Abstract
Background
Animal studies suggest that brain apolipoprotein E (apoE) levels influence amyloid-β (Aβ) deposition and thus risk for Alzheimer's disease (AD). We have previously demonstrated that deletion of the ATP-binding cassette A1 transporter (ABCA1) in mice causes dramatic reductions in brain and cerebrospinal fluid (CSF) apoE levels and lipidation. To examine whether polymorphisms in ABCA1 affect CSF apoE levels in humans, we measured apoE in CSF taken from 168 subjects who were 43 to 91 years old and were either cognitively normal or who had mild AD. We then genotyped the subjects for ten previously identified ABCA1 single nucleotide polymorphisms (SNPs).
Results
In all subjects, the mean CSF apoE level was 9.09 μg/ml with a standard deviation of 2.70 μg/ml. Levels of apoE in CSF samples taken from the same individual two weeks apart were strongly correlated (r2 = 0.93, p < 0.01). In contrast, CSF apoE levels in different individuals varied widely (coefficient of variation = 46%). CSF apoE levels did not vary according to AD status, APOE genotype, gender or race. Average apoE levels increased with age by ~0.5 μg/ml per 10 years (r2 = 0.05, p = 0.003). We found no significant associations between CSF apoE levels and the ten ABCA1 SNPs we genotyped. Moreover, in a separate sample of 1225 AD cases and 1431 controls, we found no association between the ABCA1 SNP rs2230806 and AD as has been previously reported.
Conclusion
We found that CSF apoE levels vary widely between individuals, but are stable within individuals over a two-week interval. AD status, APOE genotype, gender and race do not affect CSF apoE levels, but average CSF apoE levels increase with age. Given the lack of association between CSF apoE levels and genotypes for the ABCA1 SNPs we examined, either these SNPs do not affect ABCA1 function or if they do, they do not have strong effects in the CNS. Finally, we find no evidence for an association between the ABCA1 SNP rs2230806 and AD in a large sample set.
Background
Alzheimer's disease (AD) is an age-related progressive neurodegenerative disorder that causes impairments in memory and thinking. The strongest genetic risk factor for AD is apolipoprotein E (APOE) genotype [1]. In comparison to people who are homozygous for the common ε3 allele, people who carry the ε4 allele are at higher risk for AD and generally have an earlier age of onset, while people who carry the ε2 allele are at lower risk and have a later age of onset [2-6]. ApoE is a chaperone for amyloid-β (Aβ) peptide, which deposits in the brain and is thought to initiate a cascade of events that causes AD [7,8]. Mouse models have shown that the time of onset and amount of Aβ deposition depends not only on APOE genotype but also on apoE levels. Interestingly, higher expression of mouse apoE increases the amount of Aβ deposition [9,10], while higher expression of the human ε3 isoform of APOE knocked into the mouse Apoe locus decreases levels of amyloid deposition [11]. Additionally, expression of human apoE in mice delays the onset of Aβ deposition in an isoform-specific fashion, with ε2 expression decreasing Aβ deposition the most and ε4 expression decreasing Aβ deposition the least [12,13].
Despite evidence from animal studies suggesting that apoE levels affect Aβ deposition, there is no consensus regarding levels of apoE expression and its effects on Aβ deposition in human studies. The examination of whether apoE levels affect AD risk in humans has focused on APOE promoter polymorphisms. Over 50 studies listed on the Alzforum website tested for an association between AD and one or more polymorphisms within the APOE promoter [14]. Meta-analyses on this website support the notion that APOE promoter variation is associated with risk for AD. However, it is unclear whether this association is due to linkage disequilibrium with the coding polymorphisms or whether there are independent effects on risk due to the level of APOE expression. Some studies have examined the effect of APOE promoter polymorphisms on APOE expression in vitro [15,16]. More recently, allele specific gene expression has been used in post-mortem brain samples to measure the relative expression of APOE ε3 and ε4 isoforms [17]. However, even these studies do not directly examine the effect of the promoter polymorphisms on levels of apoE protein.
Previous studies of CSF apoE levels in humans have reached varying conclusions. Some report that CSF apoE levels are lower in AD subjects than in control subjects [18-20], other studies find no association between CSF apoE levels and AD [21,22], and one study shows that CSF apoE levels are higher in AD subjects than in control subjects [23]. Multiple studies found that the APOE genotype was not associated with differing CSF apoE levels [19-22]. In contrast, plasma apoE levels are clearly dependent on APOE genotype [24,25], which suggests that apoE is metabolized differently in the CSF and plasma. Gender and age do not appear to affect CSF apoE levels [22].
Recently, our laboratory and others reported that apoE levels were greatly reduced in mice lacking functional ATP-binding cassette A1 transporter (ABCA1) [26-28]. Within the CNS of ABCA1 knock-out mice, CSF apoE was 2% of normal levels and apoE in the cortex was 20% of normal levels [26]. ABCA1 transfers cholesterol and phospholipids from the cell membrane to apolipoproteins (including apoE) to form nascent high density lipoproteins (HDL). In the rare case that both copies of ABCA1 are non-functional, as occurs in Tangier's disease, apoE and other lipoproteins do not receive normal amounts of lipid and are rapidly degraded [29]. Multiple studies have shown that levels of plasma HDL-C and associated apolipoproteins are affected by single nucleotide polymorphisms (SNPs) in ABCA1 [30-34]. In particular, studies have implicated the following SNPs in affecting levels of plasma HDL-C: rs2230806 (R219K) [33], rs2066718 (V771M) [31,32], rs2066715 (V825I) [31], rs4149313 (I883M) [34], rs2230808 (R1587K) [31]. Since ABCA1 appears to have a similar role in the CNS and in the periphery, we hypothesized that these ABCA1 SNPs would also have an effect on CSF apoE levels since apoE is the major apoprotein component of HDL produced in the CNS. Additionally, studies by others have reported that the ABCA1 SNP rs2230806 (R219K) affects risk for AD [35-38]. This is particularly interesting because ABCA1 falls within a region of chromosome 9 that is linked to late-onset AD [39-43]. The profound effect of ABCA1 levels on CNS apoE levels in mice, in addition to reports that an ABCA1 SNP may affect risk for AD, suggested that ABCA1 may be involved in the genetic control of CNS apoE levels in humans.
