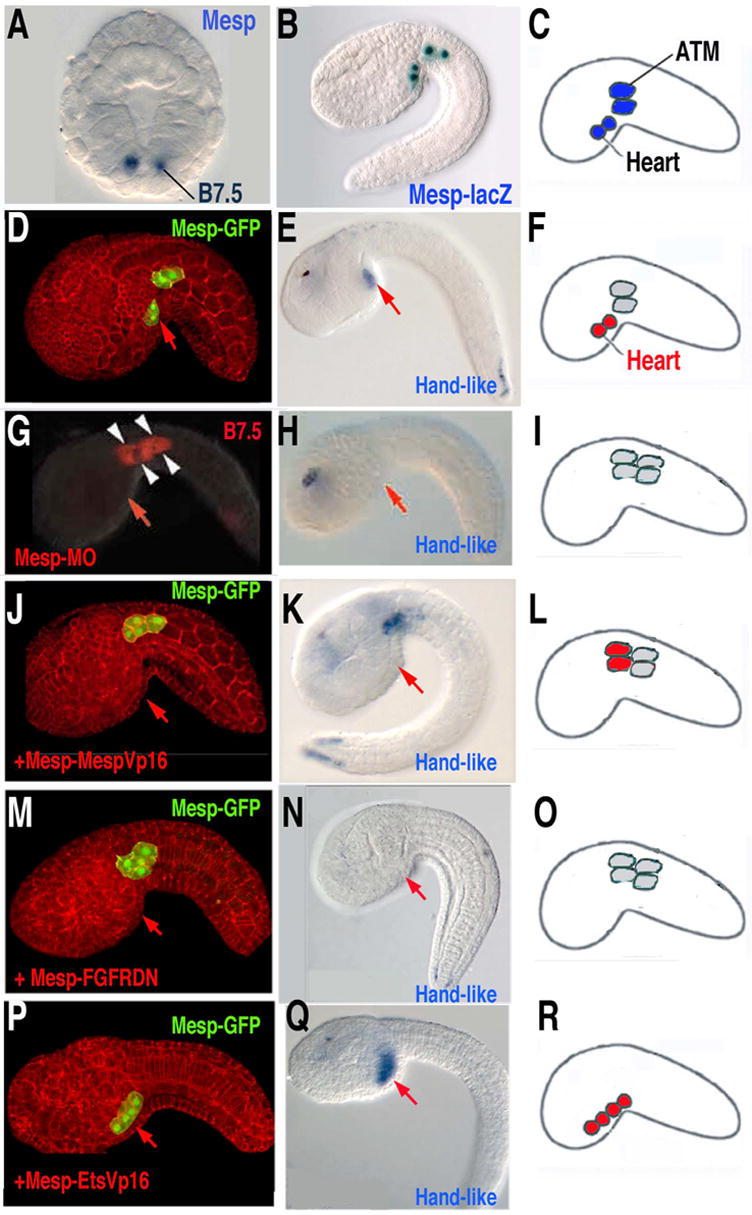
(A) Mesp expression in the B7.5 cells, gastrula stage, anterior is up. (B) X-gal staining of transgenic Mesp-lacZ embryos shows normal position of B7.5 lineage cells, as diagrammed in (C). (D–F) Control embryos show heart cell migration (D) and specification (Hand-like gene expression)(E), as diagrammed in (F). (G–I) Knockdown of Mesp (through injection of anti-sense morpholinos) blocks heart cell migration (G) and specification (Hand-like gene expression) (H), as diagrammed in (I). (J–L) Targeted expression of constitutively active MespVP16 in the B7.5 lineage cells blocks heart cell migration (J) but not specification (Handlike expression)(K), as diagrammed in (L). (M–O) Targeted expression of a dominant negative FGF receptor in the B7.5 lineage cells blocks heart cell migration (M) and specification (Handlike expression)(N), as diagrammed in (O). (P–R) Targeted expression of constitutively active EtsVp16 in the B7.5 lineage cells causes all B7.5 lineage cells to migrate (P) and express heart lineage genes (Hand-like expression)(Q), as diagrammed in (R). (Portions of this figure are modified from [24], [31] and [30].)
