Abstract
Gene amplification is one of the molecular mechanisms resulting in the up-regulation of gene expression. In non-Hodgkin's lymphomas, such gene amplifications have been identified rarely. Using comparative genomic hybridization, a technique that has proven to be very sensitive for the detection of high-level DNA amplifications, we analyzed 108 cases of B-cell neoplasms (42 chronic B-cell leukemias, 5 mantle cell lymphomas, and 61 aggressive B-cell lymphomas). Twenty-four high-level amplifications were identified in 13% of the patients and mapped to 15 different genomic regions. Regions most frequently amplified were bands Xq26-28, 2p23-24, and 2p14-16 as well as 18q21 (three times each). Amplification of several proto-oncogenes and a cell cycle control gene (N-MYC (two cases), BCL2, CCND2, and GLI) located within the amplified regions was demonstrated by Southern blot analysis or fluorescence in situ hybridization to interphase nuclei of tumor cells. These data demonstrate that gene amplifications in B-cell neoplasms are much more frequent than previously assumed. The identification of highly amplified DNA regions and genes included in the amplicons provides important information for further analyses of genetic events involved in lymphomagenesis.
Full text
PDF



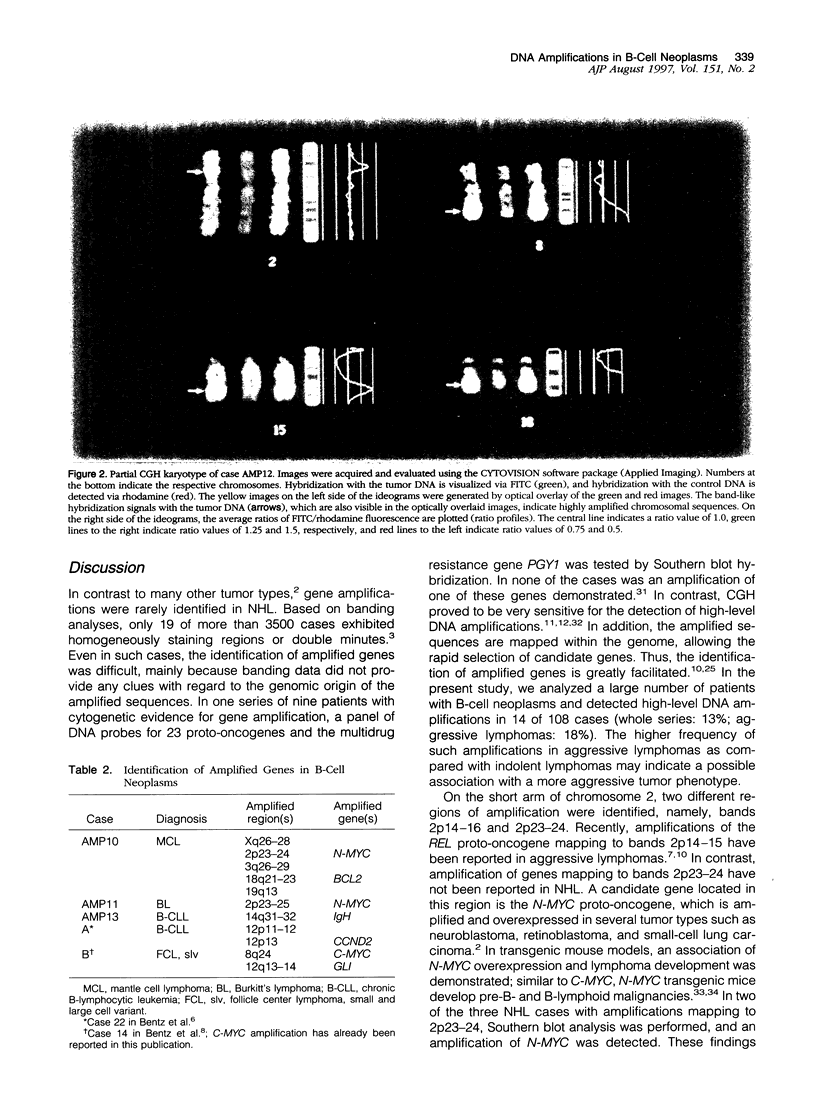



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alitalo K., Schwab M. Oncogene amplification in tumor cells. Adv Cancer Res. 1986;47:235–281. doi: 10.1016/s0065-230x(08)60201-8. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda D., Houldsworth J., Parsa N. Z., Chaganti R. S. Gene amplification in non-Hodgkin's lymphoma. Br J Haematol. 1994 Apr;86(4):792–797. doi: 10.1111/j.1365-2141.1994.tb04831.x. [DOI] [PubMed] [Google Scholar]
- Bentz M., Döhner H., Huck K., Schütz B., Ganser A., Joos S., du Manoir S., Lichter P. Comparative genomic hybridization in the investigation of myeloid leukemias. Genes Chromosomes Cancer. 1995 Mar;12(3):193–200. doi: 10.1002/gcc.2870120306. [DOI] [PubMed] [Google Scholar]
- Bentz M., Huck K., du Manoir S., Joos S., Werner C. A., Fischer K., Döhner H., Lichter P. Comparative genomic hybridization in chronic B-cell leukemias shows a high incidence of chromosomal gains and losses. Blood. 1995 Jun 15;85(12):3610–3618. [PubMed] [Google Scholar]
- Bentz M., Werner C. A., Döhner H., Joos S., Barth T. F., Siebert R., Schröder M., Stilgenbauer S., Fischer K., Möller P. High incidence of chromosomal imbalances and gene amplifications in the classical follicular variant of follicle center lymphoma. Blood. 1996 Aug 15;88(4):1437–1444. [PubMed] [Google Scholar]
- Dildrop R., Ma A., Zimmerman K., Hsu E., Tesfaye A., DePinho R., Alt F. W. IgH enhancer-mediated deregulation of N-myc gene expression in transgenic mice: generation of lymphoid neoplasias that lack c-myc expression. EMBO J. 1989 Apr;8(4):1121–1128. doi: 10.1002/j.1460-2075.1989.tb03482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner H., Fischer K., Bentz M., Hansen K., Benner A., Cabot G., Diehl D., Schlenk R., Coy J., Stilgenbauer S. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood. 1995 Mar 15;85(6):1580–1589. [PubMed] [Google Scholar]
- Döhner H., Stilgenbauer S., James M. R., Benner A., Weilguni T., Bentz M., Fischer K., Hunstein W., Lichter P. 11q deletions identify a new subset of B-cell chronic lymphocytic leukemia characterized by extensive nodal involvement and inferior prognosis. Blood. 1997 Apr 1;89(7):2516–2522. [PubMed] [Google Scholar]
- Forus A., Flørenes V. A., Maelandsmo G. M., Meltzer P. S., Fodstad O., Myklebost O. Mapping of amplification units in the q13-14 region of chromosome 12 in human sarcomas: some amplica do not include MDM2. Cell Growth Differ. 1993 Dec;4(12):1065–1070. [PubMed] [Google Scholar]
