Summary
Background
The plant pathogen Pseudomonas syringae injects 20–40 different proteins called effectors into host plant cells, yet the functions and sites of action of these effectors in promoting pathogenesis is largely unknown. Plants in turn defend themselves against P. syringae by activating the salicylic acid (SA)-mediated signaling pathway. The P. syringae-specific HopI1 effector has a putative chloroplast targeting sequence and a J domain. J domains function by activating 70-kDa heat shock proteins (Hsp70).
Results
HopI1 is an ubiquitous P. syringae virulence effector that acts inside plant cells. When expressed in plants, HopI1 localizes to chloroplasts, the site of SA synthesis. HopI1 causes chloroplast thylakoid structure remodeling and suppresses SA accumulation. HopI1’s C terminus has bona fide J-domain activity that is necessary for HopI1-mediated virulence and thylakoid remodeling. Furthermore, HopI1-expressing plants have increased heat tolerance, establishing that HopI1 can engage the plant stress response machinery.
Conclusions
These results strongly suggest that chloroplast Hsp70 is targeted by the P. syringae HopI1 effector to promote bacterial virulence by suppressing plant defenses. The targeting of Hsp70 function through J-domain proteins is known to occur in a mammalian virus, SV40. However, this is the first example of a bacterial pathogen exploiting a J-domain protein to promote pathogenesis through alterations of chloroplast structure and function.
Introduction
Plant and human pathogens cause disease by interfering with host defense responses and altering host signaling and metabolism to create an environment favorable for their survival. Gram-negative bacteria often use a type III secretion system (T3SS) to inject effector proteins directly into host cells [1]. The T3SS is essential for virulence in many pathogens [2]. Plant pathogens with individual effector gene mutations usually exhibit only a modestly reduced ability to cause disease and/or grow on plants. Some effectors, called Avirulence (Avr) proteins, can have more dramatic effects on pathogen fitness due to their recognition by resistant plant hosts, although some only cause a modest reduction in bacterial growth on plants [3]. Disease resistance genes confer the ability to recognize Avr effectors, events that lead to the activation of defenses accompanied by a programmed cell death called the hypersensitive response [4]. In susceptible plants, cell death occurs at a late stage of pathogenesis and is important for symptom formation and possibly disease spread.
Some virulence effectors suppress plant defense responses as a way of promoting pathogen growth [5]. For example, AvrPtoB (now called HopAB2PtoDC3000 [6]) is a cell death inhibitor [7] and suppressor of basal defenses [8]. Several effectors suppress cell wall-based defenses in a manner that requires a major defense signal called salicylic acid (SA) [9]. However, some defense-suppressing effectors are SA-independent [10].
We previously performed a genetic screen with Pseudomonas syringae pv. maculicola strain PmaES4326 that identified novel type III effectors [11]. HopI1 (previously named HopPmaI) is of particular interest because the original transposon mutant had attenuated virulence [11]. Interestingly, HopI1 and many other effectors have features of chloroplast-targeted proteins [11]. HopI1 also has 4 copies of a 37/38 amino acid proline- and glutamine-(P/Q)-rich repeat region and a C-terminal region with homology to a J domain (Fig. 1A) [11]. J domains are conserved ~70 amino acid modules found in Hsp70 cochaperones such as Hsp40 (DnaJ). J domain-containing proteins (J proteins) interact with Hsp70, activate its ATPase activity and protein folding (reviewed in [12]). J proteins also play a role in the growth of several mammalian pathogens. For example, the small and large T antigen J proteins from the SV40 virus are vital for viral replication and tumorogenesis [13].
Fig. 1.
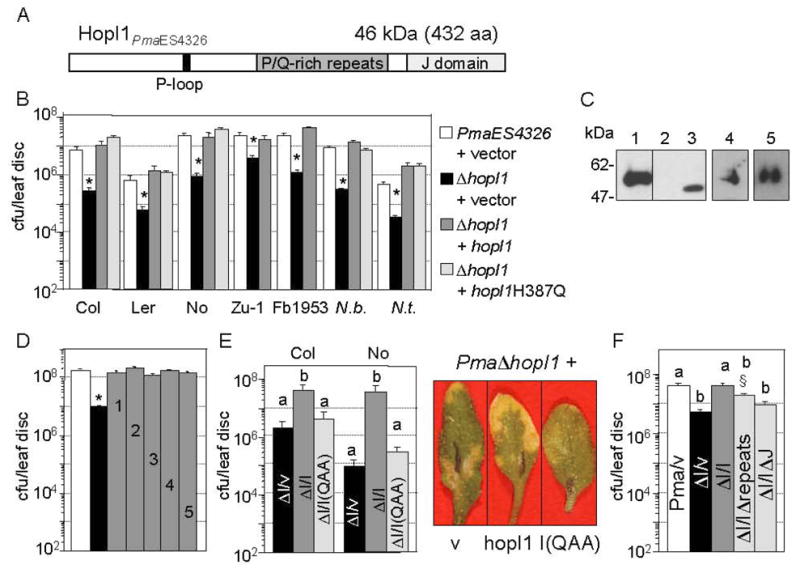
Importance of HopI1 and its J domain for P. syringae virulence. Data represent the means of 8 samples with standard errors.
(A) Schematic structure of HopI1PmaES4326 showing different protein regions.
(B) The ΔhopI1 strain with the empty vector pCKTR (black bar) grew significantly less than PmaES4326 with empty vector (white bar) on many A. thaliana accessions and two tobacco species (*P<0.05). This phenotype was complemented by hopI1PmaES4326 (JJ19, dark-gray bar) or hopI1PmaES4326 with a point mutation in J domain (hopI1H387Q, JJ77, light-gray bar) expressed from the native promoter and integrated into the chromosome.
(C) Epitope-tagged versions of HopI1 were expressed in the ΔhopI1 PmaES4326 strain from native promoter in conditions that promote effector protein production. Proteins were detected with anti-HA antibody by SDS-PAGE and Western blot analysis: (1) JJ19=hopI1-HA-Myc-His, (2) pCKTR, (3) JJ78= hopI1-HA, (4) JJ77=hopI1H/Q-HA-Myc-His, (5) JJ207=hopI1HPD/QAA-HA-Myc-His. Lines 1–3 are from one gel.
(D) The ΔhopI1 strain carrying an HA-tagged hopI1 allele (gray bar) from any of several P. syringae strains expressed from the nptII promoter grew significantly more on Columbia (Col) plants than ΔhopI1 strain carrying the control vector pME6012 (black bar) (*P<0.0001): (1) hopI1PmaES4326; JJ78; (2) hopI1PtoDC3000, JJ152; (3) hopI1PsyB728a; JJ151; (4) hopI1PsyCit7, JJ153; (5) hopI1Psy61, JJ154. White bar, wild type PmaES4326 strain with empty vector.
(E) Left panel: The J-domain triple mutant HPD/QAA of HopI1 integrated into the chromosome under native promoter (JJ207) did not complement the growth defect of ΔhopI1 strain in Col and Nossen (No). Strains with different letters showed significant differences in their growth (P<0.001, Fisher’s protected least significant difference measure (PLSD), a post hoc, multiple t-test). ΔI,ΔhopI1 PmaES4326 strain; v, vector pCKTR; I, full-length HopI1 (JJ19); I(QAA), HPD/QAA mutant of HopI1 (JJ207). Right panel: representative Col leaves from the infection with the Δ carrying vector (v), full length HopI1 (hopI1) or the QAA mutant shown in the left panel.
(F) The growth of the ΔhopI1 strain was not rescued by hopI1Δ (JJ193) or hopIΔJ (JJ194) integrated into the chromosome under native promoter. However, the ΔhopI1 strain containing hopI1Δ showed a trend of being partially complemented compared to the vector control (§P<0.056, Fisher’s PLSD). Strains with different letters show significant differences in growth (P<0.02, Fisher’s PLSD).
