Abstract
Regulation of spatio-temporal gene expression in diverse cell and tissue types is a critical aspect of development. Progression through Caenorhabditis elegans vulval development leads to the generation of seven distinct vulval cell types (vulA, vulB1, vulB2, vulC, vulD, vulE, and vulF), each with its own unique gene expression profile. The mechanisms that establish the precise spatial patterning of these mature cell types are largely unknown. Dissection of the gene regulatory networks involved in vulval patterning and differentiation would help us understand how cells generate a spatially defined pattern of cell fates during organogenesis. We disrupted the activity of 508 transcription factors via RNAi and assayed the expression of ceh-2, a marker for vulB fate during the L4 stage. From this screen, we identified the tailless ortholog nhr-67 as a novel regulator of gene expression in multiple vulval cell types. We find that one way in which nhr-67 maintains cell identity is by restricting inappropriate cell fusion events in specific vulval cells, namely vulE and vulF. nhr-67 exhibits a dynamic expression pattern in the vulval cells and interacts with three other transcriptional regulators cog-1 (Nkx6.1/6.2), lin-11 (LIM), and egl-38 (Pax2/5/8) to generate the composite expression patterns of their downstream targets. We provide evidence that egl-38 regulates gene expression in vulB1, vulC, vulD, vulE, as well as vulF cells. We demonstrate that the pairwise interactions between these regulatory genes are complex and vary among the seven cell types. We also discovered a striking regulatory circuit that affects a subset of the vulval lineages: cog-1 and nhr-67 inhibit both one another and themselves. We postulate that the differential levels and combinatorial patterns of lin-11, cog-1, and nhr-67 expression are a part of a regulatory code for the mature vulval cell types.
Author Summary
During development, in which the single-celled egg generates a whole organism, cells become different from each other and form patterns of types of cells. It is these spatially defined fate patterns that underlie the formation of complex organs. Regulatory molecules called transcription factors influence the fate patterns that cells adopt. Understanding the role of these transcription factors and their interactions with other genes could tell us how cells establish a certain pattern of cell fates. This study focuses on studying how the seven cell types of the Caenorhabditis elegans vulva arise. This organ is one of the most intensively studied, and while the signaling network that initiates vulval development and sets the gross pattern of cell differentiation is well understood, the network of transcription factors that specifies the final cell fates is not understood. Here, we identify nhr-67, a new transcription factor that regulates patterning of cell fates in this organ. Transcription factors do not necessarily act alone, and we explore how NHR-67 works with three other regulatory factors (each with human homologs) to specify the different properties of the vulval cells. We also demonstrate that the interconnections of these transcription factors differ between these seven diverse cell types, which may partially account for how these cells acquire a certain pattern of cell fates.
Introduction
Complex gene regulatory networks operating in diverse cell types and tissues are crucial for development. Diverse intercellular signals and transcription factor networks control gene expression within individual cell types, acting on cis-regulatory modules of target genes [1]. Understanding such regulation first requires documenting all the regulatory inputs and outputs from each gene [2]. This information allows circuit diagrams to be constructed that provide a global perspective on how diverse cell types acquire their identity. Gene regulatory networks have been well studied in a wide range of biological model systems such as endomesoderm specification in the sea urchin embryo [3], dorso-ventral patterning in the Drosophila embryo [4], and mesoderm specification in Xenopus [5]. The common themes that might emerge from these studies would advance our understanding of organogenesis in vertebrates.
The Caenorhabditis elegans vulva is postembryonically derived from six vulval precursor cells P3.p–P8.p. The central three vulval precursor cells P5.p–P7.p are induced to adopt 1° (primary) and 2° (secondary) vulval fates via epidermal growth factor (EGF) and Notch signaling, whereas the remaining precursors fuse with the hypodermal syncytium hyp7 [6]. The vulva is composed of seven distinct cell types, each with its own set of expressed genes and morphogenetic migrations [7–9]. The P6.p 1° lineages generate the vulE and vulF cells, while the P5.p and P7.p 2° lineages generate the vulA, vulB1, vulB2, vulC, and vulD cells. The signals that induce 1° versus 2° fates in the primordial vulval precursor cells are known. However, the processes that govern patterning and differentiation of the mature vulval cell types are largely unknown [6]. Both Ras and Wnt pathways are required for the precise spatial patterning of the 1° vulE and vulF cells [10], and both Wnt/Ryk and Wnt/Frizzled signaling pathways are necessary for patterning the P7.p 2° vulA–vulD cells [11–13].
Genes expressed in the mature vulval cell types include some with known functions and many others without known physiological roles. lin-3 (EGF) is expressed in vulF and is required to signal from vulF to uterine uv1 cells [14,15]. egl-17 encodes a fibroblast growth factor (FGF)-like protein that is required for migration of the sex myoblasts to their precise final positions [16,17]. egl-17 is initially expressed in the 1° vulval lineages and is shut off during the L4 stage. Expression in vulC and vulD is observed during early L4 and persists throughout adulthood. The vulval expression correlates with the sites of muscle attachment. egl-26 encodes a novel protein that contains an H box/NC domain and is expressed in vulB1, vulB2, vulD, and vulE cells [18,19]. zmp-1 encodes a zinc metalloprotease and is expressed in vulD and vulE during the L4 stage and in vulA in adults [9]. ceh-2 encodes a homeodomain protein that is related to Drosophila empty spiracles and is expressed in vulB1 and vulB2 cells during the L4 stage and in vulC upon entry into L4 lethargus [9,20]. pax-2 is a recent gene duplication of the PAX2/5/8 protein EGL-38 [21] and is expressed exclusively in the vulD cells. zmp-1, ceh-2, egl-26, and pax-2 have no known function in the vulva.
Transcription factor networks in individual vulval cell types somehow generate a spatially precise pattern of cell fates [19]. Several transcription factors that regulate gene expression in the diverse vulval cell types have already been described [19,22–24]. lin-11, a LIM homeobox transcription factor, regulates gene expression in all seven vulval cell types [25,26]. The Nkx6.1/Nkx6.2 homeodomain gene, cog-1, regulates gene expression in vulB, vulC, vulD, vulE, and vulF cells [19,27]. In contrast, egl-38 encodes a PAX2/5/8 protein that appears to be the only known example of a vulval cell type–specific regulatory factor; it promotes expression of certain target genes and restricts expression of other targets exclusively in vulF cells [14,19,28]. Additional regulatory factors need to be identified to elucidate the precise spatial patterning of the mature vulval cell types.
Here, we identify nhr-67 as a component of the gene regulatory networks underlying vulval patterning and differentiation. nhr-67 is required for the accurate patterning of gene expression and regulation of cell fusion in several vulval cell types and is dynamically expressed in the vulva. nhr-67 interacts genetically with cog-1, egl-38, and lin-11 to produce the complex expression patterns of their downstream targets. We demonstrate that the pairwise interactions between these four regulatory genes vary among the diverse vulval cell types. These results indicate that nhr-67, cog-1, lin-11, and egl-38 form a part of a genetic network that generates different patterns of gene expression in each of the seven cell types.
Results
nhr-67 Regulates Gene Expression in Multiple Vulval Cell Types
An RNA interference (RNAi) screen of 508 known and putative transcription factors encoded in the C. elegans genome (see Table S1) was conducted in a ceh-2::YFP reporter background. At the time we performed the screen, this was the best available set. ceh-2 encodes a homeodomain protein orthologous to Drosophila Empty Spiracles (EMS) and vertebrate EMX1 and EMX2 and serves as a readout for vulB fate during the L4 stage [20]. Modifiers of ceh-2 expression are good candidates for genes involved in patterning and/or differentiation of 2° vulval descendents. From this screen, we identified nhr-67 as a gene necessary for negative regulation of ceh-2 expression in the 1° vulE and vulF cells (Figure 1A–1B). Reciprocal BLAST searches indicate that nhr-67 encodes an ortholog of the tailless hormone receptor, which consists of an N-terminal transactivation domain, a centrally positioned DNA-binding domain, and a C-terminal ligand-binding domain. The only other positive was the GATA-type transcription factor egl-18, which was previously shown to be involved in vulval development [29–31]. Other genes that should have been positive in the screen (lin-11 and cog-1) were not isolated from the RNAi screen, thus indicating a high false-negative rate. Analysis of the nhr-67 deletion allele ok631 revealed severe defects in early larval development (L1 lethality and/or arrest). In order to bypass this early larval arrest phenotype, we resorted to feeding young L1 larvae with nhr-67 RNAi and assayed for defects in vulval gene expression. nhr-67 was also found to be required for negative regulation of two additional L4-specific markers: egl-26 (wild-type expression in vulB, vulD, and vulE cells) (Figure 1C–1D) and egl-17 (wild-type expression in vulC and vulD cells) in the vulF cells. Thus, nhr-67 activity is necessary for the negative regulation of expression of several 2° lineage-specific genes in the 1°-derived vulval cells during the L4 stage. Consistent with previous reports, nhr-67 RNAi results in a highly penetrant protruding vulva (Pvl) and egg-laying (Egl) defective phenotype [32] (Figure S1). However, other transcription factors exhibiting a Pvl RNAi phenotype, such as fos-1, egl-43, and unc-62, have normal vulval gene expression (unpublished data).
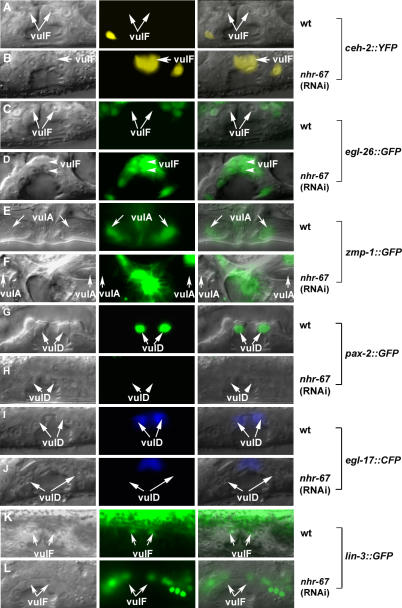
Figure 1. nhr-67 Is Required for Proper Gene Expression in Multiple Vulval Cell Types.