A clearer picture of chordate heart evolution and development requires in-depth comparative studies of heart gene networks in Ciona, amphioxus and vertebrate embryos. We now consider the genes, and emerging gene networks, governing heart development in the Ciona embryo.
5. Heart gene expression and function
Ciona cardiogenesis appears to involve the same core set of transcription factors (Nkx2.5/tinman, GATA4,5,6/pannier, Hand) required for heart formation in both vertebrates and flies [8, 31, 35]. Additionally, there is at least one transcription factor, Mesp, which appears to have a critical role in early heart specification in the chordates (Ciona and vertebrates) but not in Drosophila [24, 31, 36, 37]. The following section consists of a brief survey of the expression pattern and function of conserved heart genes in Ciona.
5.1. Mesp
There is accumulating evidence that the bHLH transcription factor Mesp plays a critical chordate-specific role in early cardiac specification. Because research on Mesp is not extensively covered in reviews of heart development, we include a thorough summary. In vertebrate genomes there are at least two Mesp genes, whose recent duplication is evidenced by their close linkage and extensive sequence similarity. These linked Mesp paralogs have been most extensively studied in mice where they play partially redundant roles in heart specification and somitogenesis. Mouse Mesp1 is initially expressed in much of the mesoderm just prior to gastrulation but is rapidly down-regulated in most non-cardiac lineages [38]. Mesp2 is predominantly expressed at later stages in paraxial mesoderm where it appears to have a key role in regulating Notch signaling during the periodic emergence of the somites[39]. In Mesp1/2 double knockout mice the mesoderm fails to undergo proper gastrulation, preventing analysis of heart development. In Mesp1/2 chimeric knockout mice, Mesp1/2 knockout cells contribute normally to almost all tissues except for heart[36]. These results were taken as evidence that Mesp1 is primarily a migration factor, first during gastrulation and subsequently in the positioning of heart precursor cells. However, it is also possible that Mesp has a more direct role in early heart specification, making the heart cells competent to respond to later specification signals (i.e.: creating the heart field) instead of merely driving proper positioning of these cells. Although Mesp1 orthologs show similar expression patterns in other vertebrates, further functional studies have not been performed. In flies, the closest ortholog to Mesp, sage, is not expressed in the cardiac mesoderm [37]. Mesp orthologs have not been characterized in any other organisms.
In Ciona, there is a single Mesp ortholog, Ci-Mesp. At the 110 cell stage, just after the B7.5 cells emerge, Ci-Mesp is expressed specifically in this pre-cardiac lineage (Fig. 5A–F)[31]. Nearly comprehensive expression surveys indicate that no other transcription factor shows this restricted pattern of expression [10]. Morpholino knockdown of Ci-Mesp specifically disrupts heart development (Fig. 5G–I)[31]. Under this treatment, the heart cells fail to properly migrate and express heart lineage markers. Treated juveniles completely lack heart tissue but otherwise appear normal. This data supports the conclusion that Mesp has a conserved role in chordate heart development but does not clarify whether this role extends beyond proper migration.
To address this question we employed the Mesp enhancer to target a constitutively active form of Mesp (MespVP16) in the B7.5 pre-cardiac lineage (Fig. 5J–L)[24]. This treatment blocked proper heart cell migration. However, the ectopically positioned heart lineage cells were able to form beating heart tissue. This study indicated that heart differentiation is independent of migration. These results also indicate that activation through Mesp is likely to mediate heart specification while repression through Mesp may be required to permit proper cell migration.
Our most recent research has begun to clarify the link between Mesp activity and early heart specification [30]. The transcription factor Ets1/2 is expressed in the B7.5 lineage shortly after Mesp. Preliminary data suggests that Mesp directly activates Ets1/2 transcription (BD unpublished). Ets1/2 expression defines the pre-cardiac mesoderm; making the B7.5 lineage cells responsive to FGF mediated heart specification. Targeted expression of a dominant negative FGF receptor or a repressor form of Ets1/2 in the B7.5 lineage blocks heart development (Fig. 5M–O). Targeted expression of a constitutively active form of Ets1/2 leads to an expanded field of heart precursor cells (Fig. 5P–R). Whether Mesp and Ets1/2 orthologs function in a related manner during vertebrate cardiogenesis remains unknown.