Here, we investigate the role, localization and activity of the P. syringae HopI1 protein. We show that HopI1 has a bona fide J domain, suppresses defenses and localizes to chloroplasts. Based on these data, we propose that HopI1 interacts with Hsp70 and inhibits defense signaling mediated by chloroplasts.
Results
HopI1 is present in all analyzed P. syringae strains
The hopI1 gene was present in the same chromosomal context of all three sequenced P. syringae strains (PtoDC3000, PsyB728a, Pph1448A) and PmaES4326 (Fig. S1A). hopI1 was not associated with any other effector, and was not a part of a genomic or pathogenicity island. Alleles of hop1I were present in all isolates of P. syringae examined: pathovars maculicola, phaseolicola, syringae, tabaci and tomato [11] as well as in strains recently isolated from diseased crops in Italy and France (Table S1). These data indicate HopI1’s early acquisition in the evolution of the pathogen.
HopI1 is a virulence factor in Arabidopsis thaliana and tobacco
PmaES4326 lacking hopI1 due to an unmarked hopI1 deletion grew normally in vitro (data not shown), but its growth was attenuated on several A. thaliana accessions as well as on Nicotiana benthamiana and N. tabacum (Fig. 1B). The virulence defect of theΔhopI1 strain was complemented with a version of HopI1PmaES4326 containing the C-terminal influenza hemagglutin (HA), c-Myc (Myc) and His epitope tags (JJ19) integrated at the hopI1 locus under the native promoter (Fig. 1B) or when constitutively expressed from nptII promoter on a multi-copy plasmid with an HA epitope tag (pJJ78, Fig. 1D). The epitope-tagged HopI1 proteins accumulated significantly in P. syringae (Fig. 1C).
Constitutively expressed hopI1 alleles from PtoDC3000 (pJJ152), PsyB728a (pJJ151), PsyCit7 (pJJ153) and Psy61 (pJJ154) also rescued theΔhopI1 strain virulence defect (Fig. 1D). These alleles (and others) had extensive variation in the number and composition of the P/Q-rich repeat region (Fig. S1B). Thus, despite the high variation in HopI1, all of the alleles examined were functional and likely act in a similar manner.
The conserved HPD loop of the J domain is important for HopI1 function
We next analyzed the importance for virulence of HopI1’s J domain and P/Q-rich repeat regions. HopI1 with a single substitution in the conserved HPD motif of the J domain expressed from the native promoter (HopI1-H387Q, JJ77) rescued the ΔhopI1 strain’s virulence defect (Fig. 1B). However, mutation of the HPD loop to QAA (JJ207) disrupted HopI1 function, as the mutant protein failed to complement the virulence defect of the ΔhopI1 strain in A. thaliana (Fig. 1E). Mutant HopI1 proteins accumulated well in P. syringae (Fig. 1C). Thus, the J domain plays an important role in HopI1 virulence function. HopI1 lacking the whole J domain (HopI1ΔJ, Δ amino acids 361–431, JJ194) did not rescue the ΔhopI1 phenotype (Fig. 1F), but also did not accumulate in bacteria (not shown).
To analyze the role of the P/Q-rich repeat region, we deleted amino acids 194–332 from HopI1-HA-Myc-His. Expression of HopI1Δrepeats from the native promoter (JJ193) had a trend of partially complementing the ΔhopI1 mutant strain (Fig. 1F). However, the level of the protein in P. syringae was reduced or undetectable (not shown). Thus, the P/Q-rich repeat region may aid in the proper folding and/or stability of HopI1 in bacteria.
HopI1 functions inside plant cells and is phosphorylated
To determine where HopI1 might function, we infected the ΔhopI1 strain on A. thaliana transformed with HopI1PmaES4326-HA-Myc-His (JJ30). These plants rescued the virulence defect of the ΔhopI1 strain (Fig. 2A), indicating that HopI1 acts from inside plant cells. HopI1-transformed plants were as resistant as control plants to the attenuated T3SS-deficient strain PmaES4326 hrcC− [11] and the avirulent strain PmaES4326/avrRpt2 [14], respectively (Fig. 2A). Thus, HopI1 acts in the context of a virulent infection.
Fig. 2.
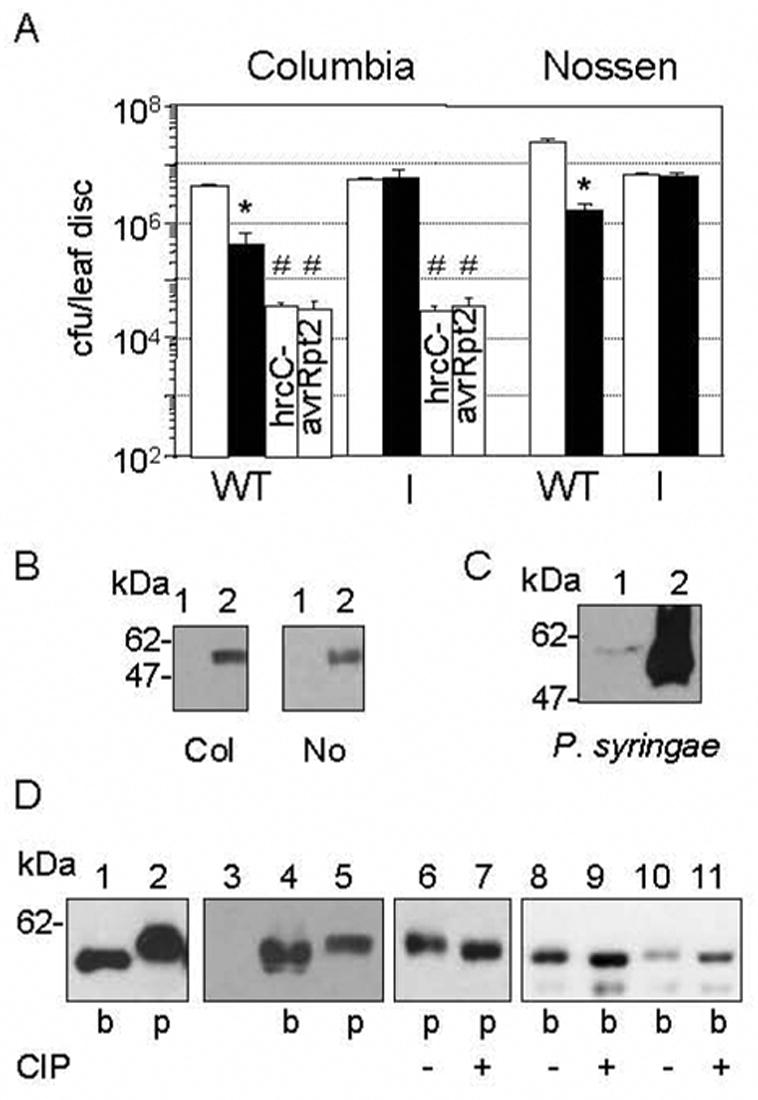
HopI1 functions inside plant cells and is phosphorylated.
(A) Col and No plants constitutively expressing HopI1PmaES4326 (I, JJ30) rescued the virulence defect of the ΔhopI1 PmaES4326 strain (black bar). Col with or without HopI1 had the same susceptibility to hrcC− PmaES4326 or PmaES4326/avrRpt2 (#P>0.4). (*The growth of the ΔhopI1 strain was lower than PmaES4326 strain (white bar) on wild-type plants, P<0.0001). Similar results were obtained with 2 additional, independently derived transgenic HopI1-expressing Col and No lines (not shown).
(B) HopI1 was expressed in transgenic A. thaliana Col (left panel) and No (right panel). Proteins were detected by SDS-PAGE and Western blot analysis with anti-HA antibody. Lines: (1) Wild-type plants, (2) Transgenic plants constitutively expressing HopI1 (JJ30=hopI1PmaES4326-HA-Myc-His).
(C) HopI1-HA-Myc-His expressed from the native promoter and detected with anti-HA antibody exhibited an apparent increase in molecular mass when PmaES4326/JJ19 was grown in A. thaliana (1) compared to hrp-inducing conditions in vitro (2) for 16 h.