Lateral images of the developing vulva during the L4 (A–D and G–L) and the adult stage (E and F). (A–L) Nomarski (left), fluorescence (center), and overlaid (right). Expression of vulval cell fate markers in wild-type (A, C, E, G, I, and K) and nhr-67 RNAi–treated animals (B, D, F, H, J, and L). In nhr-67 RNAi–treated animals, the vulval morphology is abnormal compared to wild-type; namely, the migration of vulF cells is defective. (A) In wild-type animals, syIs55 [ceh-2::YFP] expression is off in the 1° vulF cells (arrows).
(B) syIs55 animals treated with nhr-67 RNAi show ectopic ceh-2 expression in the 1° vulF cells (arrow).
(C) kuIs36 [egl-26::GFP] expression is completely absent in the vulF cells (arrows).
(D) nhr-67 RNAi results in misexpression of egl-26 in the vulF lineages (arrowheads).
(E) Wild-type zmp-1::GFP (syIs49) expression is observed in the vulA cells (arrows).
(F) In contrast, nhr-67 RNAi abolishes the vulA-specific expression (arrows) of zmp-1.
(G) guEx64 [pax-2::GFP] is expressed exclusively in vulD cells (arrows) in wild-type animals.
(H) pax-2 expression in vulD is abolished in an nhr-67 (RNAi) background (arrows).
(I) Wild-type egl-17::GFP (syIs59) expression is observed in the vulD cells (arrows).
(J) nhr-67 RNAi results in the loss of egl-17 expression in the vulD cells (arrows).
(K) In wild-type animals, lin-3::GFP (syIs107) is expressed solely in vulF cells (arrows).
(L) lin-3 expression in vulF cells is eliminated when treated with nhr-67 RNAi (arrows).
In addition to its negative regulatory role, we also found that nhr-67 is necessary for promoting expression of specific genes. For example, nhr-67 is necessary for zmp-1 expression in vulA during the adult stage (Figure 1E–1F). nhr-67 is also required for vulD-specific expression of pax-2 and egl-17 during the L4 stage (Figure 1G–1J). These examples show that nhr-67 positively regulates gene expression in the secondary vulA and vulD cells. nhr-67 is also required for positively regulating gene expression in the 1° vulval cells, namely vulF-specific expression of lin-3, an EGF-like protein (Figure 1K and 1L). Therefore, nhr-67 regulates gene expression in at least four of the seven vulval cell types.
In the L3 stage, the early 1° and 2° vulval cell fates can be distinguished by the patterns of cell division of their descendents. The 1° fated cell typically gives rise to four granddaughters that divide transversely (left-right axes); whereas a subset of the granddaughters derived from a 2° cell divide longitudinally (anterior-posterior axes). To determine if nhr-67-dependent alterations in gene expression are a consequence of fate transformations in the early 1° and 2° vulval lineages, we monitored the pattern of the vulval cell divisions in an nhr-67 RNAi background. In the absence of nhr-67, the vulval cell lineages appear wild-type in terms of both cell number and orientation of cell division (unpublished data). Thus, the perturbations in gene expression caused by reduced nhr-67 function are not the result of gross abnormalities in the early vulval cell lineages.
nhr-67 Prevents Inappropriate Fusion Events between the 1° Vulval Cells
During the L4 stage, the seven vulval cell types invaginate cooperatively to assume a characteristic morphology. The similar cell types subsequently fuse, generating toroid rings that line the vulval cavity [8]. We wanted to ascertain if the observed cell fate transformations in nhr-67(RNAi) animals were possibly due to improper fusion events between the wrong cell types. Cell fusion defects can be assayed using ajm-1::GFP (an adherens junction marker) to visualize the cell number and architecture of the vulval toroids. When observing the mid-sagittal plane of wild-type animals, ajm-1::GFP appears as dots between cells. The eight dots on either side correspond to the seven distinct vulval cell types (Figure 2A). Most nhr-67 RNAi–treated animals do not exhibit dramatic defects in cell fusion (Figure 2B). The 2° vulval lineage–derived cells (vulA, vulB1, vulB2, vulC, and vulD) consistently generate mature toroids. However, inappropriate fusion often occurs (65%, n = 17) between the presumptive vulE and vulF cells (indicated by the missing dots at the top of the vulval invagination) (Figure 2C). Since nhr-67 regulates gene expression in vulval cells other than vulE and vulF, improper cell fusion events cannot fully account for all its altered gene expression patterns.
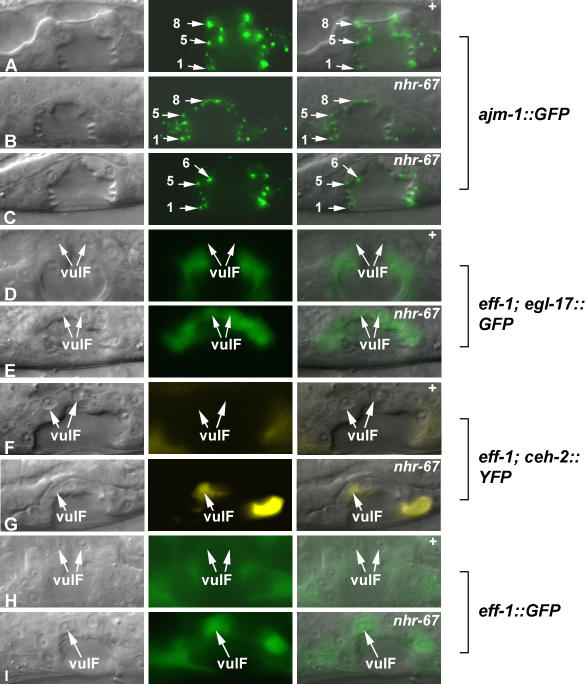
Figure 2. nhr-67 Prevents Inappropriate Fusion Events between the 1° Vulval Cells.
(A–I) Nomarski (left), fluorescence (center), and overlaid (right). The adherens junction marker ajm-1::GFP is used to visualize the cell number and architecture of the vulval toroids in wild-type (A) and nhr-67 RNAi–treated animals (B and C). When observing a mid-sagittal optical section of L4 hermaphrodites, ajm-1::GFP appears as dots between the vulval cells. Loss of adherens junction expression signifies a reduction in the cell number due to a cell fusion defect. (A) In wild-type animals, the eight dots on either side correspond to the seven distinct vulval cell types (arrows). The overall vulval morphology of nhr-67 RNAi–treated animals appears abnormal compared to wild-type.
(B) In some cases, the number of adherens junctions is normal in nhr-67 RNAi–treated animals (arrows).
(C) Reduction of nhr-67 sometimes results in the loss of dots at the top of the vulval invagination (which indicates an inappropriate fusion event between the vulE and vulF cells) (arrows). However, the altered gene expression observed in an nhr-67 RNAi background does not appear to be dependent on cell fusion defects.
(D) In the absence of eff-1-mediated fusion, ayIs4 [egl-17::GFP] expression is completely absent in the vulF cells (arrows).
(E) In contrast, depletion of nhr-67 activity in an eff-1 mutant background is sufficient to cause derepression of egl-17 in the 1° vulF cells (arrows).
(F) In eff-1 mutants, ceh-2 expression is absent in vulF cells (arrows).
(G) Reduction of nhr-67 activity in an eff-1 mutant background results in ectopic ceh-2 expression in vulF cells (arrow).
(H) In wild-type animals, eff-1::GFP is not expressed in vulF cells (arrows).
(I) eff-1 levels are elevated in vulF cells when nhr-67 activity is compromised (arrow).
We then wanted to determine if the altered gene expression occurring in the 1° vulval cells was dependent on these improper fusion events. We attempted to address this question using two approaches: (a) by analyzing the effect of nhr-67 RNAi on the expression of egl-17 and ceh-2 transgenes in an eff-1(hy21) background, and (b) by monitoring the vulval expression levels of eff-1 in animals with reduced nhr-67 activity. eff-1 is a type I membrane protein necessary for cell fusion [33]. Disruption of nhr-67 function in an eff-1-deficient background is still sufficient to cause upregulation of both egl-17 (Figure 2D and 2E) and ceh-2 (Figure 2F and 2G) in the 1° vulval cells. Thus, the nhr-67-dependent alterations in gene expression are not dependent on eff-1-mediated cell fusion. We also observed that eff-1 levels (strong expression in vulA and vulC cells, weak expression in vulF cells) are highly elevated in vulD and vulF cells when nhr-67 gene activity is compromised (Figure 2H and 2I). However, we also note that eff-1 is not sufficient to rescue the vulE-vulF fusion defects observed in nhr-67 (RNAi) background (unpublished data). One possibility is that eff-1(hy27) is a temperature-sensitive allele that fails to completely eliminate cell fusion. Another possibility is that in addition to eff-1, nhr-67 negatively regulates other target genes that mediate cell fusion.
