Abstract
The HIV-1 envelope glycoprotein is expressed on the viral membrane as a trimeric complex, formed by three gp120 surface glycoproteins non-covalently associated with three membrane-anchored gp41 subunits. The labile nature of the association between gp120 and gp41 hinders the expression of soluble, fully cleaved, trimeric gp140 proteins for structural and immunization studies. Disruption of the primary cleavage site within gp160 allows the production of stable gp140 trimers, but cleavage-defective trimers are antigenically dissimilar from their cleaved counterparts. Soluble, stabilized, proteolytically cleaved, trimeric gp41 proteins can be generated by engineering an intermolecular disulphide bond between gp120 and gp41 (SOS), combined with a single residue change, I559P, within gp41 (SOSIP). We have found that SOSIP gp140 proteins based on the subtype A HIV-1 strain KNH1144 form particularly homogenous trimers compared to a prototypic strain (JR-FL, subtype B). We now show that the determinants of this enhanced stability are located in the N-terminal region of KNH11144 gp41 and that, when substituted into heterologous Env sequences (e.g., JR-FL and Ba-L) they have a similarly beneficial effect on trimer stability. The stabilized trimers retain the epitopes for several neutralizing antibodies and related agents (CD4-IgG2, b12, 2G12, 2F5 and 4E10) and the CD4-IgG2 molecule, suggesting that the overall antigenic structure of the gp140 protein has not been adversely impaired by the trimer-stabilizing substitutions. The ability to increase the stability of gp140 trimers might be useful for neutralizing antibody-based vaccine strategies based on the use of this type of immunogen.
Keywords: HIV-1, env stabilization, trimers, gp41 N-terminus, cleavage
INTRODUCTION
The ability of human immunodeficiency virus type 1 (HIV-1) to enter its target cell and establish an infection is dependent on interactions between functional envelope glycoprotein (Env) complexes on the virus and receptors on the host cell. The HIV-1 Env complex is initially synthesized as the polyprotein precursor gp160, which undergoes oligomerization, disulfide bond formation and extensive glycosylation in the endoplasmic reticulum (Earl, Moss, and Doms, 1991), and is then proteolytically cleaved into the surface (gp120) and transmembrane (gp41) subunits by furin-like endo-proteases in the Golgi network (Fields, 1996; Hunter and Swanstrom, 1990). The resulting Env complex is a trimer, with three gp120 proteins associated non-covalently with three gp41 subunits. During the entry process, gp120 interacts with the CD4 receptor, which triggers conformational changes that facilitate gp120 binding to a coreceptor, CCR5 or CXCR4 (Berger, Murphy, and Farber, 1999; Rizzuto et al., 1998). These interactions promote extensive conformational changes in the gp41 subunit that drive the insertion of the hydrophobic gp41 N-terminal region (fusion peptide) into the host cell membrane. Subsequently, formation of the six-helix bundle configuration of the three gp41 ectodomains forces the juxtaposition of the viral and cell membranes, promoting their fusion (Doms and Moore, 2000; Jones, Korte, and Blumenthal, 1998; Melikyan et al., 2000; Moore and Doms, 2003; Sattentau and Moore, 1991; Sullivan et al., 1998; Wu et al., 1996; Zhang et al., 1999).
The trimeric nature of the Env complex has been confirmed by various lines of evidence (Blacklow, Lu, and Kim, 1995; Center et al., 2002; Center et al., 2001; Chan et al., 1997; Chan and Kim, 1998; Lu, Blacklow, and Kim, 1995; Zhu et al., 2003), most recently by cryo-electron microscopy (Zanetti et al., 2006; Zhu et al., 2006). The trimer is held together by labile, non-covalent inter-subunit interactions. The weak interactions between gp120 and gp41, and between individual gp41 subunits, are probably necessary for the conformational changes that drive membrane fusion, but they complicate the generation of soluble forms of Env trimers that are suitable for vaccine research and structural studies. To obtain soluble Env trimers, the transmembrane (TM) region and the cytoplasmic tail (CT) are routinely deleted from gp41 to create gp140 proteins that contain gp120 and the gp41 ectodomain (gp41ECTO). There are different ways to stabilize the non-covalent inter-subunit interactions: Some groups mutate the cleavage site within gp140 to make uncleaved oligomers (Chakrabarti et al., 2002; Srivastava et al., 2002; Yang et al., 2000; Yang et al., 2002; Zhang et al., 2001); our approach has been to engineer an inter-subunit disulphide bond (SOS) (Binley et al., 2000) between gp120 and gp41, and an isoleucine to proline substitution at position 559 (I559P) in the N-terminal heptad region of gp41 ectodomain (SOSIP) (Sanders et al., 2002) to promote gp41-gp41 association. We recently characterized a soluble, cleaved SOSIP gp140 protein from the subtype A virus KNH1144 (Beddows et al., 2006). Compared to other SOSIP gp140s we have studied, particularly from JR-FL, the KNH1144 protein formed more homogenous trimers; the fewer monomers, dimers, tetramers and aggregates present suggest the trimer form was unusually stable (Beddows et al., 2006). Here, we have studied the molecular determinants of enhanced trimer stability. We find that they lie within the N-terminal region of gp41ECTO, an area with a well-documented role in gp41-gp41 interactions (Center, Kemp, and Poumbourios, 1997; Poumbourios et al., 1997; Shugars et al., 1996). Specifically, we show that five amino acid changes based on the KNH1144 sequence have a trimer-stabilizing effect on heterologous gp140 proteins. The introduction of these changes did not impair the exposure of various neutralizing antibody epitopes on the resulting gp140 proteins, suggesting the overall antigenic structure of the trimer is not adversely affected.