5.2. Nkx2.5/tinman
In flies, tinman is expressed broadly throughout the mesoderm during gastrulation. Subsequently, tinman expression is restricted to the cardiac mesoderm where it plays an essential, well-defined role in heart development[40]. In vertebrates there are a number of orthologs to tinman (Nkx2.5 and paralogs) that appear to function in a semi-redundant fashion during early heart development[41]. In Ciona there is a single ortholog to tinman/Nkx2.5, Ci-Nk4 (also referred to as Ci-Nkx). Sequence alignments demonstrate that the Ci-Nk4 homeodomain is highly conserved with those of vertebrate Nkx2 family genes (>90%). These alignments also indicate that Ci-Nk4 contains a N-terminal TN domain, linker and NK domains [42] along with a potential C-terminal tyrosine enriched domain or YRD [43]. Ci-Nk4 does not contain an Nkx2.5 box, but this domain is apparently specific to vertebrate orthologs (it is also not found in the Nkx2.5 ortholog from the invertebrate chordate, amphioxus, Amphi-Nk2Tin [26]). More significantly, Ci-Nk4 lacks the highly conserved GIRAW domain that appears to mediate interaction with co-repressors in all other chordate Nkx2.5 orthologs. This domain is also absent in Drosophila tinman and other non-chordate orthologs [43].
As with its vertebrate orthologs, Ci-Nk4 is first expressed in the cardiac mesoderm after gastrulation but also shows expression in neighboring ectodermal and endodermal regions (Fig. 6B,B’)[8]. However, expression of Ci-Nk4 in the cardiac mesoderm is down-regulated in the maturing tadpole and is then up-regulated in the differentiating heart after metamorphosis. This temporal complexity in Ci-Nk4 expression is probably associated with the arrest of heart differentiation in the swimming Ciona tadpole. Although Nkx2.5 is assumed to have a central role in vertebrate cardiogenesis, its complex regulation has been difficult to decipher and multiple paralogs make functional studies difficult as well. Thus, studies of Ci-Nk4 will help clarify the regulation and function of this central heart transcription factor.
Fig. 6.
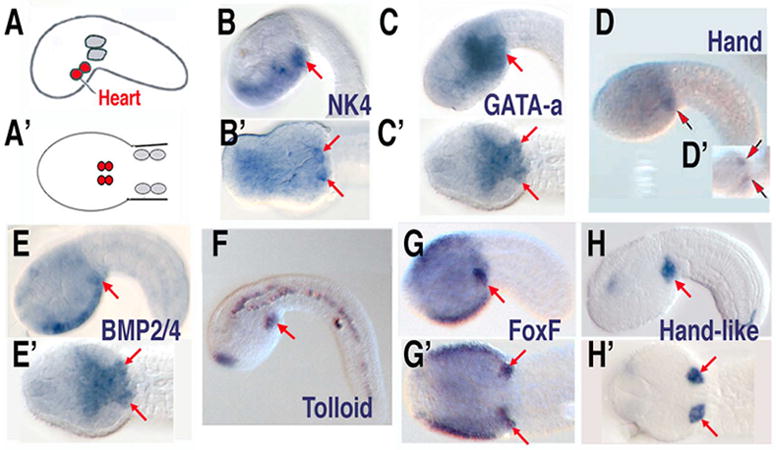
(A, A’) Lateral and ventral diagrams of B7.5 lineages, heart in red, anterior tail muscle in grey. (B–H, B’–H’) Expression patterns of heart lineage genes, (B–H) lateral views, (B’–H’) ventral views. D modified from [31]. F, G and G’ are images from the expression database (http://hoya.zool.kyoto-u.ac.jp/).