nhr-67 Is Dynamically Expressed in Multiple Vulval Cells
Previous work reported that an nhr-67 construct containing 6 kb of the promoter region directs expression in several head neurons [34]. We generated several additional transcriptional reporter constructs that tested the entire nhr-67 coding region, introns and the 3′ noncoding region for enhancer activity using the Δpes-10 basal promoter [35]. An 8-kb fragment that consisted of 1-kb 5′ sequence, the entire coding region and introns, and 2 kb of the 3′ noncoding region yielded expression in the vulva, the hyp7 epidermal syncytium, late stage embryos, and the male tail (Figures 3 and 4A). This nhr-67 construct exhibits a dynamic expression pattern in the vulval cells. During the late L4 stage, nhr-67 is first observed in vulA cells (Figure 3A) (and occasionally in vulB1), and this expression is maintained throughout adulthood. Expression in vulC is only seen upon entry into L4 lethargus and persists in adults (Figure 3B). Strong vulB1 and vulB2 expression (and occasional vulD expression) is observed only in young adults (Figure 3C). A 4.5-kb reporter construct that spans from the fourth intron to the 3′ noncoding region is sufficient to drive expression in the same tissues as seen with the 8-kb fragment (Figure 4B). No expression is seen in the vulC, vulD, vulE, and vulF cells during the L4 stage unless nhr-67 or cog-1 activity is eliminated (see below). Thus, the cis-elements driving the vulval expression of nhr-67 appear to be located in the region spanning the fourth intron to the 3′ noncoding region. We then wanted to confirm if these regulatory elements were capable of interacting with the endogenous promoter of nhr-67 in order to promote its transcription in the vulva. To test this, we generated an nhr-67 transcriptional reporter driven by 1 kb of its native promoter and containing regulatory sequences downstream of the fourth exon in their normal context. The nhr-67 transcriptional construct containing the endogenous promoter recapitulated the vulval and embryonic expression pattern observed with the nhr-67::Δpes-10 constructs (Figure 4C). We also examined whether the upstream regulatory sequences of nhr-67 interact with the downstream regulatory elements to influence its vulval expression. This test was accomplished by coinjecting a transcriptional green fluorescent protein (GFP) construct that contains a 6-kb upstream sequence of nhr-67 (Figure 4D) with the 8-kb nhr-67::Δpes-10 construct (Figure 4A) described above. We find that in the presence of the 6-kb promoter region, the vulval expression is identical to that of the 8-kb nhr-67::Δpes-10 constructs. Besides the previously reported expression in head neurons, we observed expression in the anchor cell (AC) (during mid–late L3 stage) in hermaphrodites and the linker cell in males (Figure S2A and S2B).
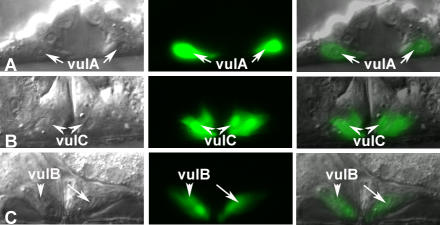
Figure 3. nhr-67 Is Dynamically Expressed in Multiple Vulval Cell Types.
(A–C) Nomarski (left), fluorescence (center), and overlaid (right). All animals displayed carry the syEx716 transgene in their background. (A) nhr-67 is robustly expressed in the vulA cells (arrows) during the L4 stage.
(B) vulC expression (arrowheads) is visible upon entry into L4 lethargus.
(C) High levels of nhr-67 expression in vulB1 and vulB2 cells (arrowhead and arrow) are detectable in young adults.
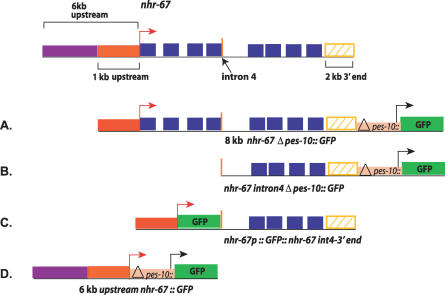
Figure 4. The Regulatory Element(s) Driving the Vulval Expression of nhr-67 Is Present in the Region That Spans from the Fourth Intron to the 3′ Noncoding Region.
Several transcriptional reporter constructs containing the nhr-67 coding exons (blue rectangles), introns (black lines), and the 3′ noncoding region (yellow hatched rectangle) were generated. The red arrow indicates the presumptive promoter of nhr-67 and the black arrow is proximal to the minimal Δpes-10 promoter. The yellow rectangle includes 2 kb of the 3′ noncoding region. The orange vertical bar indicates the junction between the fourth exon and fourth intron. Construct (A) consists of 1 kb upstream promoter sequence (red rectangle), the entire coding region (blue rectangles) and introns (black lines), and 2 kb of the 3′ noncoding region (yellow hatched rectangle) attached to minimal Δpes-10::GFP.
Construct (B) spans from the fourth intron (gene sequence downstream of the orange vertical bar) to the 3′ noncoding region (yellow rectangle) fused to minimal Δpes-10::GFP.
Construct (C) is an nhr-67::GFP transcriptional reporter driven by 1 kb of the native promoter region (red rectangle) and contains regulatory sequences 3′ of the fourth exon (sequences downstream of the orange vertical bar) and the native 3′ noncoding region (yellow rectangle).
Construct (D) contains 6-kb sequence upstream of the predicted first ATG of nhr-67 (purple and red rectangles) appended to minimal Δpes-10::GFP.
Regulation of egl-17 and ceh-2 Expression in the 1° Vulval Cells
We attempted to understand the trans-regulation of vulval expression in the diverse cell types by analyzing the regulation of two target genes in detail: egl-17 and ceh-2. To dissect the trans-regulation of these target genes, we constructed various double and triple mutant/RNAi combinations and assayed for alterations in gene expression in the 1° vulval cells.
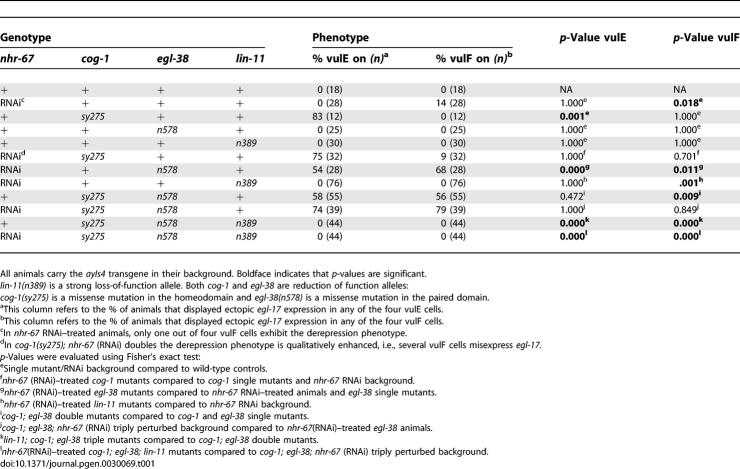
During the L4 stage, the egl-17 transcriptional reporter is expressed solely in vulC and vulD, being absent in both vulE and vulF (Figure 5 and Table 1). nhr-67 RNAi in an otherwise wild-type background results in an increase of egl-17 expression in the vulF cells (Figure 5 and Table 1). In those nhr-67 RNAi animals, only one of the four vulF cells exhibits this ectopic egl-17 expression during the L4 stage. egl-17 expression is consistently absent in the vulF cells of cog-1 and egl-38 hypomorphic alleles (Figure 5 and Table 1). In comparison, cog-1 animals treated with nhr-67 RNAi are qualitatively enhanced (i.e., several vulF cells misexpress egl-17), whereas egl-38 animals treated with nhr-67 RNAi displayed a qualitatively and quantitatively higher egl-17 expression in the vulF cells (Figure 5 and Table 1). cog-1 is necessary for negatively regulating egl-17 expression in the vulE cells and acts redundantly with egl-38 to negatively regulate egl-17 in the vulF cells [19] (Figure 5 and Table 1). We also observed frequent egl-17 upregulation in the vulE cells of egl-38; nhr-67 (RNAi) doubly perturbed hermaphrodites (Table 1), which is invariably absent in either singly perturbed background. Our study provides the first example of egl-38 modulating gene expression in the vulE cell type. Hence, egl-38, nhr-67, and cog-1 act together to negatively regulate egl-17 expression in the 1° vulval lineages during the L4 stage. Loss of lin-11 function leads to complete abolition of egl-17 gene expression in all vulval cells [26] (Figure 5 and Table 1). Lastly, the ectopic egl-17 expression visualized in the 1° descendents of cog-1-, egl-38-, and nhr-67-depleted backgrounds is dependent on lin-11 activity (Figure 5 and Table 1). Loss of nhr-67 in combination with lin-11 yields rare egl-17 expression in apparently random vulval cell types (∼4% of animals).
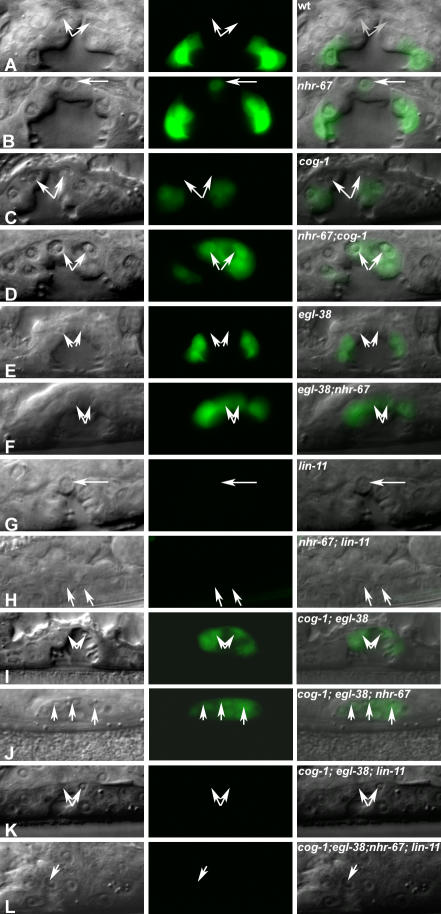
Figure 5. Regulation of egl-17 Expression in the 1° Vulval Cells.