(D) HopI1 phosphorylation in bacteria and plants. HopI1PmaES4326-HA-Myc-His expressed in bacteria (b, PmaES4326/JJ19) and transgenic plants (p, Col/JJ30) was detected with anti-HA antibody. Lanes: (1–5) HopI1 expressed in plants had an apparent greater mass than in bacteria. (3) Control A. thaliana Col; (4) The different masses of HopI1 in plants did not result from abundant plant protein displacing the HopI1 species, since when PmaES4326/JJ19 extract was mixed with Col extract (4), HopI1 mass appeared lower than when expressed in plants (5, Col/JJ30). (6–11) HopI1 was phosphorylated in plants and probably in bacteria. Extracts from plants (p) and bacteria (b) were incubated without (−) or with (+) CIP phosphatase. Dephosphorylation of HopI1 results in faster migration in the polyacrylamide gel. (10,11) PmaES4326/JJ19 extract mixed with Col extract. The shift in the HopI1 species in the plant extract was not caused by dephoshorylation of a comigrating abundant plant protein because bacterial-expressed protein incubated with CIP had the same apparent mass both in bacterial protein extract alone (9) or bacterial protein extract mixed with wild-type Col extract (11).
Transgenic plants looked indistinguishable from untransformed plants and accumulated HopI1 protein (Fig. 2B). However, HopI1-HA-Myc-His expressed in A. thaliana migrated more slowly on SDS-polyacrylamide gels than the same protein expressed in P. syringae (Fig. 2B, 2D). Interestingly, the apparent molecular mass of HopI1 also differed before and after infection (Fig. 2C). This shift in mobility was likely due to its phosphorylation in the host cells, since incubation with CIP phosphatase noticeably reduced the apparent mass of HopI1-HA-Myc-His (Fig. 2D). The mass of the bacterial-expressed protein was also reduced by CIP, but the difference was less pronounced than in plants. Therefore, HopI1 is phosphorylated in P. syringae, and it may undergo another modification in plant cells.
The J domain of HopI1 exhibits bona fide J-domain activity
In yeast, the J domain of Hsp40 (known as Ydj1) stimulates the ATPase activity of Hsp70, which is essential for growth at high temperature [15]. Substitution of the Ydj1 J domain with the predicted HopI1 J domain (amino acids 352–424, I-YDJ1, JJ204) rescued the growth of a yeast ydj1 null mutant at 26, 30 and 35°C (Fig. 3). Mutation to QAA of the HPD loop of the HopI1 J domain (I(QAA)-YDJ1, JJ206) resulted in a protein that accumulated in yeast, but did not alter the growth characteristics of the ydj1 mutant (Fig. 3). Thus, HopI1’s J domain is functional and, like other J proteins, requires an intact HPD to interact productively with Hsp70.
Fig. 3.
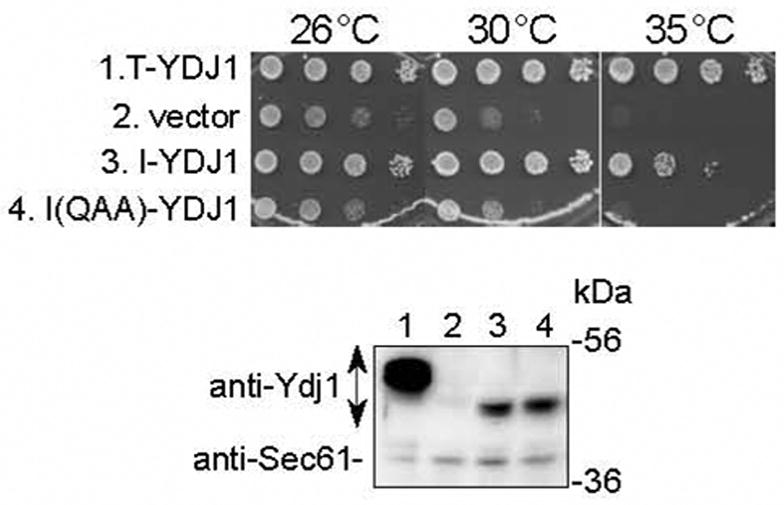
The HopI1 J domain can substitute functionally for the J domain of Ydj1 and rescues the slow growth phenotype of the ydj1 yeast mutant.
Ten-fold serial dilutions of ydj1 yeast containing the indicated constructs were grown for 4 days at 26, 30 and 35°C (upper panel). Ydj1 with its J domain replaced by the HopI1 J domain (I-YDJ1, JJ204) complemented the ydj1 null mutation. The HPD/QAA mutant of the HopI1 J domain (I(QAA)-YDJ1, JJ206) did not complement the yeast mutant phenotype. The SV40 T-antigen::Ydj1 chimera (T-YDJ1) and empty vector TEF414 were used as positive and negative controls, respectively. Ydj1 chimeras were expressed in yeast, as shown by SDS-PAGE and Western blot analysis with anti-Ydj1 antibody (lower panel): (1) T-YDJ1; (2) vector TEF414; (3) I-YDJ1, JJ204; (4) I(QAA)-YDJ1, JJ206. Detection of Sec61 (lower band) served as a loading control.
HopI1 confers heat shock tolerance to A. thaliana
The rescue of the yeast ydj1 mutant with the HopI1::YDJ1 chimera indicates that HopI1 acts like other J-domain proteins to activate Hsp70. To provide further support for this idea, we exposed A. thaliana control and HopI1-expressing plants (JJ30) to 45°C for 35 minutes. HopI1-expressing A. thaliana were more tolerant to a heat shock than control plants (Fig. S2, Table 1), as evidenced by the greater percentage of stems recovering from stress. This suggests that HopI1 engages the plant stress response machinery.
Table 1.
HopI1-expressing A. thaliana is more tolerant to heat shock. Plants were evaluated 2 days after heat shock.
| Plant Genotype | Surviving stems | |
|---|---|---|
| Col (wild type) | 7 % (n=84) | |
| Col expressing HopI1 | JJ30-6 | 72 % (n=79) |
| JJ30-15 | 86 % (n=77) | |
| JJ30-7 | 71 % (n=38) |
HopI1 is localized in the plant cell chloroplast
To gain further insight into how and where HopI1 may function, we used immunoelectron microscopy (IEM) to localize epitope-tagged HopI1. We were unable to detect HopI1 in plant cells during infection, due to the low level and/or the short window of time during which the protein was injected into plant cells. HopI1 was targeted specifically to chloroplasts, as shown with anti-HA or anti-Myc antibodies (Fig. 4A, B) in 5 independent A. thaliana lines expressing HopI1-HA-Myc-His (JJ30, Fig. 4C). Both control and HopI1-expressing plants had background staining in vacuoles. HopI1-HA-Myc-His (JJ30) also localized to chloroplasts in transiently transformed N. benthamiana and N. tabacum leaves (Fig. 4B), indicating that this effector can target mature chloroplasts. HopI1 localization in transgenic A. thaliana (JJ30) showed that the protein was present in the stroma of isolated chloroplasts (Fig. 4D).
Fig. 4.
