Abstract
Nuclear hormone receptors activate gene transcription through ligand-dependent association with coactivators. Specific LXXLL sequence motifs present in these cofactors are sufficient to mediate these ligand-induced interactions. A thyroid hormone receptor (TR)-binding protein (TRBP) was cloned by a Sos-Ras yeast two-hybrid system using TRβ1-ligand binding domain as bait. TRBP contains 2063 amino acid residues, associates with TR through a LXXLL motif, and is ubiquitously expressed in a variety of tissues and cells. TRBP strongly transactivates through TRβ1 and estrogen receptor in a dose-related and ligand-dependent manner, and also exhibits coactivation through AP-1, CRE, and NFκB-response elements, similar to the general coactivator CBP/p300. The C terminus of TRBP binds to CBP/p300 and DRIP130, a component of the DRIP/TRAP/ARC complex, which suggests that TRBP may activate transcription by means of such interactions. Further, the association of TRBP with the DNA-dependent protein kinase (DNA-PK) complex and DNA-independent phosphorylation of TRBP C terminus by DNA-PK point to a potential connection between transcriptional control and chromatin architecture regulation.
Hormone-regulated gene activation mediated by nuclear receptors relies on high-affinity interactions between their ligand-binding domains (LBDs) and transcriptional coactivators. Upon ligand induction, coactivators harboring LXXLL motifs are recruited to the target gene promoter as a preformed complex (1–3). The coactivator complex includes CBP/p300 (4–6), pCAF (7, 8), steroid receptor coactivator 1 (SRC-1) family members (9, 10), an RNA coactivator SRA (11), and an arginine transmethylase CARM1 (12). A biochemically purified DRIP/TRAP/ARC complex containing mediator/srb subunits is also recruited by nuclear receptors in a ligand-dependent manner (13–16). The histone acetylase-containing coactivator and vitamin D receptor-interacting protein (DRIP) complexes then act in concert to remodel chromatin and to regulate gene activation. We report here the identification and characterization of a ubiquitous LXXLL-containing protein, thyroid hormone receptor (TR)-binding protein (TRBP), that acts as a general coactivator. The association of TRBP with a DRIP component and CBP/p300, as well as the DNA-dependent protein kinase (DNA-PK) complex, suggests an important link between the coactivator complex and DNA-PK function (17).
Materials and Methods
Yeast Two-Hybrid Screen and TRBP cDNA Cloning.
The Sos-Ras yeast two-hybrid system was used to screen a rat pituitary cDNA library, as previously described (18), except that screen was performed in the presence of 20 μM 3,3′,5-triiodothyroacetic acid (Triac; Sigma), a hydrophilic 3,3′,5-triiodo-l-thyronine (T3) analog. Rat TRβ1-LBD (292–461) was PCR amplified and inserted in-frame with the C terminus of human Sos as bait. The cDNA sequence derived from the screen was used to search the GenBank database with the blast program. The partial 3′ cDNA clone of human TRBP (KIAA0181, anonymous sequence) identified in the computer search was obtained from the Kazuka DNA Research Institute (19). Based on the partial sequence, the full-length cDNA encoding human TRBP was isolated from HeLa cell mRNA by 5′RACE (GIBCO).
Northern Analysis.
A Northern analysis using a human multiple tissue mRNA blot (human MTN; CLONTECH) and a random-primed labeled full-length human TRBP cDNA probe was performed. Rat cyclophilin mRNA served as an internal control.
Antibodies and Immunoblotting.
Polyclonal anti-TRBP was prepared in rabbits by immunization with glutathione S-transferase (GST)-rat TRBP-(714–999) fusion protein (Covance; Denver, PA). TRBP antibodies were IgG purified by protein A-Sepharose (Santa Cruz Biotechnology), according to the supplier's protocol. Anti-Flag antibody (M5) was obtained from Kodak, and anti-DRIP130 was obtained from L. Freedman (14). Immunoblots were prepared and detected with the ECL system (Amersham Pharmacia).
Immunofluorescence Staining.
HeLa cells were fixed with methanol and stained with IgG-purified or affinity-purified TRBP antibody at a dilution of 1:50 and anti-rabbit Cy3-conjugated secondary antibody (Jackson ImmunoResearch) at a dilution of 1:200.
Cells and Transfections.