(A–L) Nomarski (left), fluorescence (center), and overlaid (right). Display animals from Table 1. Arrows indicate the position of the visible vulF cells during the mid L4 stage. All animals displayed carry the ayIs4 transgene in their background. (A) In wild-type animals egl-17 expression is absent in the vulE and vulF cells.
(B) nhr-67 RNAi typically results in ectopic egl-17 expression in one out of four vulF cells. In contrast, egl-17 remains off in the vulE cells.
(C) In cog-1(sy275) mutants, egl-17 is misexpressed in vulE cells and is absent in vulF cells.
(D) cog-1(sy275); nhr-67 RNAi doubles show a markedly stronger derepression phenotype in both 1° vulE and vulF lineages.
(E) In egl-38(n578) mutants, egl-17 expression is invariably off in both vulE and vulF cells.
(F) egl-38(n578); nhr-67 RNAi doubles show robust expression of egl-17 in both vulE and vulF cells.
(G) In lin-11(n389) animals, egl-17 expression is absent.
(H) In lin-11(n389); nhr-67 RNAi doubles, the ectopic egl-17 expression in vulF is eliminated.
(I) cog-1(sy275); egl-38(n578) mutants misexpress egl-17 in vulE and vulF cells.
(J) The cog-1 (sy275); egl-38(n578); nhr-67 RNAi triple shows complete egl-17 derepression in all the vulE and vulF descendants.
(K) In cog-1(sy275); egl-38(n578); lin-11(n389) mutants, egl-17 is completely absent.
(L) In cog-1(sy275); egl-38(n578); lin-11(n389); nhr-67 RNAi quadruples, egl-17 expression in the vulva is abolished.
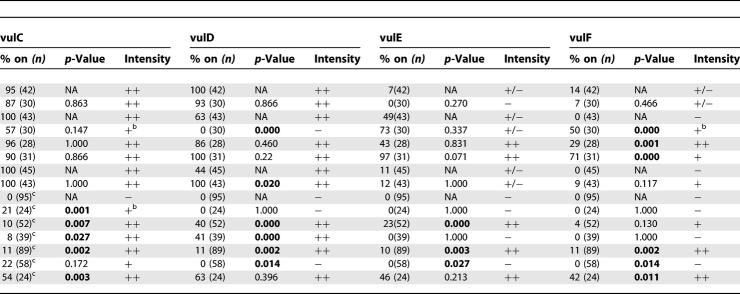
Table 1.
Regulation of egl-17 Expression in the 1° Vulval Lineages
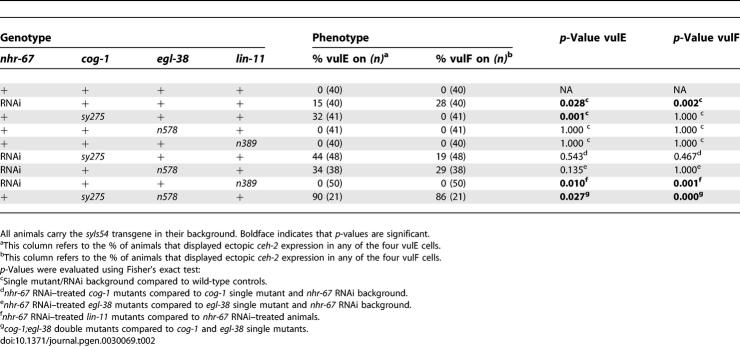
In wild-type L4 hermaphrodites, ceh-2::YFP expression is only observed in the vulB cells and is invariably absent in both vulE and vulF cells. nhr-67 RNAi results in a moderate frequency of ectopic ceh-2 expression in the vulE and vulF cells (Table 2). Eliminating lin-11 function leads to complete loss of ectopic ceh-2 expression in the 1° vulval lineages of nhr-67 RNAi animals (Table 2). ceh-2 expression is consistently absent in the 1° vulF cells of cog-1 and egl-38 single mutants (Table 2). cog-1 mutants exhibit a moderate increase of ceh-2 expression in the vulE cells [19] (Table 2). We also found that 90% of cog-1; egl-38 doubles show increased ceh-2 expression in vulE cells compared to cog-1 (32%) or egl-38 (0%) single mutants (Table 2). Thus, analysis of these double mutants provides us with a second example of egl-38 regulating gene expression in the vulE cells. As with the egl-17 reporter, simultaneous depletion of cog-1 and egl-38 activities results in a high frequency of ceh-2 misexpression in the vulF cells (Table 2). Both cog-1 and egl-38 are thus required for negative regulation of ceh-2 expression in the vulF cells.
Table 2.
Regulation of ceh-2 Expression in the 1° Vulval Lineages
Regulatory Interactions between Known Components of the Vulval Patterning Network during L4
cog-1, lin-11, and nhr-67, all of which regulate different aspects of vulval gene expression, exhibit dynamic spatial and temporal expression patterns in the developing vulva [26,27]. egl-38 expression has been observed in the vulF cells [15]. As mentioned previously, nhr-67 expression is primarily restricted to vulA (and occasionally vulB1) cells during L4 stage. Yet numerous perturbations in gene expression are observed in nhr-67 RNAi–treated animals, suggesting that nhr-67 is indeed functional during the L4 stage in other mature vulval cell types besides vulA (Figure 1). A similar observation can be made about cog-1. Wild-type animals occasionally exhibit weak cog-1 expression in vulE cells but none in vulF cells (Table 3). However, cog-1 synergistically interacts with egl-38 and nhr-67 to regulate egl-17 expression in the vulF cells (Figure 5 and Table 1). One attractive hypothesis is that levels of both these transcription factors are maintained under strict spatio-temporal control. We thus set out to investigate the interactions among these regulatory factors by assaying for alterations in the reporter gene expression in various mutant backgrounds.
Table 3.
Regulatory Interactions between Known Components of the Vulval Patterning Network during L4
Table 3.
Extended.
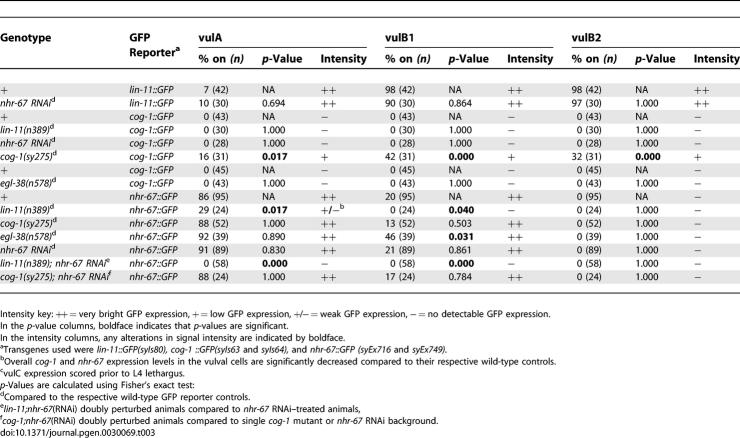
During the L4 stage, lin-11 is consistently expressed in the 2° vulB, vulC, and vulD lineages, and occasionally in the vulA and vulF cells. Neither cog-1 nor egl-38 mutations alter lin-11 vulval expression [36]. Similarly, reduction of nhr-67 gene activity also does not impact lin-11 expression in the vulva (Table 3).
The cog-1 translational reporter is strongly expressed in vulC and vulD, weakly expressed in vulE, and undetectable in vulF cells during L4 (Figure 6 and Table 3). We found that cog-1 levels are increased in the 1° vulF cells of nhr-67 RNAi–treated hermaphrodites as well as in lin-11 and egl-38 mutants (Figure 6 and Table 3). nhr-67 RNAi–treated animals also showed elevated cog-1 expression in the vulE cells (Table 3). In lin-11 mutants, cog-1 levels in vulD are completely abolished as opposed to the vulC-specific expression, which is only partially affected (∼57% of animals) (Table 3). Overall cog-1 expression levels in lin-11 loss-of-function mutants are noticeably reduced when compared to the wild-type reporter background. The frequency of vulD-specific cog-1 expression is significantly increased in egl-38 mutants (Table 3). cog-1 negatively autoregulates in vulA, vulB1, and vulB2 cells (Table 3).
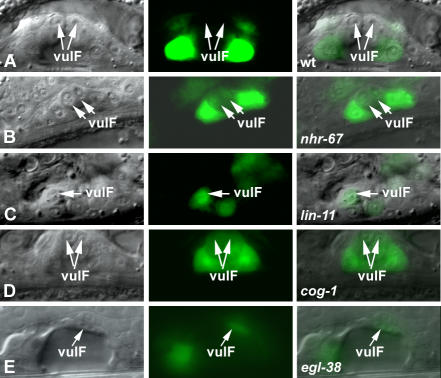
Figure 6. cog-1 Levels in the 1° VulF Cells Are Antagonized by Multiple Genes.
(A–E) Nomarski (left), fluorescence (center), and overlaid (right). Display animals from Table 3. All animals displayed carry either the syIs63 (A–D) or syIs64 (E) transgene in their background. (A) In wild-type animals, cog-1 expression is absent in the vulF cells (arrows).
(B) nhr-67 RNAi results in the derepression of cog-1 levels in the vulF cells (arrows).
(C) lin-11(n389) mutants show ectopic cog-1 expression in vulF (arrow).
(D) cog-1(sy275) mutants lose the ability to negatively autoregulate their expression levels in both vulE and vulF (arrows).
(E) cog-1 is ectopically expressed in vulF in an egl-38 mutant background (arrow).