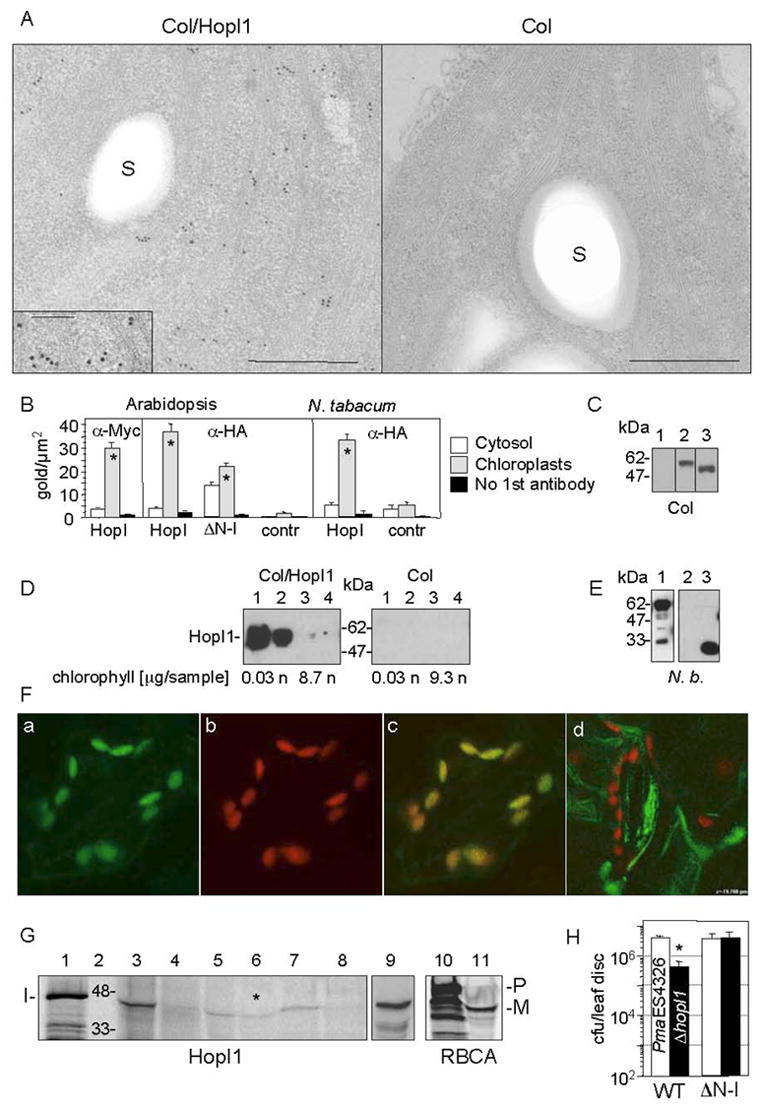
Subcellular chloroplast localization of HopI1 in transgenic HopI1 A. thaliana and transiently-transformed tobacco.
(A) Immunolocalization of constitutively expressed HopI1PmaES4326-HA-Myc-His in chloroplasts of 16 day old transgenic A. thaliana leaves (Col/HopI1=JJ30, left) and wild-type leaves (Col, right). HopI1 localized mainly to chloroplasts in transgenic plants, as visualized with anti-HA antibody. A fragment of chloroplast is shown; insert shows a higher magnification view (bar=500 nm; in the insert 200 nm); S, starch grain.
(B) Statistical analysis of the density of gold-label in HopI1-expressing plants and control plants, detected with anti-HA or anti-Myc antibodies. ΔN-HopI1PmaES4326-HA-Myc-His (JJ51) still localized to chloroplasts in transgenic A. thaliana. HopI1PmaES4326 (JJ30) localized to chloroplasts of transiently-transformed N. tabacum leaves (60 h after Agroinfection). * P<0.008, t-test. HopI, plants expressing HopI1-HA-Myc-His (JJ30). ΔN-I, plants expressing ΔN-HopI1-HA-Myc-His (JJ51). Contr, control plants (wild type (A. thaliana) or transformed with empty vector pCB302-3 (tobacco)).
(C) HopI1 alleles were expressed in transgenic A. thaliana, as shown by SDS-PAGE and Western blot analysis with anti-HA antibody: (1) A. thaliana Col, (2) JJ30=hopI1PmaES4326-HA-Myc-His, (3) JJ51=ΔN29hopI1PmaES4326-HA-Myc-His. Lines 1–3 are from one experiment.
(D) HopI1 localization was confirmed by chloroplast fractionation. HopI1-HA-Myc-His (Col/HopI1=JJ30) was detected in transgenic A. thaliana with anti-Myc antibody. Intact chloroplasts were isolated on a percoll gradient and further partitioned into stromal and membrane fractions. (1 and 2) Stroma (soluble chloroplast proteins) from chloroplasts not treated (1) or treated (2) with thermolysin. (3 and 4) Membranes (thylakoids and envelope) from chloroplasts not treated (3) or treated (4) with thermolysin. The presence of chlorophyll was used as a marker for the chloroplast membranes. HopI1 was present in chloroplast stroma: n, not analyzed; sample, amount of sample loaded on a gel.
(E) HopI1Pph1448A::GFP fusion is expressed in transiently transformed N. benthamiana, as shown with anti-GFP antibodies. Lines: (1) JJ163=hopI1Pph1448A-GFP (upper band), (2) pTA7001, (3) GFP (pAOV-GFP).
(F) Localization of HopI1Pph1448A-GFP in chloroplasts of transiently transformed N. benthamiana. GFP fluorescence (a) colocalized with chloroplast autofluorescence (b). (c) Merged image. (d) GFP was excluded from chloroplasts in N. benthamiana transiently transformed with the GFP control (pAOV-GFP).
(G) In vitro import of HopI1PmaES4326 into chloroplasts. Lanes: (1) Aliquot of HopI1 transcription/translation reaction. (2) Protein marker (kDa). (3 and 4) Supernatant (proteins outside chloroplasts) without (3) or with (4) thermolysin treatment. (5 and 6) Stroma (soluble chloroplast proteins) from chloroplasts not treated (5) or treated (6) with thermolysin after HopI1 import. Asterisk indicates that HopI1 was protected from proteolysis in stroma. (7 and 8) Membranes (thylakoids and envelope) from chloroplasts not treated (7) or treated (8) with thermolysin. (9) Mixture of (1) and (5) in 1:18 ratio. The proteins comigrate, indicating lack of transit peptide removal (10) Aliquot of preRBCA transcription/translation reaction. (11) Stroma from chloroplasts treated with thermolysin after RBCA import. A transit peptide was removed in mature RBCA protected from thermolysin. I, HopI1; P, preRBCA; M, mature RBCA. All lanes except (9) are from one gel. The gel exposure time for audioradiography was shorter for lanes 10 and 11.
(H) Col constitutively expressing HopI1PmaES4326 lacking its N-terminal 29 amino acids (ΔN-I, JJ51) rescued the virulence defect of the ΔhopI1 PmaES4326 strain. (*The growth of the ΔhopI1 strain was reduced compared to the PmaES4326 strain on wild-type plants, P<0.0001).
To confirm these results in living cells, we transiently expressed C-terminal GFP fusions of HopI1 orthologues in N. benthamiana and visualized GFP by confocal microscopy. The highest expression was seen with the HopI1::GFP fusion of the Pph1448A orthologue (pJJ163, Fig. 4E). Fluorescence from the HopI1Pph1448A::GFP chimera colocalized with chloroplasts (Fig. 4F). Leaves expressing GFP alone (pAOV-GFP) showed fluorescence in the cytosol and nucleus (Fig. 4Fd). Fluorescence was absent from leaves transformed with empty vector pTA7001.
To further test if HopI1 localized inside or associated outside chloroplasts, we performed import assays. In vitro transcription and translation reactions of untagged (pJJ90) and HA-Myc-His tagged (pJJ89) hopI1 resulted primarily in the synthesis of 46 kDa and 51 kDa protein products, respectively, confirming the predicted size of HopI1PmaES4326 (Fig. 4G). Both proteins were imported into isolated pea chloroplasts as evidenced by their protection from thermolysin digestion in the stroma fraction (Fig. 4G). Some of the HopI1 protein was associated with chloroplast membranes, but was not protected from proteolysis. This could be because a pool of HopI1 was not yet transported into stroma, or a portion of HopI1 protein might associate with the outer chloroplast membrane. We did not observe removal of a transit peptide from HopI1, as occurs when the precursor of ribulose-1,5-bisphosphate carboxylase/oxygenase activase (preRBCA) is imported (Fig. 4G). This result is consistent with our observation that the apparent size of the HopI1-HA-Myc-His-tagged protein was not reduced in plant extracts compared to bacterial extracts (Fig. 2D).