Given the contrasting results and small sample sizes used in some studies of apoE levels in human CSF, we chose to begin our study by characterizing CSF apoE levels in a relatively large sample of 168 individuals with respect to AD status, APOE genotype, gender, race and age. We next examined whether ten ABCA1 SNPs, including five SNPs shown to affect plasma HDL-C, affected levels of apoE in the CSF. Finally, in a large sample of 1225 AD cases and 1431 controls, we attempted to replicate the previously reported association between the ABCA1 SNP rs2230806 and AD.
Results
ApoE levels and stability in human CSF
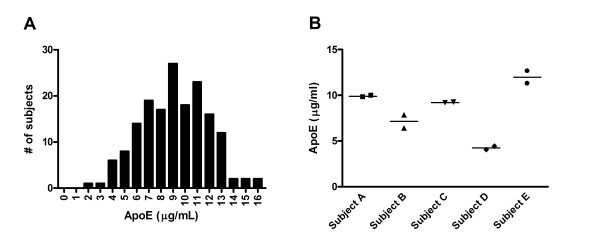
ApoE levels were measured in CSF samples from 168 subjects who were 43 to 91 years old (Table 1). We included all samples available without regard to AD status, APOE genotype, gender, race or age. ApoE values were sorted into 1 μg/ml bins and the number of subjects with apoE values within each bin from 0 to 16 μg/ml was tallied (Fig. 1A). The mean apoE level was 9.09 μg/ml with a standard deviation of 2.70 μg/ml. The number of individuals per bin was in a normal distribution according to a Kolmogorov-Smirnov test (p > 0.10).
Table 1.
Characteristics of subjects who underwent lumbar puncture.
| CDR 0, <65 | CDR 0, ≥65 | CDR 0.5 | CDR 1+ | |
| n = | 70 | 55 | 26 | 17 |
| Male | 29% | 28% | 54% | 47% |
| Female | 71% | 72% | 46% | 53% |
| Age* | 54 ± 6 | 76 ± 8 | 75 ± 8 | 76 ± 6 |
| ε2 freq. | 0.11 | 0.13 | 0.06 | 0.03 |
| ε3 freq. | 0.64 | 0.73 | 0.56 | 0.74 |
| ε4 freq. | 0.25 | 0.14 | 0.38 | 0.24 |
*Age is mean ± standard deviation
Figure 1.
Distribution of apoE levels in human CSF. A, ApoE levels were sorted into bins of 1 μg/ml and the number of subjects with apoE values within each bin was tallied. The data represents 168 subjects without division by CDR status, APOE genotype, gender, race or age. B, ApoE levels were measured in CSF samples taken two weeks apart from five different patients.
To determine the intra-individual stability of CSF apoE levels sampled over time, lumbar puncture was performed on five subjects at two different times that were two weeks apart. CSF apoE levels within the same individual were strongly correlated (r2 = 0.93, p < 0.01). In contrast, CSF apoE levels between different individuals showed large variation (coefficient of variation = 46%) (Fig. 1B). This demonstrates that CSF apoE levels are relatively stable within an individual during a short time interval, but vary widely between individuals. Furthermore, this suggests that CSF apoE levels may be influenced by stable individual differences, such as genetic sequence variation.
Effects of AD status, APOE genotype, gender or age on CSF apoE levels
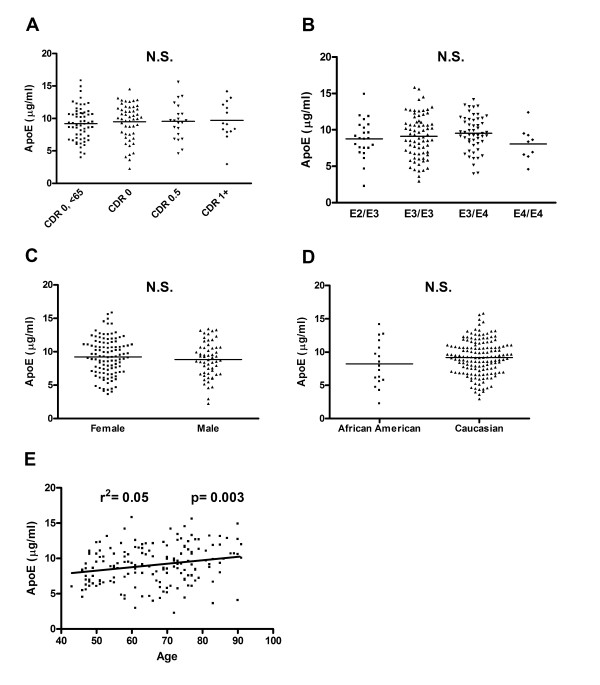
There are varying reports in the literature on whether CSF apoE levels are affected by AD status, APOE genotype, gender or age. In our relatively large sample, we investigated whether these variables, as well as race, modified CSF apoE levels. The levels of CSF apoE were not significantly different between subjects who were cognitively normal who had a clinical dementia rating (CDR) score of 0 and those who had very mild (CDR 0.5) or mild-moderate dementia believed to be due to AD (CDR 1+) (Fig. 2A). Since a recent study reported that apoE levels may be affected by APOE genotype [44], we examined whether APOE genotype affects CSF apoE levels in our sample. Despite large numbers of patients, we found no significant differences in CSF apoE levels in subjects with different APOE genotypes (Fig. 2B). Next, we looked for gender effects on CSF apoE levels and found none (Fig. 2C). We also found no significant difference in CSF apoE levels between subjects who identified themselves as Caucasians and African Americans (Fig. 2D). Additionally, we studied whether age affects CSF apoE levels (Fig. 2E). Average apoE levels increased by a small but significant extent, ~0.5 μg per 10 years (r2 = 0.05, p = 0.003). Finally, to test the possibility that AD status, APOE genotype, gender and age interact to influence apoE levels in the CSF, we performed a multivariate ANOVA and found no significant interactions. We conclude that CSF apoE levels are not greatly affected by AD status, APOE genotype, gender or race, but do increase with age.