5.3. GATA factors
In both flies and vertebrates GATA factors have a central role in heart specification, cooperating with Nkx2.5/tinman and Bmp/Dpp signals to up-regulate cardiac mesoderm specific genes. There are only two GATA family transcription factors in the Ciona genome, Ci-GATA-a and Ci-GATA-b. Sequence comparisons cannot distinguish whether these genes are more orthologous to the GATA-4,5,6 or GATA-1,2,3 families. However, Ci-GATA-a is expressed in the cardiac mesoderm and neighboring posterior endoderm, making it the likely ortholog to GATA-4,5,6 (Fig. 6C, C’). Ci-GATA-b expression is restricted to portions of the anterior ectoderm [10]. Preliminary analysis indicates that proper function of Ci-GATA-a in the cardiac mesoderm is required for heart development (BD unpublished). The ability to disrupt Ci-GATA-a function independently in the cardiac mesoderm or endoderm of Ciona embryos represents a potent method for further functional characterization of this core cardiac transcription factor.
5.4. Hand
The bHLH transcription factor Hand appears to play a conserved role in heart development, acting downstream of Nkx/GATA in both flies and vertebrates. There is a single ortholog to Hand in the Ciona genome, Ci-Hand, which is expressed specifically in the bilateral heart rudiments in tailbud stage embryos (Fig. 6D,D’)[31]. Interestingly, in a nearly complete expression screen of annotated transcription factors, only Ci-Hand is exclusively expressed in the cardiac mesoderm[10]. Preliminary data suggests that Ci-Hand is not required for heart specification, but is required for proper differentiation (Y. Satou, personal communication).
5.5. T-box factors
Several T-box transcription factors have been implicated in vertebrate heart development, including Tbx1–5, 18 and 20 [44]. Additionally, there is evidence that putative orthologs to Tbx6 (dorsocross) and Tbx20 have important roles in development of the fly dorsal vessel [45–48]. The Ciona genome contains nine annotated T-box factors, representing 7 of 8 sub-families (there is no Ciona ortholog to the vertebrate limb T-box factors, Tbx4/5) [49]. Extensive expression data is available for all characterized Ciona T-box factors excepting Tbx20. Apparent lack of embryonic expression for Ci-Tbx20 indicate that any conserved role for this gene in Ciona heart development would be restricted to later, post-larval stages. Ciona appears to have recently duplicated the locus for Ci-Tbx6. Although these genes have no direct role in heart development, they appear to have a conserved role in regulating the early heart specification factor Mesp [23](see below).
5.6. Signaling factors
A wide array of signaling pathways have been implicated in vertebrate heart specification[50]. These include early activator roles for BMP and FGF as well as a crucial but ambiguous role for Wnt signaling [51]. There are also indications for contributions of Notch and Nodal signaling. Additionally, there is a well-defined role for RA signaling in later patterning events [33]. The precise contributions of these numerous signaling pathways and how they are interpreted by heart precursor cells remain enigmatic. As with transcription factors, there is a striking lack of redundancy in Ciona signaling pathway components (for example there is only one Ciona FGF receptor vs. three in many vertebrates and six distinct FGF ligands vs. approximately 20 in vertebrate genomes). Additionally, the cellular simplicity of the Ciona embryo will permit detailed analysis of how cellular machinery (receptor localization, endosome processing, etc.) is employed during signaling integration by targeted heart precursor cells. However, it must first be determined whether any of these pathways have conserved roles in Ciona heart development. Because roles for BMP and FGF signaling in cardiac specification appear to be conserved from flies to vertebrates, they would be expected to have similar roles in Ciona as well. As detailed above, our recent research has demonstrated a central role for FGF signaling in Ciona heart specification [30]. Expression data also supports a role for BMP. The sole Ciona ortholog to BMP2 and 4 (Ci-Bmp2/4) is expressed in the ventral head epidermis and bilateral heart rudiments (similar to Ci-Nk4) and the ortholog to Tolloid is also expressed in the heart lineage (Fig. 6E,E’ and F). There is currently no data on possible roles for Wnt, Nodal or Notch signaling pathways in Ciona heart specification. As previously mentioned, the RA metabolic enzyme RALDH2 is expressed just posterior to the initial heart field, as is the case in vertebrate embryos [34].