RESULTS
Specific amino acids in the N-terminal region of gp41ECTO contribute to enhanced oligomerization of cleaved gp140 from KNH1144
We recently reported that cleaved, SOSIP gp140 proteins from the subtype A strain KNH1144 form unusually stable and homogenous trimers compared to JR-FL SOSIP gp140, which is expressed as both dimers and trimers (Sanders et al., 2002). The SOS gp140 protein from KNH1144 is also more stable than the corresponding JR-FL construct, the latter being expressed as a mixture of trimers, dimers and, predominantly, monomers (Beddows et al., 2006; Binley et al., 2000). On purification, JR-FL SOS gp140 yields mostly monomeric gp140 proteins as a result of the instability of the gp41-gp41 interactions (Binley et al., 2000). We therefore sought to determine why the KNH1144 gp140 was better able to form stable trimers.
The N-terminal region of gp41, particularly around the Heptad Repeat 1 (HR1) region, plays a role in oligomerization of gp140 proteins (Center, Kemp, and Poumbourios, 1997; Center et al., 2004; Poumbourios et al., 1997). When the N-terminal regions of gp41 from KNH144 and JR-FL were aligned, five amino acids were seen to differ, as follows (KNH1144 vs. JR-FL): I535M, Q543L, S553N, K567Q and R588G (Fig. 1). To determine, which, if any, of these five differences contributed to the enhanced stability of KNH1144 trimers, we substituted each residue in KNH11144 SOSIP gp140 with the corresponding one from JR-FL, then expressed the mutant Env proteins and studied them on BN-PAGE gels. The wild-type forms of KNH1144 SOS and SOSIP gp140 proteins were also analyzed, to allow a comparison with the trimer-stabilizing effect of the I559P substitution in the SOSIP version (Fig. 2A). In general, the amino acid substitutions described below had similar effects whether they were made in the SOS or the SOSIP gp140 background, so only a subset of the results is depicted.
Fig. 1.

(A) Schematic view of gp41 region showing the location of the fusion peptide (FP), heptad repeat regions 1 and 2 (HR1 and HR2), the transmembrane region (TM) and the cytoplasmic tail (CT). The intramolecular disulfide bond is also shown. (B) Alignment of the N-terminus regions of KNH1144, JR-FL and Ba-L gp41, highlighting the 5 amino acids (bold and shaded) in and near the HR1 region (underlined) that differ in JR-FL and B-aL when compared to KNH1144.
Fig. 2.

Trimer formation by cleaved, wild-type and mutant KNH1144 gp140 proteins. (A) SOS and SOSIP versions of KNH1144 gp140 proteins. (B) KNH1144 SOSIP gp140 mutants containing the indicated single residue substitutions in the gp41 N-terminal region, compared with the wild-type KNH1144 SOSIP gp140. (C) KNH1144 SOSIP and SOS mutant gp140s, as indicated. Each panel shows a BN-PAGE analysis, followed by western blotting using MAb CA13.
The S553N and R588G changes had little or no effect on trimer formation by KNH1144 SOSIP gp140 (Fig. 2B, lanes 3 and 5), whereas the I535M substitution enhanced trimer formation (Fig. 2B, lane 1), an observation we confirmed in a larger-scale expression and purification study (data not shown). In contrast, substitutions of glutamine and lysine at positions 543 and 567 (Q543L and K567Q) destabilized the KNH1144 SOSIP gp140 trimers (Fig. 2B, lanes 2 and 4). When all five amino acids were substituted in the KNH1144 SOS and SOSIP gp140 templates, the destabilizing effect on trimer formation was pronounced. The extent of the increase in monomer formation, compared to wild-type, was estimated to be ~45% and ~60% for the KNH1144 SOSIP and SOS gp140 mutants, respectively (Fig. 2C, lane 1, SOSIP; lane 2, SOS; compare with Fig. 2A). Hence, we conclude that the five amino acid differences between the N-terminal regions of KNH1144 and JR-FL gp41 do influence the stability of cleaved gp140 trimers.
Substitution of five amino acids from the N-terminal region of KNH1144 gp41ECTO promotes JR-FL gp140 trimer formation
We used both the SOS and SOSIP versions of JR-FL gp140 as templates on which to explore the effects of the five amino acid differences in the KNH114 gp41 ectodomain, to ensure we took into account the additional, possibly complicating, influence of the I559P change. In the first construct, a chimera, the JR-FL gp120 subunit was combined with the KNH1144 gp41 ectodomain (JR-FLgp120-1144gp41 ECTO); the second construct was a mutant JR-FL SOS gp140 in which the five varying amino acids (positions 535, 543, 553, 567 and 588) were substituted by the corresponding residues from KNH1144 (JR-FL gp41 NT 1–5); the third was another chimera in which the C-terminal region of gp41ECTO from JR-FL was replaced by the corresponding segment of KNH1144 gp41 (JR-FL-1144 gp41 CT) (Fig. 3A).
Fig. 3.