Figure 2.
ApoE levels in human CSF do not vary according to presence or absence of Alzheimer's disease, level of cognitive impairment, APOE genotype, gender or race, but do increase with age. A, Subjects were grouped by age and AD status. Subjects with a clinical dementia rating (CDR) score of 0 (cognitively normal) that were less than age 65 were placed into the first group (CDR 0, <65; n = 59). Subjects that were 65 and older with a CDR score of 0, 0.5, or 1–2 were placed into the second (CDR 0, n = 50), third (CDR 0.5, n = 21) and fourth (CDR 1+, n = 14) groups, respectively. There was no difference in CSF apoE levels by one-way ANOVA. B, Subjects were grouped by APOE genotype into four groups: E2/E3 (n = 23), E3/E3 (n = 72), E3/E4 (n = 52), and E4/E4 (n = 9). There was no difference in CSF apoE levels by one-way ANOVA. C, Subjects were divided into two groups, female (n = 109) and male (n = 57). There was no difference in CSF apoE levels by a two-tailed Student's T-test. D, Subjects were grouped by self-identified racial group: African American (n = 17) and Caucasian (n = 149). There was no difference in CSF apoE levels by a two-tailed Student's T-test. E, CSF apoE levels were graphed as a function of subject age (n = 168). The slope of the regression line was 0.05, with a 95% confidence interval of 0.02 to 0.08.
Effects of ABCA1 SNPs on CSF apoE levels and risk for AD
We sought to determine whether SNPs in ABCA1 affect CSF apoE levels. The subjects for whom we had CSF apoE data were genotyped for the following ABCA1 SNPs: rs2230806 (R219K), rs2066718 (V771M), rs2066715 (V825I), rs4149313 (I883M), rs2230808 (R1587K), rs1883025 (intron), rs2275544 (intron), rs2777799 (intron), rs3904999 (intron) and rs6479283 (intron). The numbers of subjects for which we obtained conservatively called (high quality) genotypes, as well as the frequencies of the minor and major alleles, are listed in Table 2. We found no association between CSF apoE levels and any of the ABCA1 SNPs, including the five coding SNPs that were previously associated with alterations in plasma HDL-C levels.
Table 2.
The number of subjects with high quality genotypes and the frequency of the minor and major ABCA1 SNP alleles.
| n = | minor allele freq. | major allele freq. | |
| rs2230806 | 123 | 0.309 | 0.691 |
| rs2066718 | 124 | 0.040 | 0.960 |
| rs2066715 | 144 | 0.073 | 0.927 |
| rs4149313 | 124 | 0.185 | 0.815 |
| rs2230808 | 124 | 0.315 | 0.685 |
| rs1883025 | 102 | 0.358 | 0.642 |
| rs2275544 | 122 | 0.131 | 0.869 |
| rs2777799 | 123 | 0.126 | 0.874 |
| rs3904999 | 123 | 0.203 | 0.797 |
| rs6479283 | 119 | 0.223 | 0.777 |
We also attempted to reproduce the finding, reported by some groups but not others, that the ABCA1 rs2230806 SNP is associated with altered risk for AD [35-38,45]. We combined information on 794 subjects from Washington University with 1,862 additional subjects from the University of California-San Diego and the United Kingdom to yield the maximum power. The subjects from Washington University had previously been analyzed and it was found that risk for AD in this group did not depend on the rs2230806 SNP [36]. The 1,862 additional subjects had not previously been used to examine the rs2230806 SNP. In this large group of 1225 case and 1431 control subjects, there was no effect of the rs2230806 SNP on risk for AD (Table 3). Analysis of sub-groups based on APOE genotype and gender also failed to show an effect of the rs2230806 SNP on risk for AD.
Table 3.
The distribution of the rs2230806 polymorphism in subjects with Alzheimer's disease and control subjects.
| # | # AA | # AG | # GG | freq. A | freq. G | AD vs. Control | ||
| Total | AD | 1225 | 81 | 476 | 668 | 0.260 | 0.740 | p = 0.76 |
| n = 2656 | Control | 1431 | 105 | 548 | 778 | 0.265 | 0.735 | |
| E3/E3 | AD | 437 | 31 | 170 | 236 | 0.265 | 0.735 | p = 0.93 |
| n = 1316 | Control | 879 | 63 | 351 | 465 | 0.271 | 0.729 | |
| E4/E3 | AD | 555 | 32 | 227 | 296 | 0.262 | 0.738 | p = 0.10 |
| n = 832 | Control | 277 | 18 | 92 | 167 | 0.231 | 0.769 | |
| E4/E4 | AD | 125 | 8 | 40 | 77 | 0.224 | 0.776 | p = 0.86 |
| n = 150 | Control | 25 | 1 | 9 | 15 | 0.220 | 0.780 | |
| Females | AD | 267 | 26 | 105 | 136 | 0.294 | 0.706 | p = 0.99 |
| n = 505 | Control | 238 | 23 | 94 | 121 | 0.294 | 0.706 |
p values are caculated by Chi Square tests with 2 degrees of freedom
Discussion
A notable finding in this study was that CSF apoE levels vary widely between individuals, with a range in our sample from 2 μg/ml to 16 μg/ml, but are stable within individuals during an interval of 2 weeks. This suggests the presence of stable factors within individuals, which may be genetic or environmental, that regulate CSF apoE levels. Recently, it was reported that levels of Aβ vary according the time of day and it is possible that apoE could vary in a similar fashion [46]. However, since all of our samples were obtained at the same time of day (8:00 am), any diurnal variation of apoE levels in this study should be minimal.