6. The future of heart development research in Ciona: gene networks
Future work on Ciona heart development will exploit the unique strengths of this model system, namely the cellular and genetic simplicity and the ease and rapidity of transient transgenic analysis. These qualities should permit the elucidation of a comprehensive gene regulation network governing heart specification, beginning with localized maternal determinants such as Macho-1 and culminating in the activation of cardiac specific genes in the anterior B7.5 lineage. An outline of this emerging network is already available from earlier studies (Fig. 7). Further characterization of this network will include efforts to disentangle the intersecting roles of numerous signaling pathways implicated in vertebrate heart development, including Wnt, FGF and BMP. Low redundancy in Ciona signaling pathway components combined with the ability to specifically manipulate these components in either the target or secreting cell lineages represents a powerful tool for dissecting complex signaling events.
Fig. 7.
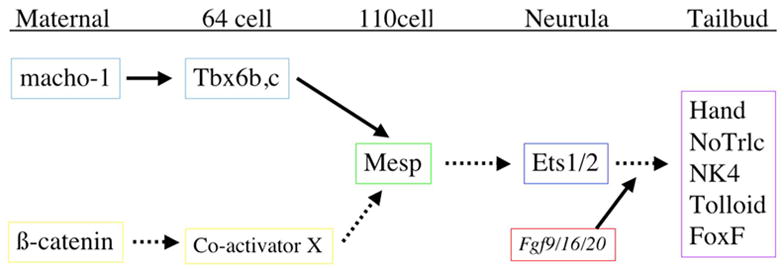
Gene network governing Ciona heart specification. Regulatory connections with experimental support indicated in solid lines, provisional connections in dotted lines.
The determination of complete gene networks depends on the isolation and characterization of associated regulatory DNAs [52]. The ease of generating transgenic embryos permits rapid and extensive analysis of Ciona enhancers. The first result of this effort was the isolation of a short (110bp) enhancer for Ci-Mesp [24]. Extensive analysis of this enhancer and the orthologous enhancer from Ciona savignyi led to the identification of a number of critical T-box binding sites which were subsequently confirmed as binding sites for transcription factors Tbx6b/c. This led to the prediction that both vertebrate Mesp genes were downstream of Tbx6. Subsequent studies in mice and teleosts confirmed that Mesp2 is directly activated by Tbx6 [53, 54]. Current efforts are focused on characterizing the enhancers for Ci-NK4, Ci-GATA-a, Ci-Hand, as well as other factors which are expressed in the Ciona heart lineage, including Tolloid, Bmp2/4, FoxF and Hand-like (Fig. 6). Detailed analysis of these enhancers coupled with identification of target genes will permit construction of a comprehensive network for Ciona heart development.
The Ciona system also offers an opportunity to investigate the cellular basis for heart formation, including directed migration of cardioblasts. Efforts are currently underway to use transgenic GFP reporter genes in conjunction with cell sorting methods to isolate individual heart cells at different phases of migration and differentiation (L. Christian, in preparation). Such sorted cell populations can be used for the comprehensive identification of cardiac factors using newly developed microarrays and tiling arrays. By characterizing heart lineage gene expression in different transgenic backgrounds, downstream morphogenetic genes can be integrated into existing heart gene networks. Furthermore, remarkably low cell numbers and fluorescently labeled lineages will permit the use of live time lapse imaging to assess the detailed behavior of normal and transgenic heart lineage cells [55]. As candidate effector genes are identified, lineage specific manipulation of these genes can be assessed through time-lapse imaging to determine their precise roles in the cellular events underlying heart morphogenesis.
Supplementary Material
Video of adult Ciona heart, stained by injection of vital dye.