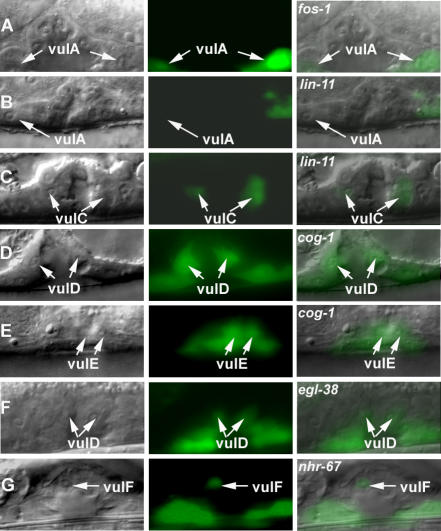
nhr-67::GFP expression is consistently observed in vulA during the L4 stage (Table 3). lin-11 mutants only partially eliminate the vulA-specific expression of nhr-67 (Figure 7 and Table 3). nhr-67 expression in vulA is completely abolished only in the absence of both lin-11 and its positive autoregulatory activity (Table 3). Overall, nhr-67 expression levels in lin-11 loss-of-function mutants are noticeably reduced when compared to a lin-11(+) background. lin-11 activity is also required for directing the ectopic nhr-67 expression in the 1° lineages when the autoregulatory loop is compromised (Table 3, see below). Also, loss of lin-11 sometimes caused premature vulC expression of nhr-67 during L4 stage, which can be interpreted either as a cell type or a temporal regulatory defect (Figure 7 and Table 3). Reduction of cog-1 function results in increased expression of nhr-67 in vulC and vulD during the L4 stage and vulE and vulF during L4 lethargus (Figure 7 and Table 3). Depletion of both cog-1 and nhr-67 activities leads to a more robust increase in nhr-67 levels in the vulF cells (Table 3). egl-38 mutants sometimes showed ectopic nhr-67 expression in vulC and vulD cells during the L4 stage (Figure 7 and Table 3) and significantly increased its frequency of expression in vulB1 cells (Table 3).
Figure 7. nhr-67 Is Differentially Regulated in the 1° and 2° Vulval Lineages.
(A–G) Nomarski (left), fluorescence (center), and overlaid (right). Display animals from Table 3. All animals displayed carry either syEx716 (A, D, E, and F) or syEx749 (B and C) [nhr-67::GFP] transgene in their background. (A) Wild-type animals treated with control fos-1 RNAi show no alteration of nhr-67 vulval expression compared to non-RNAi–treated animals. fos-1 RNAi animals exhibit abnormal vulval morphology, yet show wild-type nhr-67 expression in vulA cells (arrows).
(B) lin-11(n389) mutants partially eliminate the vulA-specific expression of nhr-67 (arrow).
(C) In a lin-11(n389) background, premature vulC expression (arrows) is observed sometimes during L4 stage.
(D) cog-1(sy275) mutants misexpress nhr-67 in the 2° vulD cells (arrows).
(E) In a cog-1(sy275) background, nhr-67 levels are highly elevated in the 1° vulE lineages (arrows).
(F) egl-38(n578) mutants show ectopic nhr-67 expression in vulD cells (arrows).
(G) nhr-67 RNAi feeding results in the robust increase in its own expression levels in vulF cells (arrow).
Negative Autoregulation of nhr-67 and cog-1 in 1° Vulval Lineages
In addition to the cross-inhibitory interactions between cog-1 and nhr-67 in both the 1° vulE and vulF cells, we also discovered that they both negatively autoregulate in the same cell types. Inhibition of nhr-67 by RNAi feeding results in the robust increase of nhr-67::GFP expression levels in both vulE and vulF cells (Figure 7 and Table 3). Elevation of nhr-67 transcriptional levels is also visible in the vulC and vulD lineages of nhr-67 (RNAi) animals during L4 stage. Upregulation of nhr-67 expression in vulC, vulD, vulE, and vulF cells is also visible with the 4.5-kb nhr-67 transcriptional reporter construct (Figure 4B) in an nhr-67(RNAi) background (unpublished data). We used fos-1 RNAi feeding as a control to exclude the possibility that the observed negative autoregulation was a nonspecific effect of inducing RNAi. fos-1 RNAi–treated animals exhibited a strong Pvl phenotype (at least in part due to its AC invasion phenotype) [37] and did not alter nhr-67 levels in the 1° lineages (Figure 7 and Table 3). Similarly, ectopic expression of cog-1::GFP in all 1° vulval descendents is consistently observed when cog-1 activity is compromised (Figure 6 and Table 3). Thus, nhr-67 and cog-1 appear to be activated in all the mature vulval cell types but are then restricted by both autoregulatory and trans-regulatory mechanisms.
Discussion
nhr-67: A Novel Regulator of Vulval Patterning in C. elegans
nhr-67 encodes a C. elegans ortholog of tailless, a crucial regulator of blastoderm patterning in the terminal pathway of Drosophila embryogenesis as well as neuronal development. We find that nhr-67 activity is required for the regulation of gene expression in several mature vulval cell types and is dynamically expressed in the vulva. For technical reasons, we have been unable to determine whether nhr-67 acts in the vulval cells for these functions. However, the expression of nhr-67 in the vulva and the complexity of the interactions are most consistent with a primarily autonomous action of nhr-67. However, given the expression of nhr-67 in the AC, it is possible that the effects (particularly on the 1° lineage) are nonautonomous. For example, the AC generates EGF and Wnt signals and is required to differentiate vulE and vulF cells, presumably via these signals [10]. Loss of vulF-specific lin-3 expression in an nhr-67 RNAi background is certainly consistent with this model. The AC also promotes 1° over 2° fate [38]. The ectopic expression of 2° lineage-specific genes ceh-2 and egl-17 in the 1° vulval cells is also consistent with this model. However, lineage analysis of nhr-67 (RNAi) hermaphrodites argues that these alterations are not full 1° to 2° cell fate transformations in the early vulval lineages. In addition, the observed effects on pax-2 and zmp-1 expression are inconsistent with this model. It remains a formal possibility that some of nhr-67 effects in the vulva are due to a role in the AC.
Our data are consistent with the function of Drosophila tailless, which facilitates proper gap gene expression at the posterior end of the blastoderm embryo via its dual activator/repressor activity [39–41]. Specifically, tailless blocks segmentation and maintains the identity of the terminal boundaries via repression of Kruppel and knirps activity and promotes hunchback expression, which is necessary for the establishment of terminal-specific structures [42,43]. tailless is also necessary for regulating gene expression during the generation of head segments as well as anterior brain development [44]. We also find that nhr-67 prohibits improper fusion events between related cell lineages, at least partly due to strict spatial regulation of the fusogen eff-1 in certain vulval cell types.
As discussed below, nhr-67 interacts genetically with three other transcriptional regulators, cog-1, egl-38, and lin-11, to produce complex patterns of gene expression, probably through trans-regulation of cell type–specific enhancers (Figure 8).
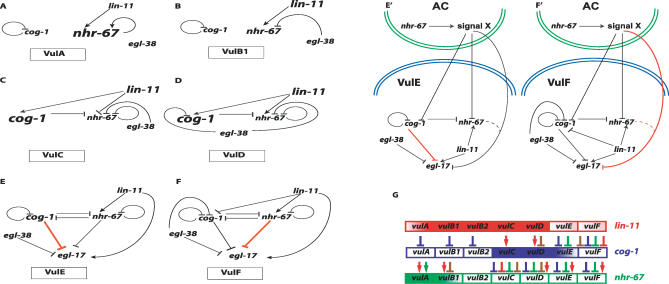
Figure 8. A Summary of the Gene Regulatory Network That Functions during Vulval Patterning and Differentiation in C. elegans .
nhr-67 activity is included along with the other vulval patterning genes: lin-11, cog-1, and egl-38. Colored arrows represent positive inputs and colored block arrows represent repressor inputs for target gene expression in the distinct vulval cell types. The role of egl-38 regulating egl-17 and ceh-2 gene expression in vulE is revealed by analysis of egl-38(n578) animals treated with nhr-67 RNAi and cog-1(sy275); egl-38(n578) double mutants.
Comparison of the Vulval Network with Other Genomic Networks
We have uncovered a novel set of genetic interactions between nhr-67, several transcription factors, and many target genes that contribute to the identity of distinct vulval cell types. For example, nhr-67 appears to be particularly important in the execution of vulF fate and maintaining its cellular identity via regulation of gene expression and fusion events between distinct cell types. Not only does nhr-67 inhibit inappropriate gene expression that is associated with the 2° vulval lineages (Figure 1), but it also promotes gene expression of the EGF protein LIN-3, which is necessary for uv1 fate specification and facilitates proper vulval-uterine connection during development [14]. The functional data obtained from numerous RNAi experiments demonstrates that nhr-67 (like its Drosophila ortholog) is a versatile regulatory gene that operates on at least four of the seven vulval cell types (vulA, vulD, vulE, and vulF). However, we have not tested whether any of these approximately ten interactions are direct.
An interesting feature of the network is our suggestion that both nhr-67 and cog-1 might negatively autoregulate in the same vulE and vulF cells. Drosophila melanogaster tailless does not regulate itself [45], suggesting that nhr-67 autoregulation is a developmental phenomenon unique to nematodes (C. elegans). This apparent divergence in tailless regulation between phyla suggests that a more precise fine-tuning of tailless levels is required for the execution of accurate patterning in the C. elegans vulva. In contrast to their different autoregulatory properties, we find that certain genetic interactions are indeed conserved between the D. melanogaster tailless and C. elegans nhr-67; namely tailless restricts the expression domain of ems in the head segments [44], which is comparable to nhr-67 repressing the worm ems ortholog ceh-2 in the inappropriate vulval cells. Additional tailless targets from other organisms [40,46,47] may also have an impact on vulval patterning. Predictions can also be made in the reciprocal direction and used to elucidate vertebrate development. For example, FGF signaling is required for both vertebrate and inverterbrate heart development [48,49]. The LIM domain protein ISL1 promotes differentiation in a subset of cardiac progenitor cells and transcriptionally activates several FGF genes in mice [50]. Our trans-regulation experiments reveal that both egl-17 and ceh-2 contain cis-regulatory elements that are directly or indirectly dependent on cog-1 (Nkx6.1/6.2), egl-38 (Pax2/5/8), nhr-67 (tll), and lin-11 (LIM) activity. These data may provide further insights into the elaborate regulation of classic developmental genes such as FGF and EMS, both of which have multiple roles in metazoan development.