CV-1 and 293 cells (DMEM/10% FCS) were transfected in six-well plates by the calcium phosphate method. Total amounts of DNA for each well were balanced by adding vector pcDNA3 (Invitrogen). Transfection efficiency was monitored by assaying β-galactosidase. The TRE (F2) and 2XERE luciferase reporters have been described (20, 21). The transfections with TRE and 2XERE reporters were performed in medium with hormone-depleted serum and were followed by treatment with serum-free medium, with or without 10 nM T3 or 1 μM estradiol, for 24 h. The 7XAP-1, 5XNFκB, and 4XCRE luciferase reporters, and protein kinase A (PKA) and MEK kinase (MEKK) plasmids were obtained from Stratagene. Human somatostatin gene promoter (nucleotides −360 to +91) was cloned from HeLa genomic DNA by PCR. Full-length human TRBP and its fragments were subcloned into pcDNA3. Mouse CREB-binding protein (CBP) and human SRC-1 expression plasmids were previously described (6, 9). Data shown represent means of triplicate transfections ± standard errors.
Recombinant Proteins and Binding Assays.
In vitro GST binding assays were performed with proteins labeled with [35S]methionine by using a TnT coupled reticulocyte-lysate in vitro translation system (Promega). GST fusions were expressed in Escherichia coli BL21(DE3), isolated by using lysis buffer (5 mM Tris, pH 7.4/5 mM NaCl/1 mM EDTA/5 mM EGTA/1% Triton X-100/2 mM PMSF/1 mM DTT), and purified with GST beads (Pharmacia). The immobilized GST protein beads (20 μl, 2 μg) were incubated either with 5 μl of TnT labeled proteins at room temperature for 1 h or with 200 μg of nuclear extracts at 4°C for 16 h in the binding buffer [20 mM Hepes, pH 7.4/50 mM NaCl/75 mM KCl/1 mM EDTA/0.05% Triton X-100/10% (vol/vol) glycerol/1 mM DTT/10 μg/ml leupeptin/10 μg/ml aprotinin/10 μg/ml trypsin inhibitor]. The beads were washed three times with the same binding buffer before being subjected to SDS/PAGE or immunoblotting analysis.
DNA-PK Kinase Assay.
Phosphorylation of TRBP recombinant fragments in vitro by DNA-PK was assayed using the SigmaTECT DNA-dependent Protein Kinase Assay System from Promega with modifications. Briefly, a purified DNA-PK catalytic subunit (cs) and regulatory subunit (Ku) complex from HeLa nuclear extract was incubated with [γ-32P]ATP, with or without linear double-stranded DNA as activator, and with GST-TRBP fusion proteins as substrates. Labeled GST proteins were washed before being resolved by SDS/PAGE and autoradiography. The amount of DNA-PK used (5–10 units) in each assay was titrated and determined with a biotinylated p53 peptide substrate bound to streptavidin-cellulose paper. Full-length GST-p53 from Santa Cruz Biotechnology was used as a positive control for DNA-dependent phosphorylation, and GST alone, which is not phosphorylated, was used as a negative control.
Protein Microsequencing.
Gel-purified proteins obtained by GST-TRBP-C-(1641–2063) affinity chromatography of HeLa nuclear extract were subjected to sequence analysis performed at the Harvard Microchemistry Facility (HMF). Proteolytic peptides were analyzed by microcapillary HPLC nano-electrospray tandem ion trap mass spectrometry. MS/MS peptide sequence interpretation was facilitated with the algorithm sequest and programs developed at the HMF.
Results
Isolation of a Ligand-Dependent TR-Interacting Coactivator by the Sos-Ras Yeast Two-Hybrid System.