Acknowledgments
I would like to thank Mike Levine, Xavier Neto and Brian Black for their comments and suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–8. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 2.Satoh N. Developmental biology of ascidians. New York: Cambridge University Press; 1994. [Google Scholar]
- 3.Nishida H. Specification of embryonic axis and mosaic development in ascidians. Dev Dyn. 2005;233:1177–93. doi: 10.1002/dvdy.20469. [DOI] [PubMed] [Google Scholar]
- 4.Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–9. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- 5.Di Gregorio A, Levine M. Analyzing gene regulation in ascidian embryos: New tools for new perspectives. Differentiation. 2002;70:132–9. doi: 10.1046/j.1432-0436.2002.700402.x. [DOI] [PubMed] [Google Scholar]
- 6.Johnson DS, Davidson B, Brown CD, Smith WC, Sidow A. Non-coding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res. 2004;14:2448–56. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munro EM, Odell GM. Polarized basolateral cell motility underlies invagination and convergent extension of the ascidian notochord. Development. 2002;129:13–24. doi: 10.1242/dev.129.1.13. [DOI] [PubMed] [Google Scholar]
- 8.Davidson B, Levine M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc Natl Acad Sci U S A. 2003;100:11469–73. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–7. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- 10.Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: Towards a comprehensive understanding of gene networks. Development. 2004;131:4047–58. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- 11.Sasakura Y, Awazu S, Chiba S, Satoh N. Germ-line transgenesis of the tc1/mariner superfamily transposon minos in Ciona intestinalis. Proc Natl Acad Sci U S A. 2003;100:7726–30. doi: 10.1073/pnas.1230736100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millar RH. Ciona. The University Press of Liverpool; 1953. [Google Scholar]
- 13.Robb JS. Comparative basic cardiology. New York: Grune & Stratton; 1965. [Google Scholar]
- 14.Oliphant LW, Cloney RA. The ascidian myocardium: Sarcoplasmic reticulum and excitation-contraction coupling. Z Zellforsch Mikrosk Anat. 1972;129:395–412. doi: 10.1007/BF00307296. [DOI] [PubMed] [Google Scholar]
- 15.Lorber V, Rayns DG. Cellular junctions in the tunicate heart. J Cell Sci. 1972;10:211–27. doi: 10.1242/jcs.10.1.211. [DOI] [PubMed] [Google Scholar]
- 16.Torrent-Guasp F, Buckberg GD, Clemente C, Cox JL, Coghlan HC, Gharib M. The structure and function of the helical heart and its buttress wrapping. I. The normal macroscopic structure of the heart. Semin Thorac Cardiovasc Surg. 2001;13:301–19. doi: 10.1053/stcs.2001.29953. [DOI] [PubMed] [Google Scholar]
- 17.Forouhar AS, Liebling M, Hickerson A, Nasiraei-Moghaddam A, Tsai HJ, Hove JR, et al. The embryonic vertebrate heart tube is a dynamic suction pump. Science. 2006;312:751–3. doi: 10.1126/science.1123775. [DOI] [PubMed] [Google Scholar]
- 18.Anderson M. Electrophysiological studies on initiation and reversal of the heart beat in Ciona intestinalis. J Exp Biol. 1968;49:363–85. doi: 10.1242/jeb.49.2.363. [DOI] [PubMed] [Google Scholar]
- 19.Kriebel ME. Electrical coupling between tunicate heart cells. Life Sci II. 1968;7:181–6. doi: 10.1016/0024-3205(68)90303-2. [DOI] [PubMed] [Google Scholar]
- 20.Tomioka M, Miya T, Nishida H. Repression of zygotic gene expression in the putative germline cells in ascidian embryos. Zoolog Sci. 2002;19:49–55. doi: 10.2108/zsj.19.49. [DOI] [PubMed] [Google Scholar]
- 21.Hirano T, Nishida H. Developmental fates of larval tissues after metamorphosis in ascidian Halocynthia roretzi. I. Origin of mesodermal tissues of the juvenile. Dev Biol. 1997;192:199–210. doi: 10.1006/dbio.1997.8772. [DOI] [PubMed] [Google Scholar]