- Harris N. L., Jaffe E. S., Stein H., Banks P. M., Chan J. K., Cleary M. L., Delsol G., De Wolf-Peeters C., Falini B., Gatter K. C. A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood. 1994 Sep 1;84(5):1361–1392. [PubMed] [Google Scholar]
- Houldsworth J., Chaganti R. S. Comparative genomic hybridization: an overview. Am J Pathol. 1994 Dec;145(6):1253–1260. [PMC free article] [PubMed] [Google Scholar]
- Houldsworth J., Mathew S., Rao P. H., Dyomina K., Louie D. C., Parsa N., Offit K., Chaganti R. S. REL proto-oncogene is frequently amplified in extranodal diffuse large cell lymphoma. Blood. 1996 Jan 1;87(1):25–29. [PubMed] [Google Scholar]
- Höglund M., Johansson B., Pedersen-Bjergaard J., Marynen P., Mitelman F. Molecular characterization of 12p abnormalities in hematologic malignancies: deletion of KIP1, rearrangement of TEL, and amplification of CCND2. Blood. 1996 Jan 1;87(1):324–330. [PubMed] [Google Scholar]
- Isola J., DeVries S., Chu L., Ghazvini S., Waldman F. Analysis of changes in DNA sequence copy number by comparative genomic hybridization in archival paraffin-embedded tumor samples. Am J Pathol. 1994 Dec;145(6):1301–1308. [PMC free article] [PubMed] [Google Scholar]
- Joos S., Falk M. H., Lichter P., Haluska F. G., Henglein B., Lenoir G. M., Bornkamm G. W. Variable breakpoints in Burkitt lymphoma cells with chromosomal t(8;14) translocation separate c-myc and the IgH locus up to several hundred kb. Hum Mol Genet. 1992 Nov;1(8):625–632. doi: 10.1093/hmg/1.8.625. [DOI] [PubMed] [Google Scholar]
- Joos S., Otaño-Joos M. I., Ziegler S., Brüderlein S., du Manoir S., Bentz M., Möller P., Lichter P. Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and amplification of the REL gene. Blood. 1996 Feb 15;87(4):1571–1578. [PubMed] [Google Scholar]
- Joos S., Scherthan H., Speicher M. R., Schlegel J., Cremer T., Lichter P. Detection of amplified DNA sequences by reverse chromosome painting using genomic tumor DNA as probe. Hum Genet. 1993 Feb;90(6):584–589. doi: 10.1007/BF00202475. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O. P., Sudar D., Rutovitz D., Gray J. W., Waldman F., Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992 Oct 30;258(5083):818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Kallioniemi O. P., Kallioniemi A., Piper J., Isola J., Waldman F. M., Gray J. W., Pinkel D. Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer. 1994 Aug;10(4):231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]
- Le Beau M. M. Chromosomal abnormalities in non-Hodgkin's lymphomas. Semin Oncol. 1990 Feb;17(1):20–29. [PubMed] [Google Scholar]
- Lichter P., Umeda P. K., Levin J. E., Vosberg H. P. Partial characterization of the human beta-myosin heavy-chain gene which is expressed in heart and skeletal muscle. Eur J Biochem. 1986 Oct 15;160(2):419–426. doi: 10.1111/j.1432-1033.1986.tb09989.x. [DOI] [PubMed] [Google Scholar]
- Matsuda F., Shin E. K., Nagaoka H., Matsumura R., Haino M., Fukita Y., Taka-ishi S., Imai T., Riley J. H., Anand R. Structure and physical map of 64 variable segments in the 3'0.8-megabase region of the human immunoglobulin heavy-chain locus. Nat Genet. 1993 Jan;3(1):88–94. doi: 10.1038/ng0193-88. [DOI] [PubMed] [Google Scholar]
- Monni O., Joensuu H., Franssila K., Knuutila S. DNA copy number changes in diffuse large B-cell lymphoma--comparative genomic hybridization study. Blood. 1996 Jun 15;87(12):5269–5278. [PubMed] [Google Scholar]
- Ohno H., Takimoto G., McKeithan T. W. The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell. 1990 Mar 23;60(6):991–997. doi: 10.1016/0092-8674(90)90347-h. [DOI] [PubMed] [Google Scholar]