We examined whether AD status, APOE genotype, gender, race or age affected CSF apoE levels, but only age was significantly correlated. It is interesting that levels of apoE are not elevated in carriers of the ε2 allele. ApoE3 and apoE4 both bind with high affinity to LDLR resulting in receptor-mediated endocytosis and degradation of apoE. ApoE2 does bind to LDLR, but much less effectively than apoE3 and apoE4 [47]. In mice, the decreased affinity of apoE2 for LDLR leads to elevated levels of CSF apoE in mice in which the human APOE ε2 gene is knocked-in to the mouse Apoe gene locus [48]. The lack of a difference in apoE levels according to genotype in human CSF samples suggests that LDLR may not have as large of an effect on human CSF apoE levels. It will be important to assess this issue in future studies in APOE ε2 homozygous individuals as there may be a much smaller effect in individuals with one copy of the APOE ε2 gene.
We hypothesized that genetic variation in certain genes may contribute to CSF apoE levels and examined whether SNPs in ABCA1, especially SNPs that have been reported to affect plasma HDL-C levels, affect CSF apoE levels. We did not find a significant association between CSF apoE levels and any of the ten ABCA1 SNPs we examined, including the five coding SNPs thought to be associated with altered HDL-C levels. Perhaps this is because the metabolism of apoE is different in the plasma and CSF. Alternatively, these changes in ABCA1 may not affect HDL in the CNS as much as occurs as with HDL in the plasma. This may be due to apoAI being the main apoprotein in plasma HDL whereas apoE is the most abundant apoprotein produced in the CNS in CSF HDL. The effects of the SNPs may also be too small to significantly affect CSF apoE levels. However, it remains possible that rare sequence variations that strongly influence ABCA1 function could contribute to variation in CSF apoE levels. Recent studies demonstrate that several rare polymorphisms in ABCA1 collectively affect overall levels of plasma HDL-C in the population [30,31]. Since ABCA1-mediated lipid transport is critical in the formation of both HDL-C in plasma and apoE-containing lipoproteins in CSF, it is possible that the same rare ABCA1 polymorphisms that have large effects on plasma HDL-C levels would also affect CSF apoE levels.
Additionally, we failed to replicate the finding of other groups that the ABCA1 rs2230806 SNP is associated with altered risk for AD [35-38]. We suggest three possible reasons for the differing results: 1) the ABCA1 rs2230806 SNP does affect risk for AD, but the effect is small so that the association cannot be reproduced regularly in samples of ~2500 subjects; or 2) the population we examined was genetically different from the populations in the other studies assessed; or 3) the ABCA1 rs2230806 SNP does not affect risk for AD. Since the populations that we and others examined are similar and consisted primarily of Caucasians with Northern European heritage, we believe that it is most likely that the ABCA1 rs2230806 SNP contributes either a very small amount or not at all to overall risk for AD.
It seems likely that many different genes modulate levels of apoE in the CSF. Studies suggest that LDLR and LRP influence levels of CSF apoE in mice [48,49]. Given the animal data, it is possible that variations in LDLR or LRP could affect CSF apoE levels in humans, but this has not yet been examined. Further investigation of the genetic control of apoE levels in the CNS could uncover new information on apoE metabolism. This research would not only be relevant to AD, but also to a number of other neurological diseases that may be modulated by apoE such as stroke [50,51], multiple sclerosis [52] and traumatic brain injury [53]. Ultimately, an understanding of the regulation of CSF apoE levels could lead to novel apoE-based treatments for AD and other neurological disorders.
Conclusion
We found that CSF apoE levels vary widely between individuals, but are stable within individuals over a two-week interval. Secondly, AD status, APOE genotype, gender and race do not affect CSF apoE levels, but CSF apoE levels do increase with age. Additionally, ABCA1 SNPs that have been reported to affect plasma HDL-C levels do not affect CSF apoE levels in our sample. Finally, any association that exists between the ABCA1 SNP rs2230806 and AD is very weak.
Methods
Subjects
Subjects in the Washington University sample were community-living participants in the Alzheimer's Disease Research Center (ADRC) registry. All research subjects underwent a clinical evaluation to determine their Clinical Dementia Rating (CDR), as well as a 2-hour psychometric test battery. A medical history was taken to exclude participants that might have confounding medical disorders. Details of the assessment have been described previously [54-56]. Additional case control DNA samples were from the University of California-San Diego and the United Kingdom.
CSF was obtained via lumbar puncture (L.P.) from 168 subjects at Washington University in the General Clinical Research Center after obtaining informed consent. The study protocol was approved by the Human Studies Committee at Washington University. All L.P.s were performed at 8 am after an overnight fast with a 22 gauge atraumatic needle. 25–30 ml of CSF was obtained from each subject and was free of blood contamination. After collection, CSF samples were briefly centrifuged at 1,000 × g to pellet any cell debris, frozen, and stored in polypropylene tubes at -80°C in 0.5 ml aliquots until analysis.