Trimer formation by cleaved, wild-type and mutant JR-FL SOS gp140 proteins. (A) Design of various chimeric and mutant JR-FL gp140s. The intermolecular disulfide bond (SOS) and the Ile to Pro substitution at position 559 (I559P; SOSIP) are shown. (B) The indicated wild-type and mutant/chimeric gp140 proteins were analyzed using BN-PAGE and western blotting with MAb CA13. The designation NT 1–5 refers to substitution of the 5 amino acids M535I, L543Q, N553S, Q567K and G588R, in the gp41 N-terminus region. (C) The JR-FLgp120-1144gp41(ECTO) SOS gp140 chimera and the JR-FL gp41 NT 1–5 SOS gp140 mutant were analyzed by SDS-PAGE and western blotting, followed by detection with MAb B13. The − and + symbols indicate the absence and presence of DTT.
The various chimeric and mutant envelope glycoproteins were expressed in HEK293T cells and analyzed on BN-PAGE gels (Fig. 3B). JR-FL SOS gp140 was predominantly monomeric whereas the SOSIP gp140 formed dimers and trimers (Fig. 3B, lanes 1 and 2). The insertion of gp41ECTO from KNH1144 into the JR-FL SOS gp140 template stabilized the trimeric form, with a reduction in the amount of monomers present (Fig. 3B, lane 3). The same change, but made in the JR-FL SOSIP gp140 context, had a lesser effect (Fig. 3B, compare lanes 2 and 4). Swapping the C-terminal region of gp41ECTO in either SOS or SOSIP JR-FL gp140 had no visible effect on oligomerization (Fig. 3B, compare lane 1 to lane 5 and lane 2 to lane 6). In contrast, oligomer formation by JR-FL SOS or SOSIP gp140 was increased by the substitution of the five varying amino acids in gp41ECTO with the corresponding residues from KNH1144 (M535I, L543Q, N553S, Q567K and G588R) (Fig. 3B, compare lane 7 to lane 1 and lane 8 to lane 2). Taken together, these results confirm that the trimer-promoting determinants of KNH1144 are located in the N-terminal region of gp41. Moreover, the greater stability of KNH1144 gp140 trimers can be conferred upon a heterologous gp140, JR-FL, by altering the specific residues that differ between the two proteins. Further studies showed that all five changes were necessary for creating an optimally stable and homogenous JR-FL gp140 trimer; various combinations of the five changes had negligible or partial effects (data not shown).
To ascertain whether the chimeric/mutant proteins were fully cleaved, we analyzed the JR-FLgp120-KNH1144gp41(ECTO) SOS gp140 chimera and the JR-FL gp41 NT 1–5 SOS gp140 mutant, using SDS-PAGE (Fig. 3C). Under denaturing conditions, both gp140 proteins were resolved as monomers (lanes 1 and 3). When DTT was added to reduce the SOS disulphide bond, release of the gp120 subunit was complete (lanes 2 and 4), along with gp41ECTO (which is not detectable by the b13 MAb used for blotting). Hence aberrantly linked, uncleaved products do not contribute to the enhanced oligomerization of JR-FL gp140 conferred by substitution of the residues from the KNH1144 gp41 N-terminal region. The stabilized, mutant proteins are fully cleaved.
To further assess the formation and stability of JR-FL gp41 NT 1–5 SOS gp140 trimers, this protein and JR-FL SOS gp140 were purified using lectin-affinity and size-exclusion chromatography (SEC) techniques. The SEC-fractionated aliquots were then resolved by BN-PAGE (Fig. 4). The JR-FL SOS gp140 protein was predominantly a monomer (Fig. 4A) whereas a much greater proportion of the Env species present in JR-FL gp41 NT 1–5 SOS migrated as well-resolved trimers (Fig. 4B; compare lanes 11–16 with the corresponding lanes in Fig. 4A). A densitometric analysis of the resolved gp140 trimer, dimer and monomer fractions on BN-PAGE was combined with BCA quantification of the pooled SEC fractions (trimer, dimer and monomer), to estimate the change in each gp140 species. Compared to the wild-type JR-FL SOS gp140 protein, trimer formation by JR-FL gp41 NT 1–5 SOS gp140 was increased by ~20% and dimer formation by ~10%, whereas the monomer content was reduced by ~50%.
Fig. 4.
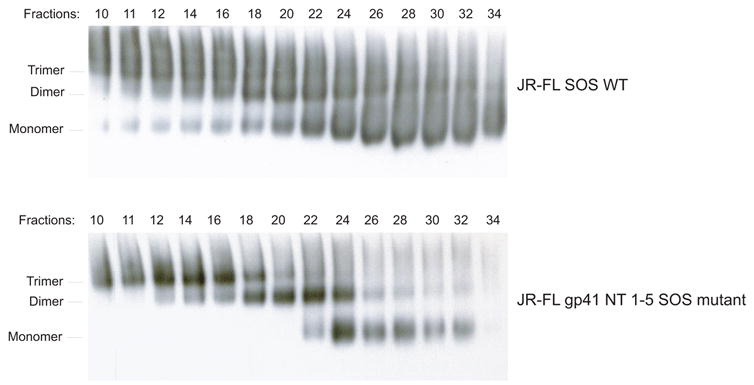
(A) The wild-type JR-FL SOS gp140 and (B) the JR-FL gp41 NT 1–5 SOS gp140 mutant were analyzed by size-exclusion chromatography followed by BN-PAGE and western blotting with MAb CA13. The mutant protein is predominantly trimeric, the wild-type protein mostly monomeric.