- Reifenberger G., Liu L., Ichimura K., Schmidt E. E., Collins V. P. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res. 1993 Jun 15;53(12):2736–2739. [PubMed] [Google Scholar]
- Reifenberger G., Reifenberger J., Ichimura K., Meltzer P. S., Collins V. P. Amplification of multiple genes from chromosomal region 12q13-14 in human malignant gliomas: preliminary mapping of the amplicons shows preferential involvement of CDK4, SAS, and MDM2. Cancer Res. 1994 Aug 15;54(16):4299–4303. [PubMed] [Google Scholar]
- Rosenbaum H., Webb E., Adams J. M., Cory S., Harris A. W. N-myc transgene promotes B lymphoid proliferation, elicits lymphomas and reveals cross-regulation with c-myc. EMBO J. 1989 Mar;8(3):749–755. doi: 10.1002/j.1460-2075.1989.tb03435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröck E., Thiel G., Lozanova T., du Manoir S., Meffert M. C., Jauch A., Speicher M. R., Nürnberg P., Vogel S., Jänisch W. Comparative genomic hybridization of human malignant gliomas reveals multiple amplification sites and nonrandom chromosomal gains and losses. Am J Pathol. 1994 Jun;144(6):1203–1218. [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Amler L. C. Amplification of cellular oncogenes: a predictor of clinical outcome in human cancer. Genes Chromosomes Cancer. 1990 Jan;1(3):181–193. doi: 10.1002/gcc.2870010302. [DOI] [PubMed] [Google Scholar]
- Sherr C. J. Mammalian G1 cyclins. Cell. 1993 Jun 18;73(6):1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- Speicher M. R., Prescher G., du Manoir S., Jauch A., Horsthemke B., Bornfeld N., Becher R., Cremer T. Chromosomal gains and losses in uveal melanomas detected by comparative genomic hybridization. Cancer Res. 1994 Jul 15;54(14):3817–3823. [PubMed] [Google Scholar]
- Taniwaki M., Nishida K., Ueda Y., Misawa S., Nagai M., Tagawa S., Yamagami T., Sugiyama H., Abe M., Fukuhara S. Interphase and metaphase detection of the breakpoint of 14q32 translocations in B-cell malignancies by double-color fluorescence in situ hybridization. Blood. 1995 Jun 1;85(11):3223–3228. [PubMed] [Google Scholar]
- Telenius H., Carter N. P., Bebb C. E., Nordenskjöld M., Ponder B. A., Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992 Jul;13(3):718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- Virgilio L., Narducci M. G., Isobe M., Billips L. G., Cooper M. D., Croce C. M., Russo G. Identification of the TCL1 gene involved in T-cell malignancies. Proc Natl Acad Sci U S A. 1994 Dec 20;91(26):12530–12534. doi: 10.1073/pnas.91.26.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visakorpi T., Hyytinen E., Koivisto P., Tanner M., Keinänen R., Palmberg C., Palotie A., Tammela T., Isola J., Kallioniemi O. P. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995 Apr;9(4):401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Menninger J., Beach D., Ward D. C. Molecular cloning and chromosomal mapping of CCND genes encoding human D-type cyclins. Genomics. 1992 Jul;13(3):575–584. doi: 10.1016/0888-7543(92)90127-e. [DOI] [PubMed] [Google Scholar]
- Yang E., Korsmeyer S. J. Molecular thanatopsis: a discourse on the BCL2 family and cell death. Blood. 1996 Jul 15;88(2):386–401. [PubMed] [Google Scholar]
- Ye B. H., Lista F., Lo Coco F., Knowles D. M., Offit K., Chaganti R. S., Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993 Oct 29;262(5134):747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- du Manoir S., Speicher M. R., Joos S., Schröck E., Popp S., Döhner H., Kovacs G., Robert-Nicoud M., Lichter P., Cremer T. Detection of complete and partial chromosome gains and losses by comparative genomic in situ hybridization. Hum Genet. 1993 Feb;90(6):590–610. doi: 10.1007/BF00202476. [DOI] [PubMed] [Google Scholar]