ApoE ELISA
ApoE ELISAs were performed on CSF apoE as previously described [48]. Briefly, plates were coated overnight with WUE4, a monoclonal antibody to human apoE [57]. The plates were washed, blocked with 1% dry milk and washed again. ApoE standards were purified from human β-VLDL (BioDesign, Sako, ME). Standards and samples were diluted and loaded onto the plate, then incubated overnight. The plate was washed and incubated with a polyclonal goat anti-apoE antibody (Calbiochem, San Diego CA). The plate was washed again and incubated with anti-goat-HRP (Vector Laboratories, Burlingame, CA). The plate was washed once more, then developed with TMB (Sigma, St. Louis, MO).
Genotyping
The following SNPS in ABCA1 were genotyped in the Washington University sample of 168 subjects: rs2230806 (R219K), rs2066718 (V771M), rs2066715 (V825I), rs4149313 (I883M), rs2230808 (R1587K), rs1883025 (intron), rs2275544 (intron), rs2777799 (intron), rs3904999 (intron) and rs6479283 (intron). Genotyping was performed using a modified single nucleotide extension reaction with allele detection by mass spectrometry (Sequenom MassArray system; Sequenom, San Diego, CA, USA). PCR primers, termination mixes and multiplexing capabilities were determined with Sequenom Spectro Designer software v2.00.17. Genotyping of rs2230806 in the large group of 2,656 subjects was performed using allele specific real-time PCR [58]. For all SNPs, genotypes were tested and found to be in Hardy-Weinberg equilibrium.
Statistical analyses
Frequency distributions, correlation analysis, ANOVAs, T-tests and Kolmogorov Smirnov tests of normality were performed using GraphPad Prism, Version 4.00 (GraphPad, San Diego, CA). Multivariate ANOVAs were performed using SAS Version 9.0 for Windows XP (SAS Institute Inc., Cary, NC).
Abbreviations
Aβ, amyloid-β peptide; ABCA1, ATP-binding cassette transporter A1; AD, Alzheimer's disease; apoE, apolipoprotein E; CDR, clinical dementia rating; CNS, central nervous system; CSF, cerebrospinal fluid; ELISA, enzyme-linked immunosorbent assay; HDL, high density lipoprotein; LDLR, low density lipoprotein receptor; LP, lumbar puncture; LRP, low density lipoprotein related protein; SNP, single nucleotide polymorphism.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
SEW performed the primary writing and editing of the manuscript and was involved in experimental design, genotyping and data analysis. ARS processed CSF samples and assayed them for levels of apoE. AMF was involved in coordinating CSF collection and experimental design. SS, AG, KM, and HJ were involved in genotyping and experimental design.
JSKK and AH were involved in experimental design and statistical analysis. LJT provided samples from the UCSD collection. AMG and DMH were involved in experimental design, data analysis, and manuscript writing. All authors approved the manuscript.
Acknowledgments
Acknowledgements
The authors gratefully acknowledge the Genetics, Clinical, Psychometric, and Biostatistics Cores of the Washington University ADRC for subject APOE genotyping and clinical, cognitive and psychometric evaluation and data management. We also acknowledge the contributions of our LP physicians at Washington University (Dept. of Neurology): David Holtzman, MD; Randall Bateman, MD; David Brody, MD, PhD; B. Joy Snider, MD, PhD; and Beth Ann Ward, MD. Grants: This work was supported by grants from the National Institute on Aging (P01 AG03991, P01 AG026276, P50 AG05681, RO1 AG16208), a pilot grant from the Genome Sequencing Center at Washington University, and the Washington University General Clinical Research Center funded by the US Public Health System (M01 RR00036). J.S.K.K. is funded by a Ford Foundation Predoctoral Fellowship.
Contributor Information
Suzanne E Wahrle, Email: wahrles@msnotes.wustl.edu.
Aarti R Shah, Email: shaha@neuro.wustl.edu.
Anne M Fagan, Email: fagana@neuro.wustl.edu.
Scott Smemo, Email: ssmemo@uchicago.edu.
John SK Kauwe, Email: keoni@icarus.wustl.edu.
Andrew Grupe, Email: Andrew.Grupe@celeradiagnostics.com.
Anthony Hinrichs, Email: tony@silver.wustl.edu.
Kevin Mayo, Email: kmayo@WUSTL.EDU.
Hong Jiang, Email: jiangh@neuro.wustl.edu.
Leon J Thal, Email: lthal@ucsd.edu.
Alison M Goate, Email: goate@icarus.wustl.edu.
David M Holtzman, Email: holtzman@neuro.wustl.edu.