- 22.Shirae-Kurabayashi M, Nishikata T, Takamura K, Tanaka KJ, Nakamoto C, Nakamura A. Dynamic redistribution of vasa homolog and exclusion of somatic cell determinants during germ cell specification in Ciona intestinalis. Development. 2006;133:2683–93. doi: 10.1242/dev.02446. [DOI] [PubMed] [Google Scholar]
- 23.Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, Taketo MM. Mesp1: A novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. 1996;122:2769–78. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- 24.Davidson B, Shi W, Levine M. Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis. Development. 2005;132:4811–8. doi: 10.1242/dev.02051. [DOI] [PubMed] [Google Scholar]
- 25.de Selys-Longchamps M. Dévelopment du coeur, du péricarde, et des épicardes chez Ciona intestinalis. Arch Biologie. 1900;17:499–542. [Google Scholar]
- 26.Holland ND, Venkatesh TV, Holland LZ, Jacobs DK, Bodmer R. Amphink2-tin, an amphioxus homeobox gene expressed in myocardial progenitors: Insights into evolution of the vertebrate heart. Dev Biol. 2003;255:128–37. doi: 10.1016/s0012-1606(02)00050-7. [DOI] [PubMed] [Google Scholar]
- 27.Raffin M, Leong LM, Rones MS, Sparrow D, Mohun T, Mercola M. Subdivision of the cardiac Nkx2. 5 expression domain into myogenic and nonmyogenic compartments. Dev Biol. 2000;218:326–40. doi: 10.1006/dbio.1999.9579. [DOI] [PubMed] [Google Scholar]
- 28.Berril NJ. The tunicata. London: Ray Society; 1950. [Google Scholar]
- 29.MacLean DW, Meedel TH, Hastings KE. Tissue-specific alternative splicing of ascidian Troponin I isoforms. Redesign of a protein isoform-generating mechanism during chordate evolution. J Biol Chem. 1997;272:32115–20. doi: 10.1074/jbc.272.51.32115. [DOI] [PubMed] [Google Scholar]
- 30.Davidson B, Shi W, Beh J, Christiaen L, Levine M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev. 2006;20:2728–38. doi: 10.1101/gad.1467706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satou Y, Imai KS, Satoh N. The ascidian Mesp gene specifies heart precursor cells. Development. 2004;131:2533–41. doi: 10.1242/dev.01145. [DOI] [PubMed] [Google Scholar]
- 32.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–35. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 33.Hochgreb T, Linhares VL, Menezes DC, Sampaio AC, Yan CY, Cardoso WV, et al. A caudorostral wave of Raldh2 conveys anteroposterior information to the cardiac field. Development. 2003;130:5363–74. doi: 10.1242/dev.00750. [DOI] [PubMed] [Google Scholar]
- 34.Simoes-Costa MS, Vasconcelos M, Sampaio AC, Cravo RM, Linhares VL, Hochgreb T, et al. The evolutionary origin of cardiac chambers. Dev Biol. 2005;277:1–15. doi: 10.1016/j.ydbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 35.Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- 36.Kitajima S, Takagi A, Inoue T, Saga Y. Mesp1 and Mesp2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–26. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- 37.Moore AW, Barbel S, Jan LY, Jan YN. A genomewide survey of basic helix-loop-helix factors in drosophila. Proc Natl Acad Sci U S A. 2000;97:10436–41. doi: 10.1073/pnas.170301897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. Mesp1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–47. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 39.Morimoto M, Takahashi Y, Endo M, Saga Y. The Mesp2 transcription factor establishes segmental borders by suppressing notch activity. Nature. 2005;435:354–9. doi: 10.1038/nature03591. [DOI] [PubMed] [Google Scholar]