Unexpectedly, HopI1 lacking its predicted N-terminal chloroplast targeting signal expressed in transgenic A. thaliana (Δ 29HopI1, JJ51, Fig. 4C) rescued the virulence defect of ΔhopI1 strain (Fig. 4H). The Δ 29HopI1 product predominantly localized to the chloroplasts (Fig. 4B), suggesting that a non-canonical import mechanism is utilized by HopI1 to enter chloroplasts.
HopI1 alters thylakoid ultrastructure
Chloroplasts of plants infected with high or low doses of PmaES3426 (or the ΔhopI1 strain complemented with HopI1) had reduced length and increased height of thylakoid grana and thicker individual granal thylakoids compared to chloroplasts in plants infected with the ΔhopI1 strain (Fig. 5B, Table 2C). Importantly, HopI1 and ΔN29HopI1 were sufficient to cause these changes in transgenic A. thaliana (Fig. 5A, Table 2A). Similar alterations were also observed in N. benthamiana (Fig. 5A, Table 2B) and N. tabacum (data not shown) transiently transformed with HopI1. Thus, HopI1 can affect thylakoid ultrastructure after chloroplast biogenesis. The J-domain activity of HopI1 was important for triggering these changes, since HopI1 containing the QAA mutation transiently expressed in N. benthamiana (JJ202) or delivered to A. thaliana leaves from P. syringae during infection (JJ207) did not alter chloroplast morphology (Table 2B, C). No changes were observed after transformation with control vectors. HopI1-expressing A. thaliana mesophyll cells had similar chloroplast numbers per cell, with a similar number of starch grains, but the area of starch per chloroplast section was reduced by half in HopI1-expressing plants (Table 2A).
Fig. 5.
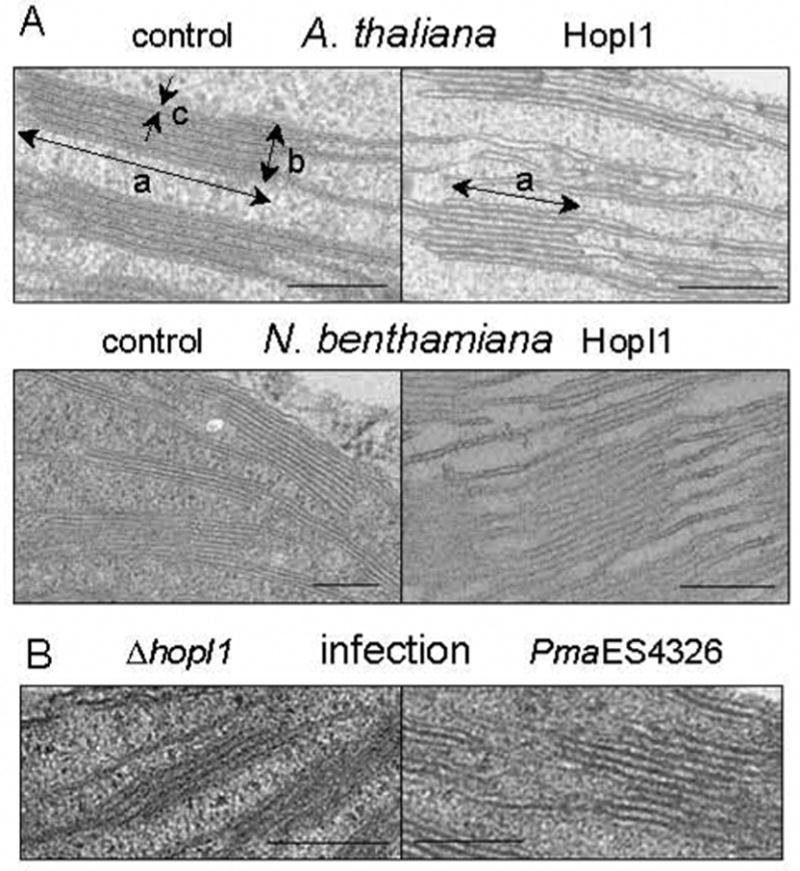
HopI1-mediated changes in thylakoid structure.
(A) Thylakoid grana morphology in wild-type A. thaliana, vector transformed N. benthamiana (control), and HopI1 transgenic leaves (HopI1=JJ30). Note the morphological alterations of grana stack/granal thylakoid length (a), grana height (b) and granal thylakoid thickness (c) in HopI1 transgenic leaves. Bar = 200 nm.
(B) Remodeling of A. thaliana thylakoid structure during infection with wild-type PmaES4326, but not the ΔhopI1 strain, was similar to that observed after HopI1 expression in planta. Observations were made 18 h after infection at OD600 0.3 (similar changes were seen 2 and 3 days after inoculation with OD600 0.001, see Table 2C). Bar=200 nm.
Table 2.
Structural changes in chloroplasts caused by HopI1PmaES4326. Mean value +/− standard error is shown. P, t-test; dpi, days post inoculation; v, vector; chl, chloroplast. See Fig. 4A for the structures measured for a, b, c. Experiments in A and C were done on separate occasions and thus cannot be directly compared.
| A. 16-day old A. thaliana plants | |||||||
|---|---|---|---|---|---|---|---|
| Col | Col/HopI1 | P-value | |||||
| chloroplast/cell cross section | 8.9±0.4 (n=15 cells) | 8.7± 0.5 (n=15 cells) | 0.7 | ||||
| starch grains/chloroplast section | 2.7±0.2 (n=24 chl.) | 2.4± 0.3 (n=30 chl.) | 0.5 | ||||
| area of starch/chloroplast | 47±1% (n=24 chl.) | 23±3% (n=30 chl.) | 0.003 | ||||
| length of grana (a) | 382±12 nm (n=26) | 335±13 nm (n=27) | 0.001 | ||||
| height of grana (b) | 167±9 nm (n=42) | 187±5 nm (n=27) | 0.004 | ||||
| Thickness of thylakoid (c) | 9.3±0.4 nm (n=20) | 11±0.5 nm (n=20) | 0.01 | ||||
|
| |||||||
| B. Transiently transformed N. benthamiana, 2 dpi with A. tumefaciens at OD600=0.6. | |||||||
|
| |||||||
| vector | HopI1 | HopI1(QAA) | n | P-value | P-value | P-value | |
| pCB302-3 | JJ30 | JJ202 | v-JJ30 | v-JJ202 | JJ30–JJ202 | ||
|
| |||||||
| a [nm] | 414±8 | 369±11 | 416±10 | 26 | 0.002 | 0.90 | 0.01 |
| b [nm] | 212±6 | 246±14 | 208±8 | 29 | 0.04 | 0.71 | 0.009 |
| c [nm] | 17.5±0.7 | 21.3±1.3 | 17.0±0.8 | 20 | 0.03 | 0.68 | 0.047 |
|
| |||||||
| C. A. thaliana infected with ΔhopI1 PmaES4326 carrying indicated constructs at OD600=0.001, 3 dpi | |||||||
| ΔhopI1 | +vector | +HopI1 | +HopI1(QAA) | n | P-value | P-value | P-value |
|
| |||||||
| pCKTR | JJ19 | JJ207 | v-JJ19 | v-JJ207 | JJ19–JJ207 | ||
|
| |||||||
| a [nm] | 377± 9 | 317±9 | 373±12 | 23 | 0.0002 | 0.77 | 0.002 |
| b [nm] | 99±5 | 124±6 | 105±5 | 28 | 0.001 | 0.34 | 0.001 |
| c [nm] | 9.1± 0.3 | 10.6±0.5 | 9.4±0.4 | 21 | 0.003 | 0.55 | 0.033 |
Although the appearance of thylakoids in HopI1-expressing A. thaliana and N. tabacum was similar, only the latter plants showed localized cell death with apoptotic-like features (see Supplemental Results and Fig. S3). In both plants, the thylakoids resembled those in plants with alterations in photosynthetic balance caused by changes in the photosystem II-to-I ratio or state transitions [16] or defects in chloroplast lipid biosynthesis [17]. However, photosynthetic yield was the same in wild type and HopI1-expressing A. thaliana, indicating their similar photosynthetic capacity (Fig. S4A). HopI1 did not affect the lipid content or the composition of abundant plant fatty acids (Fig. S4B).