References
- Raber J, Huang Y, Ashford JW. ApoE genotype accounts for the vast majority of AD risk and AD pathology. Neurobiol Aging. 2004;25:641–650. doi: 10.1016/j.neurobiolaging.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Mayeux R, Stern Y, Ottman R, Tatemichi TK, Tang MX, Maestre G, Ngai C, Tycko B, Ginsberg H. The apolipoprotein epsilon 4 allele in patients with Alzheimer's disease. Ann Neurol. 1993;34:752–754. doi: 10.1002/ana.410340527. [DOI] [PubMed] [Google Scholar]
- Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, Pericak-Vance MA, Goldgaber D, Roses AD. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:9649–9653. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg SM, Rebeck GW, Vonsattel JP, Gomez-Isla T, Hyman BT. Apolipoprotein E epsilon 4 and cerebral hemorrhage associated with amyloid angiopathy. Ann Neurol. 1995;38:254–259. doi: 10.1002/ana.410380219. [DOI] [PubMed] [Google Scholar]
- Wisniewski T, Ghiso J, Frangione B. Biology of A beta amyloid in Alzheimer's disease. Neurobiol Dis. 1997;4:313–328. doi: 10.1006/nbdi.1997.0147. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Bales KR, Verina T, Cummins DJ, Du Y, Dodel RC, Saura J, Fishman CE, DeLong CA, Piccardo P, Petegnief V, Ghetti B, Paul SM. Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 1999;96:15233–15238. doi: 10.1073/pnas.96.26.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Fagan AM, Mackey B, Tenkova T, Sartorius L, Paul SM, Bales K, Ashe KH, Irizarry MC, Hyman BT. Apolipoprotein E facilitates neuritic and cerebrovascular plaque formation in an Alzheimer's disease model. Ann Neurol. 2000;47:739–747. doi: 10.1002/1531-8249(200006)47:6<739::AID-ANA6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- DeMattos RB. Apolipoprotein E dose-dependent modulation of beta-amyloid deposition in a transgenic mouse model of Alzheimer's disease. J Mol Neurosci. 2004;23:255–262. doi: 10.1385/JMN:23:3:255. [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Bales KR, Tenkova T, Fagan AM, Parsadanian M, Sartorius LJ, Mackey B, Olney J, McKeel D, Wozniak D, Paul SM. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2000;97:2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan AM, Watson M, Parsadanian M, Bales KR, Paul SM, Holtzman DM. Human and murine ApoE markedly alters A beta metabolism before and after plaque formation in a mouse model of Alzheimer's disease. Neurobiol Dis. 2002;9:305–318. doi: 10.1006/nbdi.2002.0483. [DOI] [PubMed] [Google Scholar]
- Alzforum [www.alzforum.org/res/com/gen/alzgene].
- Artiga MJ, Bullido MJ, Frank A, Sastre I, Recuero M, Garcia MA, Lendon CL, Han SW, Morris JC, Vazquez J, Goate A, Valdivieso F. Risk for Alzheimer's disease correlates with transcriptional activity of the APOE gene. Hum Mol Genet. 1998;7:1887–1892. doi: 10.1093/hmg/7.12.1887. [DOI] [PubMed] [Google Scholar]
- Artiga MJ, Bullido MJ, Sastre I, Recuero M, Garcia MA, Aldudo J, Vazquez J, Valdivieso F. Allelic polymorphisms in the transcriptional regulatory region of apolipoprotein E gene. FEBS Lett. 1998;421:105–108. doi: 10.1016/S0014-5793(97)01543-3. [DOI] [PubMed] [Google Scholar]
- Bray NJ, Jehu L, Moskvina V, Buxbaum JD, Dracheva S, Haroutunian V, Williams J, Buckland PR, Owen MJ, O'Donovan MC. Allelic expression of APOE in human brain: effects of epsilon status and promoter haplotypes. Hum Mol Genet. 2004;13:2885–2892. doi: 10.1093/hmg/ddh299. [DOI] [PubMed] [Google Scholar]
- Blennow K, Hesse C, Fredman P. Cerebrospinal fluid apolipoprotein E is reduced in Alzheimer's disease. Neuroreport. 1994;5:2534–2536. doi: 10.1097/00001756-199412000-00032. [DOI] [PubMed] [Google Scholar]
- Lehtimaki T, Pirttila T, Mehta PD, Wisniewski HM, Frey H, Nikkari T. Apolipoprotein E (apoE) polymorphism and its influence on ApoE concentrations in the cerebrospinal fluid in Finnish patients with Alzheimer's disease. Hum Genet. 1995;95:39–42. doi: 10.1007/BF00225071. [DOI] [PubMed] [Google Scholar]
- Landen M, Hesse C, Fredman P, Regland B, Wallin A, Blennow K. Apolipoprotein E in cerebrospinal fluid from patients with Alzheimer's disease and other forms of dementia is reduced but without any correlation to the apoE4 isoform. Dementia. 1996;7:273–278. doi: 10.1159/000106892. [DOI] [PubMed] [Google Scholar]
- Mulder M, Ravid R, Swaab DF, de Kloet ER, Haasdijk ED, Julk J, van der Boom JJ, Havekes LM. Reduced levels of cholesterol, phospholipids, and fatty acids in cerebrospinal fluid of Alzheimer disease patients are not related to apolipoprotein E4. Alzheimer Dis Assoc Disord. 1998;12:198–203. doi: 10.1097/00002093-199809000-00012. [DOI] [PubMed] [Google Scholar]
- Lefranc D, Vermersch P, Dallongeville J, Daems-Monpeurt C, Petit H, Delacourte A. Relevance of the quantification of apolipoprotein E in the cerebrospinal fluid in Alzheimer's disease. Neurosci Lett. 1996;212:91–94. doi: 10.1016/0304-3940(96)12774-9. [DOI] [PubMed] [Google Scholar]
- Lindh M, Blomberg M, Jensen M, Basun H, Lannfelt L, Engvall B, Scharnagel H, Marz W, Wahlund LO, Cowburn RF. Cerebrospinal fluid apolipoprotein E (apoE) levels in Alzheimer's disease patients are increased at follow up and show a correlation with levels of tau protein. Neurosci Lett. 1997;229:85–88. doi: 10.1016/S0304-3940(97)00429-1. [DOI] [PubMed] [Google Scholar]