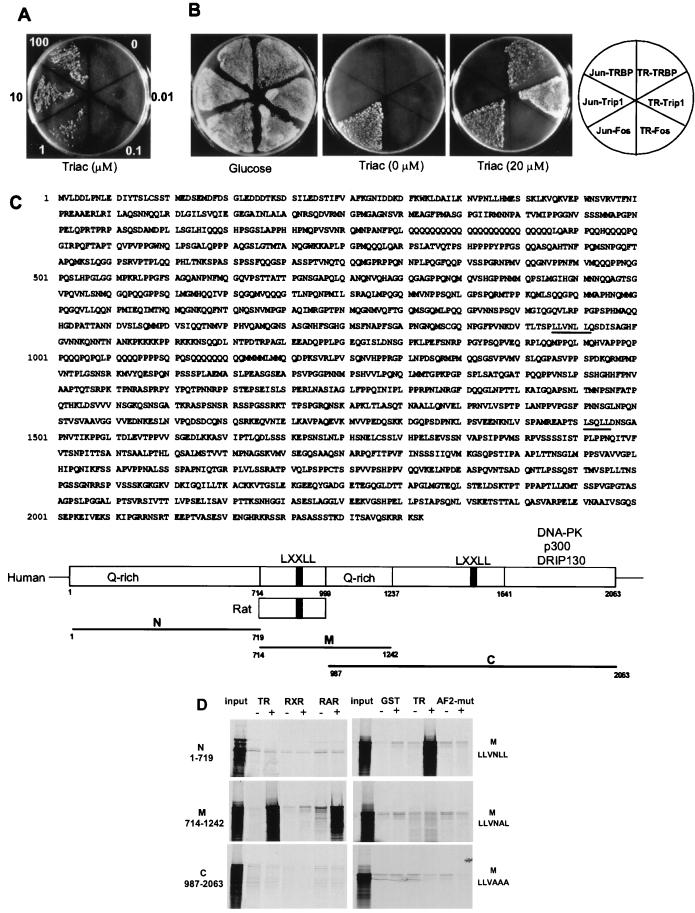
To identify coactivators that associate with liganded nuclear receptors, we performed a Sos-Ras yeast two-hybrid screen using TRβ1-LBD as bait. This two-hybrid system detects protein interactions in cytoplasm with the advantage of bypassing a transcriptional readout and eliminating the false-positive interference by nuclear factors (18). The TRBP clone isolated in the presence of ligand showed ligand-dependent interaction (Fig. 1A). When TRBP, Trip1 (a known TR-interacting clone obtained from our screen) (22), and c-Fos were compared by using TR-LBD or Jun-DNA-binding domain (DBD) as bait, TRBP and Trip1 bound to TR-LDB but not Jun-DBD. As a control, c-Fos bound Jun-DBD but not TR-LDB. In addition, the binding of TRBP was ligand-dependent by comparison with the Jun–Fos interaction (Fig. 1B). Clearly, these results suggest that the binding of TRBP to TR-LDB in yeast is specific and ligand dependent.
Figure 1.
Isolation of a ligand-dependent TR-interacting coactivator by the Sos-Ras yeast two-hybrid system. (A) Growth of TRBP clone coexpressing hSos/TR-LDB at indicated concentrations of Triac. (B) Bait- and ligand-dependent interaction of TRBP in yeast. TRBP, a positive control Trip1, and a negative control c-Fos, were tested by using hSos/TR-LDB and hSos/Jun-DBD as baits in the presence or absence of 20 μM Triac on galactose plates. A glucose plate was used as control. (C) Deduced human TRBP amino acid sequence with LXXLL motifs underlined and schematic representation of TRBP with its deletion fragments. The rat TRBP sequence (714–999) derived from the two-hybrid screen is also depicted. (D) TRBP binds to liganded TR and retinoic acid receptor (RAR) in vitro. GST fusion nuclear receptor LDBs [TR, retinoid X receptor (RXR), RAR, and TR AF-2 mutant] were incubated with in vitro translated, 35S-labeled TRBP fragments (N, M, and C) shown in C, with or without cognate ligands. Bound TRBP fragments were resolved by SDS/PAGE and detected by autoradiography. The AF-2 mutant of TR was E457A, and alanine-to-leucine substituted M-region LXXLL mutants of TRBP are as indicated. M/LLVNLL is wild type.
The library-derived TRBP clone encodes a novel 286-aa rat sequence with an in-frame LXXLL motif. blast search against the GenBank database revealed similarity to an anonymous partial 3′ human cDNA clone (KIAA0181), isolated and sequenced from KG-1 lymphocytic cells (19). The deduced primary sequences of human and rat TRBP possess 92% identity and the LXXLL sequence motif is conserved, which suggests the functional importance of the motif. The full-length human cDNA for TRBP was completed by using 5′RACE, yielding a Kozak consensus translational start site (ACCATGG) and three in-frame stops in the 5′-untranslated region preceding the large open reading frame. This TRBP cDNA predicts a protein encoding 2,063 amino acids with two LXXLL motifs and several Q-rich regions (Fig. 1C). GST pull-down analysis showed that only the LXXLL motif obtained from the two-hybrid screen was responsible for the association of TRBP to the liganded TR- or retinoic acid receptor (RAR)-LBDs. In addition, mutagenesis studies suggested that this interaction is AF-2 as well as LXXLL dependent (Fig. 1D). Further database analysis revealed that TRBP is previously unidentified, with similarities to an uncharacterized Caenorhabditis elegans protein and the C-terminal activation domain of CBP/p300. This latter finding, along with TRBP's interactions with liganded nuclear receptors, suggests that TRBP serves as a coactivator. The TRBP gene (KIAA0181) is located on human chromosome 20q11.22–23.