HopI1 suppresses salicylic acid-dependent defenses
Since HopI1 localizes to chloroplasts, it could affect the production of a chloroplast-produced defense signal such as SA. The virulence defect of the ΔhopI1 strain was suppressed in nahG transgenic A. thaliana that is impaired in SA accumulation due to the SA catabolic activity of NahG (Fig. 6A). The growth defect of the ΔhopI1 strain was also largely suppressed in sid2 mutant plants that exhibit impaired SA synthesis (Fig. 6A).
Fig. 6.
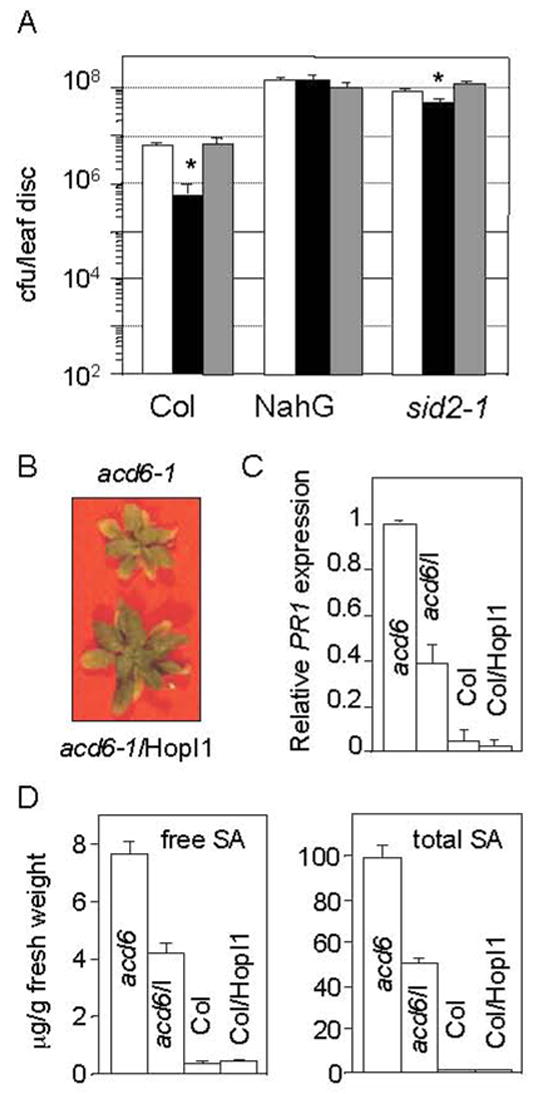
HopI1 interference with salicylic acid-dependent defenses.
(A) Growth of the ΔhopI1 strain was not attenuated in SA-deficient Col (NahG). The growth defect of the ΔhopI1 strain was largely suppressed in the SA-deficient Col sid2-1 mutant. The ΔhopI1 strain grows slightly better with the hopI1-complementing clone in sid2-1 (*P<0.05): white bar, PmaES4326 with empty vector pCKTR; black bar, the ΔhopI1 strain with the empty vector; gray bar, ΔhopI1 with the hopI1 gene (JJ19). Data represent the means of 8 samples with standard errors.
(B) acd6-1 plants expressing HopI1 were larger than the acd6-1 plants alone. Four week-old plants were photographed.
(C) PR1 mRNA accumulation was lower in acd6-1 plants expressing HopI1 than in acd6-1 plants alone, as determined by quantitative real time RT-PCR. HopI1 did not change PR1 expression in Col. Values are relative to the PR1 level in acd6-1 normalized to EF1α. The mean value of 3 independent experiments, each containing triplicates of 2–4 lines/genotype is shown with standard error.
(D) The free (left panel) and total (right panel) SA level is lower in 23 day old acd6-1 plants expressing HopI1 than in acd6-1 plants alone. HopI1 did not change the SA level in Col. Error bars show standard error (n=3). This experiment was repeated with 5 week-old plants with similar results.
To test whether HopI1 was sufficient to suppress SA accumulation, we expressed HopI1 (JJ30) in the constitutive gain-of-function defense mutant acd6-1 (accelerated cell death 6-1) that has high SA levels. acd6-1 has reduced stature, constitutive defenses and spontaneous cell death patches; these phenotypes require SA accumulation or signaling [18–20]. Homozygous acd6-1 plants expressing HopI1 were almost twice the size and showed less cell death than acd6-1 plants (Fig. 6B, Table 3). HopI1 expression resulted in a 60% decrease in the level of the SA-inducible PR1 (pathogenesis-related 1) gene transcript and in ~50% lower free and total SA levels (Fig. 6C, D). The change in size and SA levels were comparable to what has been observed in acd6-1 in the presence of the ald1 and pad4 defense signaling mutations [20]. These observations are consistent with HopI1 being sufficient to suppress SA accumulation and SA-dependent defenses.
Table 3.
Effect of HopI1 on acd6-1 plant size. Diameter of 3-week old plants and weight of 5-week old plants is shown. acd6-1 plants expressing HopI1 are larger than acd6-1 plants alone. Col expressing HopI1 and wild-type Col are the same size. P, t-test; ±, standard error.
| acd6-1 | acd6-1/HopI1 | P value | Col | Col/HopI1 | P value | |
|---|---|---|---|---|---|---|
| Rosette | 1.20±0.06 | 2.13±0.12 | 4.03±0.09 | 3.93±0.08 | 0.9 | |
| <0.0001 | ||||||
| [cm] | n=14 | n=14 | n=15 | n=15 | ||
| Weight | 6.67±0.87 | 19.57±2.41 | ||||
| <0.0001 | ||||||
| [mg] | n=15 | n=7 |
Discussion
Many P. syringae type III effectors have N-terminal regions that resemble chloroplast targeting signals [11]. However, HopI1 is the first effector whose main subcellular location (and probably its main site of action) is the chloroplast. HopI1 is present in all examined P. syringae strains. Interestingly, several divergent hopI1 alleles can function equivalently to promote P. syringae growth and disease. We found no evidence that any of the HopI1 orthologues could restrict the host range. However, HopI1 induced cell death on N. tabacum, possibly as part of the disease process. The result of HopI1’s presence in plants is a suppression of chloroplast-mediated defenses (SA) and a remodeling of the thylakoids. HopI1’s J-domain function is essential for virulence during infection, for thylakoid remodeling and for stimulating Hsp70’s ATPase activity in yeast. In plants, Hsp70-family proteins are found in chloroplasts and in other organelles and in the cytoplasm [21]. Based on these data, we suggest that HopI1’s J domain interacts with the chloroplast form of Hsp70 and that this interaction is critical for HopI1’s effects in suppressing the host defense response and altering the thylakoid ultrastructure.
HopI1 enters the chloroplast by a non-canonical mechanism and does not appear to be processed. Of note, some endogenous plant proteins also transit into the chloroplast without processing [22]. In any event, the targeting of chloroplast functions may be a common virulence mechanism for P. syringae. For example, some strains produce tagetotoxin that dramatically alters chloroplast morphology, resulting in large vacuole-like structures within the chloroplasts [23]. Another P. syringae phytotoxin, the polyketide coronatine, localizes to chloroplasts [24] and affects stomatal closure, providing a route for bacteria to gain access to underlying mesophyll cells [25].
HopI1 harbors a P/Q-rich repeat region that contributes to its stability in bacteria and may be important for virulence. Since many protein interaction modules are often proline-rich [26], we speculate that this region may be involved in protein-protein interaction. Interestingly, HopI1 has multiple predicted phosphorylation sites in the N terminus, in the P/Q-rich repeat region and in the J domain (ScanProsite, http://us.expasy.org). Indeed, HopI1 is phosphorylated in both host cells and bacteria. Further investigation will be important to determine whether this modification is important for HopI1 function, as has been shown for other effectors from plant and human pathogens [27].