- Schiele F, De Bacquer D, Vincent-Viry M, Beisiegel U, Ehnholm C, Evans A, Kafatos A, Martins MC, Sans S, Sass C, Visvikis S, De Backer G, Siest G. Apolipoprotein E serum concentration and polymorphism in six European countries: the ApoEurope Project. Atherosclerosis. 2000;152:475–488. doi: 10.1016/S0021-9150(99)00501-8. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Holtzman DM. Differential metabolism of ApoE isoforms in plasma and CSF. Exp Neurol. 2003;183:4–6. doi: 10.1016/S0014-4886(03)00185-7. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, Holtzman DM. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J Biol Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- Hirsch-Reinshagen V, Zhou S, Burgess BL, Bernier L, McIsaac SA, Chan JY, Tansley GH, Cohn JS, Hayden MR, Wellington CL. Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain. J Biol Chem. 2004;279:41197–41207. doi: 10.1074/jbc.M407962200. [DOI] [PubMed] [Google Scholar]
- Koldamova R, Staufenbiel M, Lefterov I. Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. J Biol Chem. 2005;280:43224–43235. doi: 10.1074/jbc.M504513200. [DOI] [PubMed] [Google Scholar]
- Oram JF. Molecular basis of cholesterol homeostasis: lessons from Tangier disease and ABCA1. Trends Mol Med. 2002;8:168–173. doi: 10.1016/S1471-4914(02)02289-X. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Frikke-Schmidt R, Nordestgaard BG, Jensen GB, Tybjaerg-Hansen A. Genetic variation in ABC transporter A1 contributes to HDL cholesterol in the general population. J Clin Invest. 2004;114:1343–1353. doi: 10.1172/JCI200420361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst MC, Thumann H, Aslanidis C, Langmann T, Buechler C, Patsch W, Baralle FE, Dallinga-Thie GM, Geisel J, Keller C, Menys VC, Schmitz G. Screening for functional sequence variations and mutations in ABCA1. Atherosclerosis. 2004;175:269–279. doi: 10.1016/j.atherosclerosis.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Clee SM, Zwinderman AH, Engert JC, Zwarts KY, Molhuizen HO, Roomp K, Jukema JW, van Wijland M, van Dam M, Hudson TJ, Brooks-Wilson A, Genest J, Jr., Kastelein JJ, Hayden MR. Common genetic variation in ABCA1 is associated with altered lipoprotein levels and a modified risk for coronary artery disease. Circulation. 2001;103:1198–1205. doi: 10.1161/01.cir.103.9.1198. [DOI] [PubMed] [Google Scholar]
- Wang J, Burnett JR, Near S, Young K, Zinman B, Hanley AJ, Connelly PW, Harris SB, Hegele RA. Common and rare ABCA1 variants affecting plasma HDL cholesterol. Arterioscler Thromb Vasc Biol. 2000;20:1983–1989. doi: 10.1161/01.atv.20.8.1983. [DOI] [PubMed] [Google Scholar]
- Wollmer MA, Streffer JR, Lutjohann D, Tsolaki M, Iakovidou V, Hegi T, Pasch T, Jung HH, Bergmann K, Nitsch RM, Hock C, Papassotiropoulos A. ABCA1 modulates CSF cholesterol levels and influences the age at onset of Alzheimer's disease. Neurobiol Aging. 2003;24:421–426. doi: 10.1016/S0197-4580(02)00094-5. [DOI] [PubMed] [Google Scholar]
- Li Y, Tacey K, Doil L, van Luchene R, Garcia V, Rowland C, Schrodi S, Leong D, Lau K, Catanese J, Sninsky J, Nowotny P, Holmans P, Hardy J, Powell J, Lovestone S, Thal L, Owen M, Williams J, Goate A, Grupe A. Association of ABCA1 with late-onset Alzheimer's disease is not observed in a case-control study. Neurosci Lett. 2004;366:268–271. doi: 10.1016/j.neulet.2004.05.047. [DOI] [PubMed] [Google Scholar]
- Katzov H, Chalmers K, Palmgren J, Andreasen N, Johansson B, Cairns NJ, Gatz M, Wilcock GK, Love S, Pedersen NL, Brookes AJ, Blennow K, Kehoe PG, Prince JA. Genetic variants of ABCA1 modify Alzheimer disease risk and quantitative traits related to beta-amyloid metabolism. Hum Mutat. 2004;23:358–367. doi: 10.1002/humu.20012. [DOI] [PubMed] [Google Scholar]
- Sundar PD, Feingold E, Minster RL, Dekosky ST, Kamboh MI. Gender-specific association of ATP-binding cassette transporter 1 (ABCA1) polymorphisms with the risk of late-onset Alzheimer's disease. Neurobiol Aging. 2006 doi: 10.1016/j.neurobiolaging.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Santoro L, Kemmerer B, Hall JL, Saunders AM, Roses AD, Small GW, Scott WK, Conneally PM, Vance JM, Haines JL. Identification of novel genes in late-onset Alzheimer's disease. Exp Gerontol. 2000;35:1343–1352. doi: 10.1016/S0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- Myers A, Wavrant De-Vrieze F, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Knowles H, Morris JC, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, Rice F, Giles P, Tunstall N, Jones L, Lovestone S, Williams J, Owen MJ, Hardy J, Goate A. Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet. 2002;114:235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, Mahoney A, Beaty T, Fallin MD, Avramopoulos D, Chase GA, Folstein MF, McInnis MG, Bassett SS, Doheny KJ, Pugh EW, Tanzi RE. Results of a high-resolution genome screen of 437 Alzheimer's disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- Scott WK, Hauser ER, Schmechel DE, Welsh-Bohmer KA, Small GW, Roses AD, Saunders AM, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA. Ordered-subsets linkage analysis detects novel Alzheimer disease loci on chromosomes 2q34 and 15q22. Am J Hum Genet. 