Antigenic properties of the wild-type JR-FL SOS and stabilized JR-FL gp41 NT 1–5 SOS mutant gp140s
To determine whether the antigenicity of the mutant JR-FL gp140 had been altered by the introduction of the trimer-stabilizing substitutions, we used SPR methods to study its binding of various antibodies, in comparison to the wild-type gp140. In these studies, biotinylated Protein G was immobilized onto a Streptavidin (SA)-coated chip, which was then used to capture various agents via their Fc regions (Fig. 5A). The CD4-IgG2 protein (used as a surrogate for CD4) and the following MAbs were all studied: b12 (neutralizing, anti-CD4BS), 2G12 (neutralizing, high-mannose epitope on the ‘silent face’), 2F5, 4E10 (both neutralizing, anti-gp41), PA-1 (non-neutralizing, anti-V3), b6 (non-neutralizing, anti-CD4BS) and 17b (non-neutralizing, CD4-induced epitope). Equal molar amounts of purified wild-type and mutant gp140 trimers (>90% purity) were then injected at 10 μl/min, to react with the immobilized MAbs.
Fig. 5.
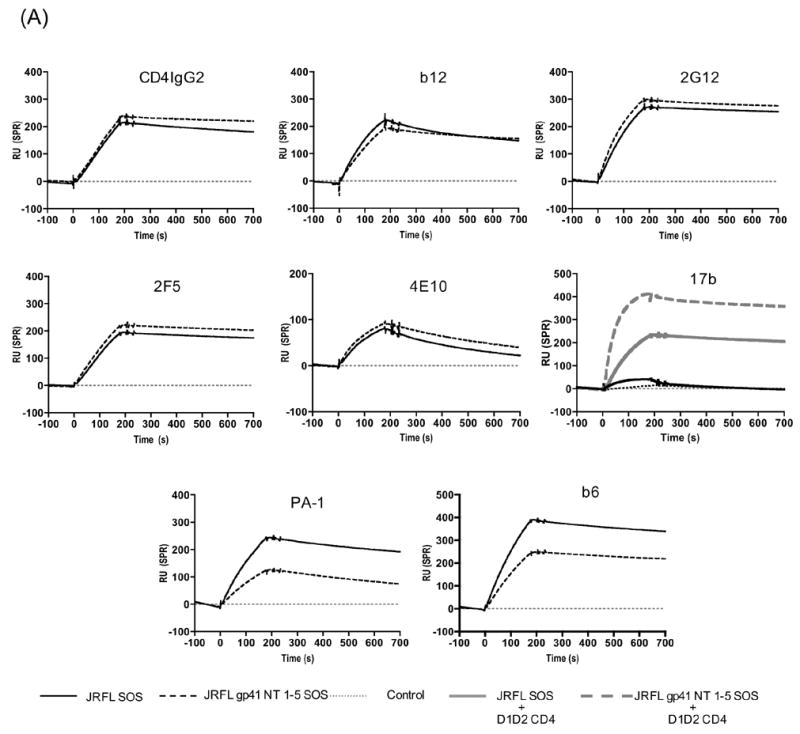

(A) Representative SPR analysis of the binding of MAbs to the JR-FL SOS gp140 and the gp41 NT 1–5 SOS gp140 mutant to the following test agents were: (I) CD4IgG2, (II) b12, (III) 2G12, (IV) 2F5, (V) 4E10, (VI) PA-1, (VII) b6 and (VIII) 17b −/+ D1D2-CD4. The y-axis shows the SPR response unit (RU), the x-axis the time in seconds (s). (B) Injected samples from the BIAcore machine were manually collected after the ligand-binding analysis, then analyzed by BN-PAGE. The wild-type JR-FL SOS gp140 and the gp41 NT 1–5 SOS gp140 mutant proteins are shown, from a representative experiment, one using the PA-1 mAb.
Both the wild-type SOS gp140 and the mutant gp41 NT 1–5 SOS gp140 bound CD4-IgG2 similarly (Fig. 5A and Table 1). Their reactivities with b12 and 2G12 were also similar, with similar response unit (RU) values at the end-of-injection time (t=180s). The two neutralizing anti-gp41 MAbs, 2F5 and 4E10, also bound similarly to the two gp140 proteins (Fig. 5A and Table 1). In the absence of sCD4 (D1D2-CD4), neither gp140 protein bound efficiently to MAb 17b, but when D1D2-CD4 was added, the 17b epitope was induced on both proteins. The extent of the induction was greater for the stabilized trimer than for the wild-type protein (25-fold compared to 5-fold respectively; Fig. 5A and Table 1). The non-neutralizing MAbs PA-1 and b6 bound less efficiently to the stabilized trimer than to its wild-type counterpart (Fig. 5A and Table 1). We are presently investigating the basis for this observation in more detailed studies that use a variety of techniques.
Table 1.
SPR binding of WT and mutant forms of trimeric JR-FL gp140 proteins to various monoclonal antibodies or CD4-IgG2.
| Test agent | WT SOS gp140 Mean RU ± SD (t=180sa) | gp41 NT 1–5 SOS gp140 mutant Mean RU ± SD (t=180sa) |
|---|---|---|
| CD4-IgG2 | 212 ± 8b | 224 ± 13b |
| b12 | 223 ± 6 | 195 ± 10 |
| 2G12 | 269 ± 7 | 278 ± 15 |
| 2F5 | 191 ± 8 | 216 ± 6 |
| 4E10 | 83 ± 4 | 94 ± 5 |
| PA-1 | 242 ± 11 | 123 ± 10 |
| b6 | 391 ± 14 | 233 ± 11 |
| 17b | 39 ± 5 | 14 ± 2 |
| 17b (+ D1D2 CD4) | 237 ± 8 | 399 ± 18 |
End-of-injection time (t) in seconds (s)
Mean RU ± SD based on three experiments, all using 5 nM of analyte
To ensure that we were comparing the MAb binding properties of two trimer variants (wild-type and stabilized), the injected gp140 samples used in the ligand-binding assays were manually collected from the BIAcore X system and analyzed using BN-PAGE. Both gp140 proteins were substantially trimeric, even after passage through the BIAcore system (Fig. 5B).