How might HopI1’s interaction with Hsp70 lead to its virulence effect? Hsp70 interactions with J proteins are known to catalyze many cellular events, including the folding of client proteins [12] (which may be recruited by the J-domain protein) or the targeting of client proteins for degradation [28]. HopI1 might stimulate the folding or assembly of a defense-suppressing protein (a negative regulator), compete with a plant J protein that activates Hsp70’s folding of a defense protein(s) or promote the degradation of a defense component. Unlike the probable virulence role of chloroplast Hsp70, cytosolic Hsp70 was found to be essential for the hypersensitive defense response and non-host resistance to P. chicorii [29].
How and why does HopI1 alter thylakoid structure? One possibility is that HopI1 interactions with Hsp70 divert Hsp70 from its functions in providing the appropriate stoichiometry of thylakoid components. In this scenario, the alterations may not be essential for the virulence effect of HopI1, but rather could be a collateral result of Hsp70 engagement. In Chlamydomonas, a chloroplast J protein together with its Hsp70 partner are important for thylakoid membrane biogenesis and integrity [30]. Alternatively, the change in thylakoid ultrastructure could be important for the virulence effect of HopI1. For example, such alterations could affect the ability of chloroplasts to produce or transport SA or they could promote the generation of a SA-antagonistic signal, such as jasmonic acid, whose precursors are chloroplast-synthesized [31]. The future isolation of HopI1-binding proteins that are Hsp70 clients should shed light on how HopI1 modulates plant defenses and the possible involvement of thylakoid alterations in this process.
Experimental Procedures
Bacteria and plant growth, infection and heat stress conditions
E. coli strains DH5α and DB3.1 (Invitrogen, Carlsbad, CA), and P. syringae pv. maculicola strain PmaES4326 and its derivatives were grown as described [32]. Agrobacterium tumefaciens C58C1/pCH32 (from R. W. Michelmore, University of California, Davis, CA) and GV3101/pMP90 were grown as described [3].
Arabidopsis thaliana plants were grown in a 16 h light/8 h dark cycle at 20°C as described [33]. N. tabacum “Burley” and N. benthamiana were grown in a 16 h light/8 h dark cycle at 24°C. For chloroplast isolation, pea (Pisum sativum) and A. thaliana were grown in a 12 h light/12 h dark cycle at 24°C and 20°C, respectively.
All infection experiments were repeated at least twice with similar results. 19–21-day-old A. thaliana and 4-week-old tobacco plants were inoculated at OD600 of 0.0001 or 0.0003 as described [34]. Eight independent samples were averaged for each genotype for the P. syringae growth experiments. Unless stated otherwise, samples were taken 3 days after infection.
For heat treatments, 5- or 6-week old A. thaliana plants were incubated for 35 min at 45°C. Pictures were taken before and 0 min, 20 min, 1h, 12h, 2 days, 5 days after treatment. This experiment was repeated 4 times using 3 independent transgenic lines constitutively expressing HopI1 (JJ30).
Complementation of yeast ydj1 mutant
The temperature sensitive ydj1 null yeast strain ACY95b [15] was complemented with wild type (pJJ204) or the HPD/QAA mutant (pJJ206) HopI1 J domain::Ydj1 chimeras expressed from the Translation Elongation Factor 1α promoter (see supplemental experimental procedures for details). Yeast transformations, growth, serial dilutions, and protein analysis were performed as described [35]. Vector pTEF414 and pT-YDJ1 were used as negative and positive controls, respectively. The experiment was repeated twice, using 2 independent colonies/construct, with the same results.
Plant gene expression analysis
Quantitative PCR was done by real time RT-PCR to measure defense-related gene transcript levels (see supplemental experimental procedures for details).
SA quantitation
Free and total SA (the sum of free and glucosyl-SA) was extracted and quantified as described previously [36]. The experiment was repeated twice, with triplicates, using 2 lines/genotype.
Protein analysis
Protein extracts from P. syringae and plants and Western blot analysis with GFP antibody were performed as described [3, 32]. HA- and HA-Myc-His-tagged effectors were detected using monoclonal HA and Myc antibodies (HA.11 and 9E10, respectively, Covance, Berkeley, CA) at a 1:1,200 dilution. Secondary horseradish peroxidase-conjugated antibodies (Pierce, Rockford, IL) were used at a 1:20,000 dilution. To determine phosphorylation, 10 μl of protein extract (without SDS) was incubated at 37°C for 1 h with 10 U of CIP phosphatase (NE Biolabs).
Statistical methods
All statistical analyses were done using the Statview statistical package 5.0.1 for MacIntosh (SAS Institute, Cary, NC, USA).
Ultrastructural analysis and immunolocalization
Leaf segments were prepared as described [37]. For each genotype or treatment, at least three replicate leaves were fixed and three sections from each leaf were analyzed. Figures show representative images. For statistical analysis at least 25 cells were used per genotype/line. For morphometric analysis of chloroplast ultrastructural changes, a random sample of micrographs (at least 20) was analyzed for each treatment. Only plant cells adjacent to bacteria were analyzed. The entire structural analysis was done twice with plants grown at different times.
Immunolocalization was done as described [38]. HopI1 tagged with HA- Myc-His epitopes was detected using monoclonal anti-HA and anti-Myc antibodies at 1:50 dilution for 2 h at room temperature and secondary 10 nm colloidal gold conjugate goat-anti-mouse IgG antibodies (Electron Microscopy Sciences, Ft. Washington, PA) at a 1:20 dilution. After washing in TBS-BSA and deionized water, specimens were stained with uranyl acetate and lead. Three blocks of wild-type, five HopI1-expressing A. thaliana lines (JJ30) and at least three transiently transformed tobacco leaves were sectioned. A scanning transmission electron microscope (Tecnai F30; FEI Company) was used at an accelerating voltage of 300 kV.
In vivo GFP localization
N. benthamiana leaves were collected 2 days after infiltration with A. tumefaciens GV3101(pMP90) containing hopI1Pph1448A-GFP fusion (pJJ163), GFP control (pAOV-GFP) or empty vector (pTA7001). GFP and chloroplast autofluorescence in live mesophyll and guard cells were observed and by a Leica TCS SP2 AOBS laser scanning confocal microscope (Leica, Mannheim, Germany) with a 63x (N.A.1.4) oil objective.
Chloroplast isolation and protein import assay
A. thaliana chloroplasts were isolated from 14-day-old seedlings grown on soil covered with cheesecloth on a two-step percoll gradient, treated with thermolysin and further fractionated to stroma and membranes as described [39]. Chloroplasts isolated from 2 independent transgenic lines expressing epitope-tagged HopI1 and control Col were used for Western blot analysis. Chlorophyll was measured by spectrophotometric analysis as described [40].
Intact chloroplasts were isolated from 8-day-old pea leaves as described [40]. [35S]Met-labeled precursor proteins were produced using the rabbit reticulocyte TNT in vitro transcription/translation kit (Promega, Madison, WI). A cDNA construct of preRBCA from Spinacia oleracea was used as an import control. hopI1 cDNA with or without HA-Myc-His tag was cloned into pBluescript SK (Stratagene, La Jolla, CA), resulting in plasmids pJJ89 and pJJ90, respectively. In vitro import assays were performed as described [40]. After incubation with radiolabeled proteins, half of the mix was treated with thermolysin, the chloroplasts were hypotonically lysed and were separated into soluble and membrane fractions for SDS-PAGE analysis. This experiment was repeated twice.
Data deposition
The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF458047, DQ401061-DQ401068).