2003;73:1041–1051. doi: 10.1086/379083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe P, Wavrant-De Vrieze F, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, Norton N, Williams H, Williams N, Lovestone S, Perez-Tur J, Hutton M, Chartier-Harlin MC, Shears S, Roehl K, Booth J, Van Voorst W, Ramic D, Williams J, Goate A, Hardy J, Owen MJ. A full genome scan for late onset Alzheimer's disease. Hum Mol Genet. 1999;8:237–245. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- Fukumoto H, Ingelsson M, Garevik N, Wahlund LO, Nukina N, Yaguchi Y, Shibata M, Hyman BT, Rebeck GW, Irizarry MC. APOE epsilon 3/ epsilon 4 heterozygotes have an elevated proportion of apolipoprotein E4 in cerebrospinal fluid relative to plasma, independent of Alzheimer's disease diagnosis. Exp Neurol. 2003;183:249–253. doi: 10.1016/S0014-4886(03)00088-8. [DOI] [PubMed] [Google Scholar]
- Burns MP, Vardanian L, Pajoohesh-Ganji A, Wang L, Cooper M, Harris DC, Duff K, Rebeck GW. The effects of ABCA1 on cholesterol efflux and Abeta levels in vitro and in vivo. J Neurochem. 2006 doi: 10.1111/j.1471-4159.2006.03925.x. [DOI] [PubMed] [Google Scholar]
- Bateman RJ, Wen G, Morris JC, Holtzman DM. Fluctuations of CSF amyloid-beta levels: implications for a diagnostic and therapeutic biomarker. Neurology. 2007;68:666–669. doi: 10.1212/01.wnl.0000256043.50901.e3. [DOI] [PubMed] [Google Scholar]
- Schneider WJ, Kovanen PT, Brown MS, Goldstein JL, Utermann G, Weber W, Havel RJ, Kotite L, Kane JP, Innerarity TL, Mahley RW. Familial dysbetalipoproteinemia. Abnormal binding of mutant apoprotein E to low density lipoprotein receptors of human fibroblasts and membranes from liver and adrenal of rats, rabbits, and cows. J Clin Invest. 1981;68:1075–1085. doi: 10.1172/JCI110330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer JD, Demattos RB, McCormick LM, O'Dell MA, Spinner ML, Bales KR, Paul SM, Sullivan PM, Parsadanian M, Bu G, Holtzman DM. The low density lipoprotein receptor regulates the level of central nervous system human and murine apolipoprotein E but does not modify amyloid plaque pathology in PDAPP mice. J Biol Chem. 2005;280:25754–25759. doi: 10.1074/jbc.M502143200. [DOI] [PubMed] [Google Scholar]
- Zerbinatti CV, Wahrle SE, Kim H, Cam JA, Bales K, Paul SM, Holtzman DM, Bu G. Apolipoprotein E and low density lipoprotein receptor-related protein facilitate intraneuronal Abeta 42 accumulation in amyloid model mice. J Biol Chem. 2006 doi: 10.1074/jbc.M604436200. [DOI] [PubMed] [Google Scholar]
- Nicoll JA, Burnett C, Love S, Graham DI, Dewar D, Ironside JW, Stewart J, Vinters HV. High frequency of apolipoprotein E epsilon 2 allele in hemorrhage due to cerebral amyloid angiopathy. Ann Neurol. 1997;41:716–721. doi: 10.1002/ana.410410607. [DOI] [PubMed] [Google Scholar]
- Greenberg SM, Vonsattel JP, Segal AZ, Chiu RI, Clatworthy AE, Liao A, Hyman BT, Rebeck GW. Association of apolipoprotein E epsilon2 and vasculopathy in cerebral amyloid angiopathy. Neurology. 1998;50:961–965. doi: 10.1212/wnl.50.4.961. [DOI] [PubMed] [Google Scholar]
- Chapman J, Vinokurov S, Achiron A, Karussis DM, Mitosek-Szewczyk K, Birnbaum M, Michaelson DM, Korczyn AD. APOE genotype is a major predictor of long-term progression of disability in MS. Neurology. 2001;56:312–316. doi: 10.1212/wnl.56.3.312. [DOI] [PubMed] [Google Scholar]
- Friedman G, Froom P, Sazbon L, Grinblatt I, Shochina M, Tsenter J, Babaey S, Yehuda B, Groswasser Z. Apolipoprotein E-epsilon4 genotype predicts a poor outcome in survivors of traumatic brain injury. Neurology. 1999;52:244–248. doi: 10.1212/wnl.52.2.244. [DOI] [PubMed] [Google Scholar]
- Morris JC, McKeel DW, Jr., Fulling K, Torack RM, Berg L. Validation of clinical diagnostic criteria for Alzheimer's disease. Ann Neurol. 1988;24:17–22. doi: 10.1002/ana.410240105. [DOI] [PubMed] [Google Scholar]
- Morris JC, Storandt M, McKeel DW, Jr., Rubin EH, Price JL, Grant EA, Berg L. Cerebral amyloid deposition and diffuse plaques in "normal" aging: Evidence for presymptomatic and very mild Alzheimer's disease. Neurology. 1996;46:707–719. doi: 10.1212/wnl.46.3.707. [DOI] [PubMed] [Google Scholar]
- Berg L, McKeel DW, Jr., Miller JP, Storandt M, Rubin EH, Morris JC, Baty J, Coats M, Norton J, Goate AM, Price JL, Gearing M, Mirra SS, Saunders AM. Clinicopathologic studies in cognitively healthy aging and Alzheimer's disease: relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch Neurol. 1998;55:326–335. doi: 10.1001/archneur.55.3.326. [DOI] [PubMed] [Google Scholar]
- Krul ES, Tikkanen MJ, Schonfeld G. Heterogeneity of apolipoprotein E epitope expression on human lipoproteins: importance for apolipoprotein E function. J Lipid Res. 1988;29:1309–1325. [PubMed] [Google Scholar]
- Germer S, Holland MJ, Higuchi R. High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res. 2000;10:258–266. doi: 10.1101/gr.10.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]