Patterning in vulE versus vulF Lineages
Previous work demonstrated that patterning of the E and F descendents of the 1° vulval lineage involves both a short-range AC-dependent signal using the Ras pathway as well as lin-17 (Wnt) signaling [10]. In the context of egl-17 gene expression, cog-1 single mutants exhibit increased levels in the vulE cells only. In contrast, nhr-67 RNAi appears to exclusively affect egl-17 expression in the vulF cells. The negative regulatory activities of cog-1 in vulF and nhr-67 in vulE only become apparent in an egl-38 mutant background (which shows no phenotype on its own). This difference suggests that cog-1-mediated negative regulation plays a greater role in vulE cells whereas nhr-67-mediated negative regulation functions primarily in vulF cells. One hypothesis is that vulF cells are biased by proximity to the AC to have higher levels of nhr-67 compared to cog-1 (Figure 9). The genetic regulatory interactions within the vulval network demonstrate that cog-1 levels are negatively regulated in vulF cells via four inputs: lin-11, egl-38, nhr-67, and cog-1. In comparison, nhr-67 expression in vulF cells is modulated by two antagonistic inputs (cog-1 and nhr-67) and one positive input (lin-11), thus possibly resulting in its higher levels. These observations are consistent with a model where nhr-67 acts as the major negative regulator in vulF cells.
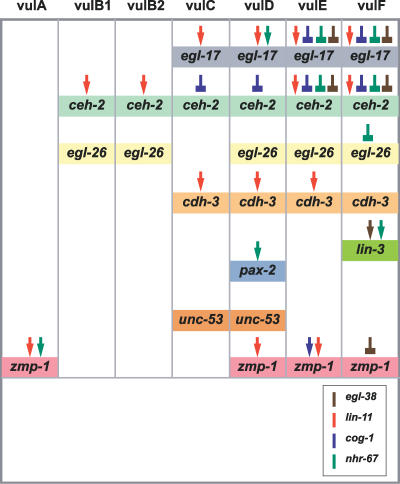
Figure 9. A Summary of the Identified Genetic Regulatory Interactions That Affect Some of the Distinct Vulval Cell Types.
In (A–F and E′ and F′), black arrows represent positive inputs and black block arrows represent repressor inputs for target gene expression that are functional within a given cell type: vulA (A), vulB (B), vulC (C), vulD (D), vulE (E and E′), and vulF (F and F′).
In (A–D), the larger font size depicts high levels of expression of the represented patterning gene within the specified cell type.
(E and F) This model assumes that the interactions mediated by nhr-67 occur within the vulE and vulF cells. Red block arrows (E and F) indicate that a specific regulatory factor (nhr-67 in vulF and cog-1 in vulE) acts as a major repressor within the specified cell type. (E′ and F′) This alternate model presumes that nhr-67 acts in the AC to differentiate between vulE and vulF cells. Signal X could be Ras, Wnt, or some other signaling pathway. Red block arrows (E′ and F′) indicate that the activity associated with a specific regulatory factor (nhr-67 in vulF and cog-1 in vulE) plays a major role in patterning gene expression with in the specified cell type.
(G) A color-coded summary of the distinct expression patterns of lin-11 (red), cog-1 (blue), and nhr-67 (green). Each box represents one of the seven vulval cell types. The colored boxes with the graded pattern represent rare weak expression of the transcriptional reporter, whereas boxes with solid colors represent robust expression. Colored arrows and block arrows are used to document all the identified regulatory interactions that occur during L4 patterning in the diverse vulval cell types: lin-11 (red), cog-1(blue), egl-38 (brown), and nhr-67 (green). This model illustrates multiple patterning differences between the seven mature vulval cell types.
nhr-67 and cog-1 cross-inhibit each other's transcriptional activities, specifically in the vulE and vulF cells, implying that a mutually antagonistic feedback loop exists that exclusively affects the cells of the 1° vulval lineages. Both cog-1 and its mammalian ortholog Nkx6.1 have been previously implicated in bistable loops that reinforce one of two possible stable end states [51,52]. The cross-inhibitory interactions between nhr-67 and cog-1 might be relevant in the specification of vulE versus vulF cell fates. The nature of the bistable loop between cog-1 and nhr-67, however, is unknown. In particular, the bistable loop may be a consequence of either direct transcriptional regulation (as implied in Figure 9) or indirect regulation through an unknown intermediate regulatory factor.
However, the above observation does not rule out the possibility that additional regulatory factors might also contribute to proper patterning of 1° lineages. These other inputs could presumably operate via several potential mechanisms such as modulating the balance between cog-1 versus nhr-67 levels, being exclusively active in one 1° cell type, and interacting at distinct cis-regulatory elements of the downstream targets.
Patterning Differences between 1° and 2° Vulval Lineages
Given the complexity of the observed vulval regulatory interactions, we propose that the network operating on each vulval cell type is unique (Figure 9). A single regulatory factor may have differential functions in terms of executing accurate spatio-temporal gene expression in diverse cells. For instance, lin-11 may upregulate cog-1 levels in the 2° vulC and vulD cells while antagonizing them in the 1° vulF cells. A similar argument can be made about the lin-11-dependent regulation of nhr-67. lin-11 may temporally regulate nhr-67 by inhibiting its vulC-specific expression during the L4 stage. In contrast, lin-11 is clearly critical for the positive regulation of nhr-67 expression in both vulE and vulF cells.
Both cog-1 and nhr-67 are present at high levels in a subset of the 2° vulval cells, yet are barely detectable in the 1° vulval cells. Nevertheless, the disruption of either factor yields obvious defects in 1° vulval cell–specific gene expression. A cross inhibition circuit, such as we propose for cog-1 and nhr-67, can be bistable, with stable states that tolerate inherent fluxes in gene expression (i.e., it would not randomly oscillate between states) [53–55]. Negative autoregulatory circuits have been shown to reduce cell–cell fluctuations in the steady-state level of transcription factors [56] and can speed up the response times of transcription networks without incurring the cost of constant protein production and turnover [57]. These two distinct circuits might enable cells to reach a developmental state with built-in flexibility, allowing rapid switching of their fate upon transient inputs (as opposed to sustained inductive inputs that are metabolically costly). In this model, dynamic levels of cog-1 and/or nhr-67 expression could correlate with particular aspects of 1° vulval cell fate execution. This might account for the elaborate autoregulatory and trans-regulatory interactions specifically seen in 1° vulval descendents, as opposed to their 2°-derived counterparts. We postulate that although all the vulval cells appear to use the same regulatory factors, their differential effects on the diverse cell types is what results in accurate gene expression.
Regulatory Code for the Seven Vulval Cell Types?
During the L4 stage, the gradient of nhr-67 expression is opposite to that of either cog-1 or lin-11. This difference in gene expression domain raises the question of whether the levels of these factors are critical for vulval development. For example, high levels of lin-11 result in misexpression of egl-17 in vulA and abnormal vulval invagination [26]. Different concentrations and combinatorial expression patterns of lin-11, cog-1, and nhr-67 might thus encode mature vulval cell types (Figure 9). For example, differentiation to the 1° vulF cell type may entail low levels of LIN-11 and NHR-67 along with lower levels of COG-1. In contrast, the 1° vulE cells require medium levels of COG-1 along with low doses of LIN-11 and NHR-67. vulA and vulB are similar to each other with respect to maintaining low COG-1 levels. However, vulA cells are characterized by their high NHR-67 levels and medium LIN-11 levels as opposed to the reverse situation in vulB1 and vulB2 cells (medium-low NHR-67, high LIN-11). Lastly, both vulC and vulD have indistinguishably high levels of LIN-11 and COG-1, and we are unable to precisely define what distinguishes these two cell types from each other. One hypothesis is the differential regulation of NHR-67 and COG-1 in both cell types: COG-1 levels are impacted by egl-38 in vulD (but not vulC), whereas NHR-67 levels are negatively regulated by lin-11 in vulC (but not vulD). An obvious limitation of this proposed regulatory code is that it does not take into account other transcription factors that may potentially mediate vulval patterning.
The intricacies of vulval organogenesis can be deconstructed by rigorously elucidating the genomic networks that operate within the seven mature vulval cell types. Deciphering this regulatory code will provide valuable information on network connections and might provide insights into other examples of organogenesis.
Materials and Methods
Microscopy.
Transgenic worms were anesthetized using 3 mM levamisole and observed using Nomarski optics (http://www.nomarski.com). Photographs were taken with a monochrome Hamamatsu digital camera (http://www.hamamatsu.com) and Improvision Openlab 4.0.4 software (http://www.improvision.com). The fluorescent images were overlaid with their respective DIC images using Adobe photoshop 7.0.1 (http://www.adobe.com). The vulval expression patterns for all strains except syIs49 were visualized during the late L4 stage. In the case of syEx716, the vulval expression was also examined during L4 lethargus and adult stage. In syIs49 animals, vulA-specific zmp-1::GFP expression was scored in adults only.
Genetics and RNAi.