Substitution of four amino acids in N-terminal region of gp41ECTO also increases the stability of SOS gp140 from HIV-1 Ba-L
To test whether the trimer-stabilizing effect of the above five gp41 amino acids was a generalized phenomenon, we studied another subtype B Env protein, Ba-L. Like JR-FL, Ba-L contains Met, Leu, Asn and Gln residues at positions 535, 543, 553 and 567, respectively. However, at position 588, Ba-L contains Arg, as does KNH1144 (Fig. 1). We therefore introduced the four non-cognate amino acids from KNH1144 into the N-terminal region of Ba-L (M535I, L543Q, N553S, Q567K) to construct a mutant Ba-L gp41 NT 1-4 SOS gp140 protein.
When expressed in HEK293T cells, the wild-type Ba-L SOS gp140, like JR-FL, was a mixture of monomers, dimers and trimers (Fig. 6A, lane 1). However, the mutant containing the above four amino acid substitutions was predominantly trimeric (Fig. 6A, lane 2), with >40% reduction in monomer formation. No individual substitution had as pronounced an effect as the quadruple combination (data not shown). The enhanced trimerization of the mutant Ba-L gp41 NT 1-4 SOS gp140 was not attributable to the presence of aberrantly cross-linked proteins, as shown by SDS-PAGE under reducing and non-reducing conditions (Fig. 6B). Thus, under denaturing conditions, in the absence of the reducing agent, the mutant protein resolved as monomeric gp140; in the presence of DTT, reduction of the disulphide bond dissociated the gp140 into its constituent subunits (as in Fig. 3C, only the gp120 component is detected in this analysis).
Fig. 6.

Stabilizing cleaved Ba-L SOS gp140 trimers. (A) The wild-type Ba-L SOS gp140 and the mutant Ba-L gp41 NT 1–4 SOS gp140 proteins were analyzed by BN-PAGE and western blotting with MAb CA13. (B) The same proteins were analyzed by SDS-PAGE and western blotting, followed by detection with MAb B13. The − and + symbols indicate the absence and presence of DTT.
Taken together, these results suggest that modifications of a few selected amino acids in the N-terminal region of gp41 can improve the stability of gp140 trimers and that the finding might be generalizable to diverse HIV-1 genotypes.
DISCUSSION
Here, we have identified residues in the N-terminal region of the gp41 ectodomain that influence the stability of trimeric forms of the HIV-1 gp140 glycoprotein, particularly the trimers that most, but of course incompletely, resemble the native form of the Env complex. The residues were found by inspection of the sequence of gp41ECTO from a subtype A SOSIP gp140 (KNH1144) that formed stable, cleaved trimers with unusual efficiency. Comparison of this sequence with that of JR-FL, a strain from which homogenous trimers are less easily made, identified five variable residues in a plausibly relevant region of gp41ECTO that lay in and around HR1. Substitution of those five residues in KNH1144 gp140 by the corresponding ones from JR-FL destabilized the resulting gp140 trimers. Conversely, and of more relevance, formation of JR-FL gp140 trimers could be considerably improved when the variable residues from KNH1144 were introduced in place of the JR-FL residues. The same approach also improved trimer formation in the context of the Ba-L sequence, suggesting that our observation is likely to be generally relevant for making stable, cleaved gp140 trimers. Substituting naturally variable amino acids may be a less invasive way to promote trimer stabilization than previously described alternatives, such as the use of heterologous trimerization domains (Yang et al., 2000; Yang et al., 2002), or the insertion of the SIV gp41 N-terminal region to make a HIV-SIV chimeric envelope glycoprotein (Center et al., 2004).
The effect of substituting the KNH1144 gp41 residues into JR-FL and Ba-L is to reduce the heterogeneity of the oligomeric forms of SOS gp140 proteins when they are expressed as unpurified culture supernatants. Thus there was a marked decrease in the amount of monomers present, lesser but sill notable decreases in dimers, tetramers and high-molecular weight aggregates and, of most relevance, an increase in the proportion of trimers. When the stabilized JR-FL SOS gp140 protein was purified by lectin-affinity and size-exclusion chromatography, the amounts of monomers, tetramers and aggregates were reduced, whereas trimers were markedly more abundant and a small increase in the amount of dimers was also apparent. We believe the dimers are likely to be dissociation products of trimers that arise during the purification process. This would not be too surprising, since the increase in trimer-stability is presumably only relative, not absolute, compared to the wild-type protein.
Structural and biophysical studies will be necessary to determine how the amino acid substitutions in the gp41 N-terminal region increase the formation or stability of gp140 oligomers, particularly trimers. We have noted that some of the amino acids in the KNH1144 N-terminal region have longer side chains than their JR-FL counterparts (KNH1144 vs. JR-FL: Q543L, K567Q and R588G). It is also noteworthy that the S553, K567 and R588 residues in KNH1144 have greater α-helix-stabilizing propensities than the corresponding residues, N553, Q567 and G588, in JR-FL. Hence alterations in the size or the nature of the side chains may strengthen localized helix-to-helix packing interactions in a way that stabilizes gp140 oligomers. As noted above, additional studies will be required to investigate this possibility, however.