Supplementary Material
Acknowledgments
We thank Changcheng Xu and Christoph Benning for lipid analysis, Stefan Richter and Gayle Lamppa for help with the chloroplast import assay, Chengbin Xiang, David Oliver and Richard Michelmore for plasmids, Laurens Mets for useful discussions and students Daniel Blumenthal and Yvonne Chan for help with the HopI1 transient expression experiments. This work was supported by subcontracts and grants to JTG from the NSF Plant Genome Program 00RA6325-DBI and USDA-NRI 2005-35319-16136, respectively, and by NIH grant GM75061 to JLB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Buttner D, Bonas U. Common infection strategies of plant and animal pathogenic bacteria. Curr Opin Plant Biol. 2003;6:312–319. doi: 10.1016/s1369-5266(03)00064-5. [DOI] [PubMed] [Google Scholar]
- 2.Mudgett MB. New insights to the function of phytopathogenic bacterial type III effectors in plants. Ann Rev Plant Biol. 2005;56:509–531. doi: 10.1146/annurev.arplant.56.032604.144218. [DOI] [PubMed] [Google Scholar]
- 3.Vinatzer BA, Teitzel GM, Lee MW, Jelenska J, Hotton S, Fairfax K, Jenrette J, Greenberg JT. The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol Microbiol. 2006;62:26–44. doi: 10.1111/j.1365-2958.2006.05350.x. [DOI] [PubMed] [Google Scholar]
- 4.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 5.Nomura K, Melotto M, He SY. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol. 2005;8:361–368. doi: 10.1016/j.pbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Lindeberg M, Stavrinides J, Chang JH, Alfano JR, Collmer A, Dangl JL, Greenberg JT, Mansfield JW, Guttman DS. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol Plant-Microbe Interact. 2005;18:275–282. doi: 10.1094/MPMI-18-0275. [DOI] [PubMed] [Google Scholar]
- 7.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 8.de Torres M, Mansfield JW, Grabov N, Brown IR, Ammouneh H, Tsiamis G, Forsyth A, Robatzek S, Grant M, Boch J. Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J. 2006;47:368–382. doi: 10.1111/j.1365-313X.2006.02798.x. [DOI] [PubMed] [Google Scholar]
- 9.DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA. 2004;101:9927–9932. doi: 10.1073/pnas.0401601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauck P, Thilmony R, He SY. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA. 2003;100:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guttman DS, Vinatzer BA, Sarkar SF, Ranall MV, Kettler G, Greenberg JT. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science. 2002;295:1722–1726. doi: 10.1126/science.295.5560.1722. [DOI] [PubMed] [Google Scholar]
- 12.Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan CS, Pipas JM. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol Mol Biol Rev. 2002;66:179–202. doi: 10.1128/MMBR.66.2.179-202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guttman DS, Greenberg JT. Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol Plant-Microbe Interact. 2001;14:145–155. doi: 10.1094/MPMI.2001.14.2.145. [DOI] [PubMed] [Google Scholar]
- 15.Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- 16.Kyle DJ, Staehelin LA, Arntzen CJ. Lateral mobility of the light-harvesting complex in chloroplast membranes controls excitation energy distribution in higher plants. Arch Biochem Biophys. 1983;222:527–541. doi: 10.1016/0003-9861(83)90551-9. [DOI] [PubMed] [Google Scholar]
- 17.Dormann P, Hoffman-Benning S, Balbo I, Benning C. Isolation and characterization of an Arabidopsis thaliana mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell. 1995;7:1801–1810. doi: 10.1105/tpc.7.11.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT. The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell. 1999;11:1695–1708. doi: 10.1105/tpc.11.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu H, Rate DN, Song JT, Greenberg JT. ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell. 2003;15:2408–2420. doi: 10.1105/tpc.015412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song JT, Lu H, McDowell JM, Greenberg JT. A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 2004;40:200–212. doi: 10.1111/j.1365-313X.2004.02200.x. [DOI] [PubMed] [Google Scholar]
- 21.Sung DY, Vierling E, Guy CL. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001;126:789–800. doi: 10.1104/pp.126.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soll J, Schleiff E. Protein import into chloroplasts. Nat Rev Mol Cell Biol. 2004;5:198–208. doi: 10.1038/nrm1333. [DOI] [PubMed] [Google Scholar]
- 23.Freeman TP, Duysen ME, Gulya TJ. Ultrastructural changes in sunflower chloroplasts following inoculation with Pseudomonas syringae pv. tagetis. Amer J Bot. 1985;72:707–714. [Google Scholar]
- 24.Zhao YF, Jones WT, Sutherland P, Palmer DA, Mitchell RE, Reynolds PHS, Damicone JP, Bender CL. Detection of the phytotoxin coronatine by ELISA and localization in infected plant tissue. Physiol Mol Plant Pathol. 2001;58:247–258. [Google Scholar]
- 25.Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 26.Kay BK, Williamson MP, Sudol P. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. Faseb J. 2000;14:231–241. [PubMed] [Google Scholar]
- 27.Anderson JC, Pascuzzi PE, Xiao F, Sessa G, Martin GB. Host-mediated phosphorylation of type III effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell. 2006;18:502–514. doi: 10.1105/tpc.105.036590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hohfeld J, Cyr DM, Patterson C. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001;2:885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanzaki H, Saitoh H, Ito A, Fujisawa S, Kamoun S, Katou S, Yoshioka H, Terauchi R. Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol Plant Pathol. 2003;4:383–391. doi: 10.1046/j.1364-3703.2003.00186.x. [DOI] [PubMed] [Google Scholar]
- 30.Liu C, Willmund F, Whitelegge JP, Hawat S, Knapp B, Lodha M, Schroda M. J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol Biol Cell. 2005;16:1165–1177. doi: 10.1091/mbc.E04-08-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 32.Vinatzer BA, Jelenska J, Greenberg JT. Bioinformatics correctly identifies many type III secretion substrates in the plant pathogen Pseudomonas syringae and he biocontrol isolate P. fluorescens SBW25. Mol Plant-Microbe Interact. 2005;18:877–888. doi: 10.1094/MPMI-18-0877. [DOI] [PubMed] [Google Scholar]
- 33.Greenberg JT. Positive and negative regulation of salicylic acid-dependent cell death and pathogen resistance in Arabidopsis lsd6 and ssi1 mutants. Mol Plant-Microbe Interact. 2000;13:877–881. doi: 10.1094/MPMI.2000.13.8.877. [DOI] [PubMed] [Google Scholar]
- 34.Greenberg JT, Silverman FP, Liang H. Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics. 2000;156:341–350. doi: 10.1093/genetics/156.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright CM, Fewell SW, Sullivan ML, JM P, Watkins SC, Brodsky JL. The Hsp40 molecular chaperone, Ydj1p, along with the protein kinase C pathway impact cell wall integrity in the yeast Saccharomyces cerevisiae. Genetics. 2007 doi: 10.1534/genetics.106.066274. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seskar M, Shulaev V, Raskin I. Endogenous methyl salicylate in pathogen-inoculated tobacco plants. Plant Physiol. 1998;116:387–392. [Google Scholar]
- 37.Yao N, Tada Y, Park P, Nakayashiki H, Tosa Y, Mayama S. Novel evidence for apoptotic cell response and differential signals in chromatin condensation and DNA cleavage in victorin-treated oats. Plant J. 2001;28:13–26. doi: 10.1046/j.1365-313x.2001.01109.x. [DOI] [PubMed] [Google Scholar]
- 38.Yao N, Greenberg JT. Arabidopsis ACCELERATED CELL DEATH2 Modulates Programmed Cell Death. Plant Cell. 2006;18:397–411. doi: 10.1105/tpc.105.036251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aronsson H, Jarvis P. A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Letters. 2002;529:215–220. doi: 10.1016/s0014-5793(02)03342-2. [DOI] [PubMed] [Google Scholar]
- 40.Lamppa GK. In vitro import of proteins into chloroplasts. In: Maliga P, Klessig DF, Cashmore AR, Gruissem W, Varner JE, editors. Methods in Plant Molecular Biology. New York: Cold Spring Harbor Laboratory Press; 1995. pp. 141–171. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