- 40.Bodmer R, Venkatesh TV. Heart development in Drosophila and vertebrates: Conservation of molecular mechanisms. Dev Genet. 1998;22:181–6. doi: 10.1002/(SICI)1520-6408(1998)22:3<181::AID-DVG1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka M, Kasahara H, Bartunkova S, Schinke M, Komuro I, Inagaki H, et al. Vertebrate homologs of tinman and bagpipe: Roles of the homeobox genes in cardiovascular development. Dev Genet. 1998;22:239–49. doi: 10.1002/(SICI)1520-6408(1998)22:3<239::AID-DVG6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 42.Evans SM. Vertebrate tinman homologues and cardiac differentiation. Semin Cell Dev Biol. 1999;10:73–83. doi: 10.1006/scdb.1999.0282. [DOI] [PubMed] [Google Scholar]
- 43.Elliott DA, Solloway MJ, Wise N, Biben C, Costa MW, Furtado MB, et al. A tyrosine-rich domain within homeodomain transcription factor Nkx2-5 is an essential element in the early cardiac transcriptional regulatory machinery. Development. 2006;133:1311–22. doi: 10.1242/dev.02305. [DOI] [PubMed] [Google Scholar]
- 44.Stennard FA, Harvey RP. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005;132:4897–910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- 45.Reim I, Mohler JP, Frasch M. Tbx20-related genes, mid and h15, are required for tinman expression, proper patterning, and normal differentiation of cardioblasts in Drosophila. Mech Dev. 2005;122:1056–69. doi: 10.1016/j.mod.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Reim I, Frasch M. The dorsocross t-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development. 2005;132:4911–25. doi: 10.1242/dev.02077. [DOI] [PubMed] [Google Scholar]
- 47.Qian L, Liu J, Bodmer R. Neuromancer Tbx20-related genes (h15/midline) promote cell fate specification and morphogenesis of the Drosophila heart. Dev Biol. 2005;279:509–24. doi: 10.1016/j.ydbio.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 48.Miskolczi-McCallum CM, Scavetta RJ, Svendsen PC, Soanes KH, Brook WJ. The Drosophila melanogaster T-box genes midline and h15 are conserved regulators of heart development. Dev Biol. 2005;278:459–72. doi: 10.1016/j.ydbio.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 49.Takatori N, Hotta K, Mochizuki Y, Satoh G, Mitani Y, Satoh N, et al. T-box genes in the ascidian Ciona intestinalis: Characterization of cDNAs and spatial expression. Dev Dyn. 2004;230:743–53. doi: 10.1002/dvdy.20082. [DOI] [PubMed] [Google Scholar]
- 50.Brand T. Heart development: Molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- 51.Eisenberg LM, Eisenberg CA. Wnt signal transduction and the formation of the myocardium. Dev Biol. 2006;293:305–15. doi: 10.1016/j.ydbio.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 52.Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev Cell. 2005;9:449–62. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 53.Yasuhiko Y, Haraguchi S, Kitajima S, Takahashi Y, Kanno J, Saga Y. Tbx6-mediated notch signaling controls somite-specific Mesp2 expression. Proc Natl Acad Sci U S A. 2006;103:3651–6. doi: 10.1073/pnas.0508238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terasaki H, Murakami R, Yasuhiko Y, Shin IT, Kohara Y, Saga Y, et al. Transgenic analysis of the medaka Mesp-b enhancer in somitogenesis. Dev Growth Differ. 2006;48:153–68. doi: 10.1111/j.1440-169X.2006.00853.x. [DOI] [PubMed] [Google Scholar]
- 55.Passamaneck YJ, Di Gregorio A, Papaioannou VE, Hadjantonakis AK. Live imaging of fluorescent proteins in chordate embryos: From ascidians to mice. Microsc Res Tech. 2006;69:160–7. doi: 10.1002/jemt.20284. [DOI] [PubMed] [Google Scholar]
- 56.Satoh N. The ascidian tadpole larva: Comparative molecular development and genomics. Nat Rev Genet. 2003;4:285–95. doi: 10.1038/nrg1042. [DOI] [PubMed] [Google Scholar]
- 57.Roule L. Phallusiadees 1st partie, monographie de la Ciona intestinalis. Recherches sur les ascidies simples des cotes de provence. 1884 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video of adult Ciona heart, stained by injection of vital dye.