We observed that both the wild-type JR-FL SOS gp140 and the stabilized JR-FL gp41 NT 1-5 SOS gp140 mutant bound similarly to neutralizing antibodies and proteins (b12, 2G12, 2F5, 4E10 and CD4-IgG2). Hence the overall antigenic structure of the stabilized gp140 trimers is not adversely influenced by the sequence changes introduced into gp41. We have also noted that the stabilized JR-FL trimers bound non-neutralizing antibodies (PA-1, b6 and 17b) to a lesser extent than the corresponding wild type trimers. More detailed studies are required to confirm, extend and understand the structural basis of this observation, but any effect must be quite subtle in nature, given the proximity and similarity of the b6 and b12 epitopes (Pantophlet et al., 2003). Whether stabilizing the conformation of gp140 trimers is advantageous for use of these proteins as vaccine immunogens is now under investigation.
MATERIALS AND METHODS
Reagents
CD4-IgG2 (PRO 542) (Allaway et al., 1995) and monoclonal antibody (MAb) PA-1 were provided by Dr. William Olson (Progenics Pharmaceuticals). Soluble D1D2-CD4 (sCD4-183, 2 domain) (Garlick et al., 1990) was obtained from the NIH AIDS Research and Reference Program. MAb CA13 (ARP3119), from Ms C. Arnold, was provided by the EU Programme EVA Centralized Facility for AIDS Reagents, NIBSC, UK (AVIP Contract Number LSHP-CT-2004-503487). MAbs 2G12 (Calarese et al., 2003; Trkola et al., 1996), 2F5 (Parker et al., 2001; Zwick et al., 2001), 4E10 (Cardoso et al., 2005; Zwick et al., 2001) were obtained from Hermann Katinger, MAb 17b (Thali et al., 1993) from James Robinson and MAb b12 (Burton et al., 1994) from Dennis Burton. The hybridoma for the production of MAb B13 (HIV-1 gp160 Hyb, Chessie 13–39.1) (Abacioglu et al., 1994) was obtained from NIH AIDS Research and Reference Program (donated by George K. Lewis).
Plasmids and construction of chimeric and mutant env genes
Various HIV-1 env genes, cloned into the high-level mammalian expression vector pPPI4, were used for expression of soluble gp140 glycoproteins as previously described. Furin was expressed from pcDNA3.1-Furin (Binley et al., 2000; Sanders et al., 2000). The HIV-1 Env subtype A clone KNH1144 (accession number AF457066) (Beddows et al., 2006) and the subtype B clones JR-FL and Ba-L have been described previously (Binley et al., 2000). In domain-swap experiments, the JR-FL gp41 ectodomain was replaced with the corresponding region of KNH1144 gp41, using EcoRI and HindIII restriction enzymes, followed by repair of the restriction sites and verification of the sequences. Specific amino acid substitutions were made using the QuikChange® II XL site-directed mutagenesis kit (Stratagene Inc., La Jolla, California) and the appropriate primers. The introduced mutations were verified by sequencing.
Transfection and expression of soluble gp140 envelope glycoproteins
The human Embryonic Kidney cell line HEK293T was used for expression of the various envelope glycoproteins by transient transfection, as previously described (Binley et al., 2000; Sanders et al., 2000; Sanders et al., 2002). HEK293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal calf serum, penicillin, streptomycin and L-glutamine. Transient transfections were performed using Polyethylenimine (PEI) (Polysciences Inc., Warrington, PA) (Boussif et al., 1995; Kirschner et al., 2006). For each small-scale transfection, 7 μg of env DNA and 3.5 μg of furin DNA were used. Five hours post-transfection, the 293T cells were washed and the media replaced with DMEM containing 0.05% bovine serum albumin (BSA), antibiotics (penicillin, streptomycin) and L-glutamine. Forty-eight hours post-transfection, the supernatant was collected and filtered using a 0.45 μm filter. A cocktail of protease inhibitors (Roche Diagnostics, Indianapolis, IN) was added before concentration of the supernatant by >20-fold using the Amicon ultracentrifugal filter system (Millipore, Billerica, MA). Aliquots of concentrated supernatant were analyzed by sodium-dodecyl sulfate polyacrylamide electrophoresis (SDS-PAGE), or stored at −80°C for future study.
Purification of soluble envelope glycoproteins
Supernatants (1 L) from transfected HEK293T cells were concentrated by >20-fold then processed by Lectin-affinity chromatography. The column eluate was then size-fractionated using an analytical Superose™ 6 column (GE Amersham Pharmacia, Piscataway, NJ) equilibrated with phosphate-buffered saline (PBS; 100 mM NaCl, 50 mM sodium phosphate, pH 7.0). The column was calibrated with protein standards of known molecular weights (HMW Gel Filtration Calibration Kit; Amersham Pharmacia, Piscataway, NJ). Fractions (200 μl) were collected and analyzed using Blue-native polyacrylamide electrophoresis (BN-PAGE) and SDS-PAGE. Quantification of proteins was carried out using the BCA quantification kit (Pierce) with known BSA standards.