C. elegans strains were cultured at 20 °C using standard protocols (Brenner, 1974). Transgenes used in this study are as follows: syIs54 [ceh-2::GFP], syIs55 [ceh-2::YFP], syIs51 [cdh-3::CFP], syIs49 [zmp-1::GFP], syIs77 [zmp-1::YFP], syIs59 [egl-17::CFP] [9], syIs78 [ajm-1::GFP] [26], syIs107 [lin-3::GFP] [58], ayIs4 [egl-17::GFP] [16], guEx64 [pax-2::GFP] (gift from Chamberlin lab), kuIs36 [egl-26::GFP] [18], syIs63 and syIs64 [cog-1::GFP] [27], syIs80 [lin-11::GFP] [59], syEx716 [8-kb nhr-67Δpes-10::GFP], syEx749 [8-kb nhr-67Δpes-10::GFP], syEx744 [nhr-67 intron4 Δpes-10::GFP], syEx925 [6 kb upstream nhr-67::GFP + 8 kb nhr-67Δpes-10::GFP], syEx865 [nhr-67p::GFP::nhr-67 int4–3′end], and syEx756 [unc-53::GFP]. Alleles used in this study: LGI, lin-11(n389); LGII, cog-1(sy275), eff-1(hy21); LGIII, unc-119(ed4); LGIV, unc-31(e169), egl-38(n578), dpy-4(e1166sd), dpy-20(e1282); LGV, him-5(e1490). A complete list of strains is included in Table S2.
Transgenic lines were generated using standard microinjection protocol that produces high-copy number extrachromosomal arrays [60]. syEx756 was generated by injecting the pNP10 construct [61] into unc-119(ed4); him-5 background using unc-119(+) [62] and pBSK+ (Stratagene, http://www.stratagene.com) as coinjection markers.
A reverse genetics screen was conducted against 508 transcription factors (Table S1) from the Ahringer library (Medical Research Council Geneservice) to assay for alterations in vulval expression patterns for the ceh-2::YFP transgene. RNAi feeding protocol is similar to that previously described [32]. Embryos were harvested by bleaching gravid adults and were placed on a lawn of Escherichia coli strain expressing double-stranded RNA at 20 °C. Animals were scored after 36 h (during the L4 stage) using Nomarski microscopy. We resorted to nhr-67 RNAi feeding for the rest of this study since the nhr-67 deletion allele (ok631) results in L1 lethality and/or arrest (International C. elegans Knockout Consortium). All subsequent nhr-67 RNAi feeding experiments were done as described above. nhr-67 RNAi feeding experiments that entailed the restriction of cell fusion (via a temperature-sensitive allele of eff-1) were conducted at 25 °C.
Generation of nhr-67 reporter transgenes.
nhr-67::Δpes-10::GFP reporter gene constructs: The pPD97–78 vector, which includes the Δpes-10 basal promoter driving GFP and the unc-54 3′ UTR (gift from Fire lab), was used as a template to generate 2-kb Δpes-10::GFP products. The primers used for amplification are 5′-GCTTGCATGCCTGCAGGCCTTG-3′ and 5′-AAGGGCCCGTACGGCCGACTAGTAGG-3′. All nhr-67 gene fragments were amplified from the C08F8 cosmid and were stitched together with the Δpes-10::GFP fragment via PCR fusion [63] and were designated as “pdd-1 constructs.” Construct (1) consists of 1-kb promoter sequence, the entire coding region, and introns and 2 kb of the 3′ noncoding region attached to minimal Δpes-10::GFP. The primers used to amplify this template are 5′-CTGCTCAAAACTTTTGCTCC-3′ (forward) and 5′-CAAGGCCTGCAGGCATGCAAGCTTAAAGAACTACTGTAGTTTTTG-3′ (reverse). Construct (2) spans from the fourth intron to the 3′ noncoding region fused to minimal Δpes-10::GFP. This product was generated using the forward primer 5′-GTTCGATCATGGATCCTCTCC-3′ and the same reverse primer as construct (1). Construct (3) is an nhr-67p::GFP reporter that contains 1 kb of the native promoter stitched in-frame with a 700-bp coding fragment of GFP (amplified from the pPD95–69 vector, a gift from Fire lab). The resulting 1.7-kb gene product was subsequently fused to 4.5 kb of nhr-67 regulatory sequences (that span from the fourth intron to the 3′ noncoding region) via PCR. Construct (4) contains 6-kb sequence upstream of the predicted first ATG of nhr-67, appended to minimal Δpes-10::GFP. The following primers were used to amplify this product: 5′-GAACCCGGCGACGTTACGGGGCTTC-3′ and 5′-CAAGGCCTGCAGGCATGCAAGCCATCTGTGAAACCGCAGTCATCAT-3′.
Reporter constructs were injected into unc-119(ed4); him-5 worms using unc-119(+) [62] and pBSK+ (Stratagene) as coinjection markers. lin-11(n389); syEx749 doubles were constructed by injecting the 8-kb nhr-67::Δpes-10::GFP construct into lin-11(n389); unc-119(ed4); him-5 background using unc-119(+) as a rescue marker.
Supporting Information
A mid-sagittal optical view of an adult nhr-67 RNAi–treated hermaphrodite.
(5.3 MB TIF)
(A and B) Nomarski (left), fluorescence (center), and overlaid (right). (A) nhr-67 is expressed in the AC in hermaphrodites and (B) in the linker cell in males.
(3.8 MB TIF)
(51 KB XLS)
(22 KB XLS)
Accession Numbers
The WormBase Gene IDs (www.wormbase.org) as well as the Refseq accession numbers (www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide) for the genes described in this study are ajm-1:WBGene00000100 (NM_077135; NM_077137; NM_077136; NM_171966); cdh-3:WBGene00000395 (NM_066286); ceh-2:WBGene00000429 (NM_059345); cog-1:WBGene00000584 (NM_182115); eff-1:WBGene00001159 (NM_001026819); egl-17:WBGene00001185( NM_075706); egl-26:WBGene00001193 (NM_061251); egl-38:WBGene00001204 (NM_069435); lin-3:WBGene00002992 (NM_171418;NM_171919;NM_171918); lin-11:WBGene00003000 (NM_060295); nhr-67: WBGene00003657 (NM_069693); pax-2:WBGene00003938 (NM_068112); unc-53: WBGene00006788 (NM_001027000;NM_001026999); and zmp-1:WBGene00006987 (NM_171138).
Acknowledgments
We thank Minqin Wang for strain constructions; Helen Chamberlin for the guEx64 strain; the Fire lab for GFP vectors; Nathalie Pujol for the pNP10 plasmid; and Andrew Udit, Mihoko Kato, Gladys Medina, and Barbara Perry for technical assistance. The nhr-67 deletion allele (ok631) and kuIs36 was provided by the C. elegans Genetic Center. We also thank Elissa Hallem, Takao Inoue, Steven Kuntz, Ted Ririe, Erich Schwarz, Adeline Seah, and Weiwei Zhong for comments on the manuscript.
Abbreviations
- AC
anchor cell
- EGF
epidermal growth factor
- Egl
egg laying
- FGF
fibroblast growth factor
- GFP
green fluorescent protein
- Pvl
protruding vulva
- RNAi
RNA interference
Footnotes
Competing interests. The authors have declared that no competing interests exist.
A previous version of this article appeared as an Early Online Release on March 19, 2007 (doi:10.1371/journal.pgen.0030069.eor).
Author contributions. JSF and PWS conceived and designed the experiments, analyzed the data, and wrote the paper. JSF performed the experiments.
Funding. The authors were supported by the Howard Hughes Medical Institute of which PWS is an investigator.