BN-PAGE, SDS-PAGE and Western Blot analysis
BN-PAGE was performed as described previously (Schulke et al., 2002). Concentrated culture supernatants or purified protein samples were diluted with an equal volume of a loading buffer containing 100 mM 4-(N-morpholino) propane sulfonic acid (MOPS), 100 mM Tris-HCl (pH 7.7), 40% glycerol, 0.1% Coomassie blue, and loaded onto a 4–12% Bis-Tris NuPAGE gel (Invitrogen). Gel electrophoresis was performed at 100 V for 3 h using 50 mM MOPS, 50 mM Tris (pH 7.7) as electrophoresis buffer. SDS-PAGE was performed as described previously (Schulke et al., 2002). Reduced and non-reduced samples were prepared in Laemmli sample buffer (62.5 mM Tris-HCl, pH6.8, 2% SDS, 25% glycerol, 0.01% DTT) and boiled for 5 min in the presence or absence of 50 mM dithiothreitol (DTT), respectively.
Western blot analyses were performed as described elsewhere (Schulke et al., 2002). Following transfer, the polyvinylidene difluoride (PVDF) membrane was destained, then probed using anti-Env MAbs CA13 (ARP3119) or B13, followed by horseradish peroxidase-labeled anti-mouse immunoglobulin G (IgG) (Kirkegaard & Perry Labs), at a final concentration of 0.2 μg/ml. The bound MAbs were detected using the Western Blot Chemiluminescence Reagent Plus system (Perkin-Elmer Life Sciences, Boston, MA). Protein mixtures containing Thyroglobulin (669 kDa), Ferritin (440 kDa), Catalase (232 kDa), Lactate dehydrogenase (140 kDa) and BSA (66 kDa) (Amersham Biosciences) were used as standard markers for native gels. For denaturing electrophoresis, the MultiMark® multi-colored standard (Invitrogen) was used.
BIAcore Surface Plasmon Resonance (SPR)
The BIAcore X system (BIAcore Inc, Uppsala, Sweden) was used for comparison of the JR-FL WT versus mutant gp140 env binding to various monoclonal antibodies. All assays were performed at 25°C using HBS-EP buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% [v/v] Surfactant P20; BIAcore, Uppsala, Sweden), which was degassed for 1 h before use. The flow-rate was maintained at 10 μl/minute. A BIAcore streptavidin (SA) chip was used for capturing ~1000 response units (RU) of biotinylated protein G (Pierce) in both the experimental and the control flow-cells. Biotin was then used to block the uncoated streptavidin surface on both flow-cells. In the experimental cell, ~1000 RU of various MAbs were directionally captured onto the surface-attached biotinylated Protein G via their Fc regions. Purified envelope glycoproteins (5 nM) were then injected for analysis of their ligand-binding properties. For study of the CD4-induced binding of MAbs directed to the CD4i-epitope, D1D2-CD4 (at a 6-molar excess concentration) was incubated with the envelope glycoprotein for 1 h at room temperature before injection. Following each run, the sensor surface was regenerated using two 10 μl injections of 10 mM Glycine-HCl, pH 3.0. For each analyte, association was measured for 180s, dissociation for a further >500 s. All binding studies were performed three times (n=3) with good reproducibility. The data were analyzed using BIAevaluation software 3.2 (BIAcore Inc). To correct for refractive index changes and instrument noise, the response data from the control flow-cell were subtracted from those obtained from the experimental flow-cell. For comparison of the antigenicity profiles of the wild-type and mutant gp140 proteins, the end-of-injection RU values +/− SD (n=3) are reported.
Acknowledgments
We thank William Olson, Hermann Katinger, James Robinson and Dennis Burton for the gifts of essential reagents. We are grateful to Min Lu for advice on gp41 structure-function relationships and critically reading the manuscript. This work was supported by NIH grants AI 36082 and AI 45463, NIH contract N01 AI 30030 (HIV Vaccine Design and Development Team) and by the International AIDS Vaccine Initiative. The Department of Microbiology and Immunology at the Weill Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abacioglu YH, Fouts TR, Laman JD, Claassen E, Pincus SH, Moore JP, Roby CA, Kamin-Lewis R, Lewis GK. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retroviruses. 1994;10(4):371–81. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- Allaway GP, Davis-Bruno KL, Beaudry GA, Garcia EB, Wong EL, Ryder AM, Hasel KW, Gauduin MC, Koup RA, McDougal JS, et al. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res Hum Retroviruses. 1995;11(5):533–9. doi: 10.1089/aid.1995.11.533. [DOI] [PubMed] [Google Scholar]
- Beddows S, Kirschner M, Campbell-Gardener L, Franti M, Dey AK, Iyer SP, Maddon PJ, Paluch M, Master A, Overbaugh J, Vancott T, Olson WC, Moore JP. Construction and Characterization of Soluble, Cleaved, and Stabilized Trimeric Env Proteins Based on HIV Type 1 Env Subtype A. AIDS Res Hum Retroviruses. 2006;22(6):569–79. doi: 10.1089/aid.2006.22.569. [DOI] [PubMed] [Google Scholar]
- Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–43. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklow SC, Lu M, Kim PS. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry. 1995;34(46):14955–62. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92(16):7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266(5187):1024–7. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300(5628):2065–71. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, Wilson IA. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22(2):163–73. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Center RJ, Kemp BE, Poumbourios P. Human immunodeficiency virus type 1 and 2 envelope glycoproteins oligomerize through conserved sequences. J Virol. 1997;71(7):5706–11. doi: 10.1128/jvi.71.7.5706-5711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center RJ, Leapman RD, Lebowitz J, Arthur LO, Earl PL, Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope protein on the virion surface. J Virol. 2002;76(15):7863–7. doi: 10.1128/JVI.76.15.7863-7867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center RJ, Lebowitz J, Leapman RD, Moss B. Promoting trimerization of soluble human immunodeficiency virus type 1 (HIV-1) Env through the use of HIV-1/simian immunodeficiency virus chimeras. J Virol. 2004;78(5):2265–76. doi: 10.1128/JVI.78.5.2265-2276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center RJ, Schuck P, Leapman RD, Arthur LO, Earl PL, Moss B, Lebowitz J. Oligomeric structure of virion-associated and soluble forms of the simian immunodeficiency virus envelope protein in the prefusion activated conformation. Proc Natl Acad Sci U S A. 2001;98(26):14877–82. doi: 10.1073/pnas.261573898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti BK, Kong WP, Wu BY, Yang ZY, Friborg J, Ling X, King SR, Montefiori DC, Nabel GJ. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol. 2002;76(11):5357–68. doi: 10.1128/JVI.76.11.5357-5368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89(2):263–73. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93(5):681–4. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- Doms RW, Moore JP. HIV-1 membrane fusion: targets of opportunity. J Cell Biol. 2000;151(2):F9–14. doi: 10.1083/jcb.151.2.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Moss B, Doms RW. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol. 1991;65(4):2047–55. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BN, Knipe DM, Howley PM, Chanock RM, Melinick JL, Monath TP, Roizman B, Straus SE. In: Fields Virology. Luciw PA, editor. Lippincott-Raven Publishers; Philadelphia: 1996. [Google Scholar]
- Garlick RL, Kirschner RJ, Eckenrode FM, Tarpley WG, Tomich CS. Escherichia coli expression, purification, and biological activity of a truncated soluble CD4. AIDS Res Hum Retroviruses. 1990;6(4):465–79. doi: 10.1089/aid.1990.6.465. [DOI] [PubMed] [Google Scholar]
- Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- Jones PL, Korte T, Blumenthal R. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J Biol Chem. 1998;273(1):404–9. doi: 10.1074/jbc.273.1.404. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Monrose V, Paluch M, Techodamrongsin N, Rethwilm A, Moore JP. The production of cleaved, trimeric human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein vaccine antigens and infectious pseudoviruses using linear polyethylenimine as a transfection reagent. Protein Expr Purif. 2006;48(1):61–8. doi: 10.1016/j.pep.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2(12):1075–82. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol. 2000;151(2):413–23. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Doms RW. The entry of entry inhibitors: a fusion of science and medicine. Proc Natl Acad Sci U S A. 2003;100(19):10598–602. doi: 10.1073/pnas.1932511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77(1):642–58. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CE, Deterding LJ, Hager-Braun C, Binley JM, Schulke N, Katinger H, Moore JP, Tomer KB. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J Virol. 2001;75(22):10906–11. doi: 10.1128/JVI.75.22.10906-10911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poumbourios P, Wilson KA, Center RJ, El Ahmar W, Kemp BE. Human immunodeficiency virus type 1 envelope glycoprotein oligomerization requires the gp41 amphipathic alpha-helical/leucine zipper-like sequence. J Virol. 1997;71(3):2041–9. doi: 10.1128/jvi.71.3.2041-2049.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280(5371):1949–53. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Schiffner L, Master A, Kajumo F, Guo Y, Dragic T, Moore JP, Binley JM. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J Virol. 2000;74(11):5091–100. doi: 10.1128/jvi.74.11.5091-5100.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76(17):8875–89. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau QJ, Moore JP. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174(2):407–15. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulke N, Vesanen MS, Sanders RW, Zhu P, Lu M, Anselma DJ, Villa AR, Parren PW, Binley JM, Roux KH, Maddon PJ, Moore JP, Olson WC. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol. 2002;76(15):7760–76. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugars DC, Wild CT, Greenwell TK, Matthews TJ. Biophysical characterization of recombinant proteins expressing the leucine zipper-like domain of the human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1996;70(5):2982–91. doi: 10.1128/jvi.70.5.2982-2991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava IK, Stamatatos L, Legg H, Kan E, Fong A, Coates SR, Leung L, Wininger M, Donnelly JJ, Ulmer JB, Barnett SW. Purification and characterization of oligomeric envelope glycoprotein from a primary R5 subtype B human immunodeficiency virus. J Virol. 2002;76(6):2835–47. doi: 10.1128/JVI.76.6.2835-2847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72(6):4694–703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67(7):3978–88. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100–8. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384(6605):179–83. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- Yang X, Florin L, Farzan M, Kolchinsky P, Kwong PD, Sodroski J, Wyatt R. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J Virol. 2000;74(10):4746–54. doi: 10.1128/jvi.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76(9):4634–42. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-Electron Tomographic Structure of an Immunodeficiency Virus Envelope Complex In Situ. PLoS Pathog. 2006;2(8) doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CW, Chishti Y, Hussey RE, Reinherz EL. Expression, purification, and characterization of recombinant HIV gp140. The gp41 ectodomain of HIV or simian immunodeficiency virus is sufficient to maintain the retroviral envelope glycoprotein as a trimer. J Biol Chem. 2001;276(43):39577–85. doi: 10.1074/jbc.M107147200. [DOI] [PubMed] [Google Scholar]
- Zhang W, Canziani G, Plugariu C, Wyatt R, Sodroski J, Sweet R, Kwong P, Hendrickson W, Chaiken I. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry. 1999;38(29):9405–16. doi: 10.1021/bi990654o. [DOI] [PubMed] [Google Scholar]
- Zhu P, Chertova E, Bess J, Jr, Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A. 2003;100(26):15812–7. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441(7095):847–52. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75(22):10892–905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