References
- Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci U S A. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH. Gene regulatory network controlling embryonic specification in the sea urchin. Curr Opin Genet Dev. 2004;14:351–360. doi: 10.1016/j.gde.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. Whole-genome analysis of Drosophila gastrulation. Curr Opin Genet Dev. 2004;14:477–484. doi: 10.1016/j.gde.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Koide T, Hayata T, Cho KW. Xenopus as a model system to study transcriptional regulatory networks. Proc Natl Acad Sci U S A. 2005;102:4943–4948. doi: 10.1073/pnas.0408125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg PW. Vulval development. The C. elegans Research Community, WormBook. 2005. doi/ 10.1895/wormbook.1.6.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans . Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sharma-Kishore R, White JG, Southgate E, Podbilewicz B. Formation of the vulva in Caenorhabditis elegans: A paradigm for organogenesis. Development. 1999;126:691–699. doi: 10.1242/dev.126.4.691. [DOI] [PubMed] [Google Scholar]
- Inoue T, Sherwood DR, Aspock G, Butler JA, Gupta BP, et al. Gene expression markers for Caenorhabditis elegans vulval cells. Mech Dev. 2002;119((Suppl 1)):S203–S209. doi: 10.1016/s0925-4773(03)00117-5. [DOI] [PubMed] [Google Scholar]
- Wang M, Sternberg PW. Patterning of the C. elegans 1° vulval lineage by Ras and Wnt pathways. Development. 2000;127:5047–5058. doi: 10.1242/dev.127.23.5047. [DOI] [PubMed] [Google Scholar]
- Inoue T, Oz HS, Wiland D, Gharib S, Deshpande R, et al. C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell. 2004;118:795–806. doi: 10.1016/j.cell.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Ferguson EL, Sternberg PW, Horvitz HR. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans . Nature. 1987;326:259–267. doi: 10.1038/326259a0. [DOI] [PubMed] [Google Scholar]
- Sawa H, Lobel L, Horvitz HR. The Caenorhabditis elegans gene lin-17, which is required for certain asymmetric cell divisions, encodes a putative seven-transmembrane protein similar to the Drosophila Frizzled protein. Genes Dev. 1996;10:2189–2197. doi: 10.1101/gad.10.17.2189. [DOI] [PubMed] [Google Scholar]
- Chang C, Newman AP, Sternberg PW. Reciprocal EGF signaling back to the uterus from the induced C. elegans vulva coordinates morphogenesis of epithelia. Curr Biol. 1999;9:237–246. doi: 10.1016/s0960-9822(99)80112-2. [DOI] [PubMed] [Google Scholar]
- Rajakumar V, Chamberlin HM. The Pax2/5/8 gene egl-38 coordinates organogenesis of the C. elegans egg-laying system. Dev Biol. 2006;301:240–253. doi: 10.1016/j.ydbio.2006.08.068. [DOI] [PubMed] [Google Scholar]
- Burdine RD, Branda CS, Stern MJ. EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans . Development. 1998;125:1083–1093. doi: 10.1242/dev.125.6.1083. [DOI] [PubMed] [Google Scholar]
- Burdine RD, Chen EB, Kwok SF, Stern MJ. egl-17 encodes an invertebrate fibroblast growth factor family member required specifically for sex myoblast migration in Caenorhabditis elegans . Proc Natl Acad Sci U S A. 1997;94:2433–2437. doi: 10.1073/pnas.94.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna-Rose W, Han M. The Caenorhabditis elegans EGL-26 protein mediates vulval cell morphogenesis. Dev Biol. 2002;241:247–258. doi: 10.1006/dbio.2001.0514. [DOI] [PubMed] [Google Scholar]
- Inoue T, Wang M, Ririe TO, Fernandes JS, Sternberg PW. Transcriptional network underlying Caenorhabditis elegans vulval development. Proc Natl Acad Sci U S A. 2005;102:4972–4977. doi: 10.1073/pnas.0408122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspöck G, Ruvkun G, Burglin TR. The Caenorhabditis elegans ems class homeobox gene ceh-2 is required for M3 pharynx motoneuron function. Development. 2003;130:3369–3378. doi: 10.1242/dev.00551. [DOI] [PubMed] [Google Scholar]
- Wang X, Greenberg JF, Chamberlin HM. Evolution of regulatory elements producing a conserved gene expression pattern in Caenorhabditis . Evol Dev. 2004;6:237–245. doi: 10.1111/j.1525-142X.2004.04029.x. [DOI] [PubMed] [Google Scholar]
- Tiensuu T, Larsen MK, Vernersson E, Tuck S. lin-1 has both positive and negative functions in specifying multiple cell fates induced by Ras/MAP kinase signaling in C. elegans . Dev Biol. 2005;286:338–351. doi: 10.1016/j.ydbio.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Newman AP, Inoue T, Wang M, Sternberg PW. The Caenorhabditis elegans heterochronic gene lin-29 coordinates the vulval-uterine-epidermal connections. Curr Biol. 2000;10:1479–1488. doi: 10.1016/s0960-9822(00)00827-7. [DOI] [PubMed] [Google Scholar]
- Cui M, Han M. Cis-regulatory requirements for vulval cell-specific expression of the Caenorhabditis elegans fibroblast growth factor gene egl-17 . Dev Biol. 2003;257:104–116. doi: 10.1016/s0012-1606(03)00033-2. [DOI] [PubMed] [Google Scholar]
- Freyd G, Kim SK, Horvitz HR. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11 . Nature. 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- Gupta BP, Wang M, Sternberg PW. The C. elegans LIM homeobox gene lin-11 specifies multiple cell fates during vulval development. Development. 2003;130:2589–2601. doi: 10.1242/dev.00500. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Inoue T, Sherwood DR, Jiang LI, Sternberg PW. Caenorhabditis elegans cog-1 locus encodes GTX/Nkx6.1 homeodomain proteins and regulates multiple aspects of reproductive system development. Dev Biol. 2002;252:202–213. doi: 10.1006/dbio.2002.0850. [DOI] [PubMed] [Google Scholar]
- Chamberlin HM, Palmer RE, Newman AP, Sternberg PW, Baillie DL, et al. The PAX gene egl-38 mediates developmental patterning in Caenorhabditis elegans . Development. 1997;124:3919–3928. doi: 10.1242/dev.124.20.3919. [DOI] [PubMed] [Google Scholar]
- Trent C, Tsuing N, Horvitz HR. Egg-laying defective mutants of the nematode Caenorhabditis elegans . Genetics. 1983;104:619–647. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Kim SK. Protruding vulva mutants identify novel loci and Wnt signaling factors that function during Caenorhabditis elegans vulva development. Genetics. 2000;156:1097–1116. doi: 10.1093/genetics/156.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Peyrot SM, Wood CG, Wagmaister JA, Maduro MF, et al. Cell fates and fusion in the C. elegans vulval primordium are regulated by the EGL-18 and ELT-6 GATA factors—apparent direct targets of the LIN-39 Hox protein. Development. 2002;129:5171–5180. doi: 10.1242/dev.129.22.5171. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:e12. doi: 10.1371/journal.pbio.0000012. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler WA, Shemer G, del Campo JJ, Valansi C, Opoku-Serebuoh E, et al. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- Gissendanner CR, Crossgrove K, Kraus KA, Maina CV, Sluder AE. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans . Dev Biol. 2004;266:399–416. doi: 10.1016/j.ydbio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Fire A, Ahnn J, Kelly WG, Harfe B, Kostas SA, et al. GFP applications in C. elegans . In: Kain MCS, editor. GFP strategies and applications. New York: John Wiley and Sons; 1998. pp. 153–168. [Google Scholar]
- Wang M. Pattern formation during Caenorhabditis elegans vulval development [PhD thesis] California Institute of Technology; 2000. [Google Scholar]
- Sherwood DR, Butler JA, Kramer JM, Sternberg PW. FOS-1 promotes basement-membrane removal during anchor-cell invasion in C. elegans . Cell. 2005;121:951–962. doi: 10.1016/j.cell.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR. Pattern formation during vulval development in C. elegans . Cell. 1986;44:761–772. doi: 10.1016/0092-8674(86)90842-1. [DOI] [PubMed] [Google Scholar]
- Weigel D, Jurgens G, Klingler M, Jackle H. Two gap genes mediate maternal terminal pattern information in Drosophila . Science. 1990;248:495–498. doi: 10.1126/science.2158673. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Pignoni F, Liaw GJ, Lengyel JA. Dual role of the Drosophila pattern gene tailless in embryonic termini. Science. 1991;254:418–421. doi: 10.1126/science.1925599. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Baldarelli RM, Steingrimsson E, Diaz RJ, Patapoutian A, et al. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell. 1990;62:151–163. doi: 10.1016/0092-8674(90)90249-e. [DOI] [PubMed] [Google Scholar]
- Casanova J. Pattern formation under the control of the terminal system in the Drosophila embryo. Development. 1990;110:621–628. doi: 10.1242/dev.110.2.621. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol. 1987;119:402–417. doi: 10.1016/0012-1606(87)90045-5. [DOI] [PubMed] [Google Scholar]
- Hartmann B, Reichert H, Walldorf U. Interaction of gap genes in the Drosophila head: tailless regulates expression of empty spiracles in early embryonic patterning and brain development. Mech Dev. 2001;109:161–172. doi: 10.1016/s0925-4773(01)00519-6. [DOI] [PubMed] [Google Scholar]
- Rudolph KM, Liaw GJ, Daniel A, Green P, Courey AJ, et al. Complex regulatory region mediating tailless expression in early embryonic patterning and brain development. Development. 1997;124:4297–4308. doi: 10.1242/dev.124.21.4297. [DOI] [PubMed] [Google Scholar]
- Hoch M, Gerwin N, Taubert H, Jackle H. Competition for overlapping sites in the regulatory region of the Drosophila gene Kruppel . Science. 1992;256:94–97. doi: 10.1126/science.1348871. [DOI] [PubMed] [Google Scholar]
- Goriely A, Stella M, Coffinier C, Kessler D, Mailhos C, et al. A functional homolog of goosecoid in Drosophila . Development. 1996;122:1641–1650. doi: 10.1242/dev.122.5.1641. [DOI] [PubMed] [Google Scholar]
- Yutzey KE, Kirby ML. Wherefore heart thou? Embryonic origins of cardiogenic mesoderm. Dev Dyn. 2002;223:307–320. doi: 10.1002/dvdy.10068. [DOI] [PubMed] [Google Scholar]
- Kirby ML. Molecular embryogenesis of the heart. Pediatr Dev Pathol. 2002;5:516–543. doi: 10.1007/s10024-002-0004-2. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci U S A. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallstedt A, Muhr J, Pattyn A, Pierani A, Mendelsohn M, et al. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron. 2001;31:743–755. doi: 10.1016/s0896-6273(01)00412-3. [DOI] [PubMed] [Google Scholar]
- Ptashne M. A genetic switch: Phage lambda revisited. Cold Spring Harbor (New York): Cold Spring Harbor Laboratory Press; 2004. 154 [Google Scholar]
- Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, et al. Programmable cells: Interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli . Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. J Mol Biol. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- Hwang BJ, Sternberg PW. A cell-specific enhancer that specifies lin-3 expression in the C. elegans anchor cell for vulval development. Development. 2004;131:143–151. doi: 10.1242/dev.00924. [DOI] [PubMed] [Google Scholar]
- Gupta BP, Sternberg PW. Tissue-specific regulation of the LIM homeobox gene lin-11 during development of the Caenorhabditis elegans egg-laying system. Dev Biol. 2002;247:102–115. doi: 10.1006/dbio.2002.0688. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringham E, Pujol N, Vandekerckhove J, Bogaert T. unc-53 controls longitudinal migration in C. elegans . Development. 2002;129:3367–3379. doi: 10.1242/dev.129.14.3367. [DOI] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans . Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A mid-sagittal optical view of an adult nhr-67 RNAi–treated hermaphrodite.
(5.3 MB TIF)
(A and B) Nomarski (left), fluorescence (center), and overlaid (right). (A) nhr-67 is expressed in the AC in hermaphrodites and (B) in the linker cell in males.
(3.8 MB TIF)
(51 KB XLS)
(22 KB XLS)