Ubiquitous Expression of Nuclear TRBP.
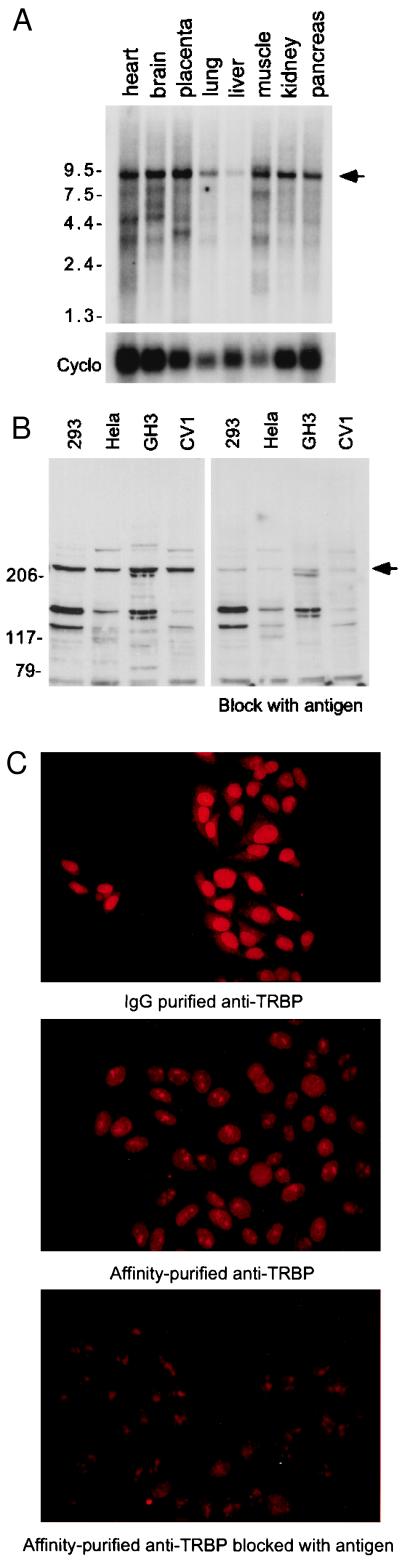
Expression of endogenous TRBP is ubiquitous (Fig. 2 A and B). A 9.0-kb TRBP mRNA transcript is expressed in all of the tissues tested (Fig. 2A). Consistent with the predicted primary structure, endogenous TRBP protein has a high apparent molecular size (Mr = 210,000), with likely low abundance as detected in several nuclear extracts by a high-affinity polyclonal TRBP-specific antibody (Fig. 2B). The antibody was characterized by immunodetection of in vitro translated TRBP. In addition, immunofluorescent staining of HeLa cells confirmed that TRBP is nuclear in localization (Fig. 2C).
Figure 2.
Ubiquitous expression of nuclear TRBP. (A) Northern analysis of human mRNAs from indicated tissues probed with a full-length human TRBP cDNA. Rat cyclophilin (Cyclo) was used as an internal control. Arrow indicates TRBP mRNA. (B) Immunoblotting with nuclear extracts derived from different indicated cell types, using a TRBP-specific polyclonal antibody, without (Left) or blocked with (Right) 500 ng/ml GST-TRBP. Arrow indicates endogenous TRBP. (C) TRBP is nuclear. Immunohistochemical analysis of endogenous TRBP in HeLa cells, using IgG-purified anti-TRBP antibody (Top), or using affinity-purified anti-TRBP antibody, without (Middle) or blocked with (Bottom) 500 ng/ml GST-TRBP as antigen. (×400.)
TRBP Is a Unique General Coactivator.
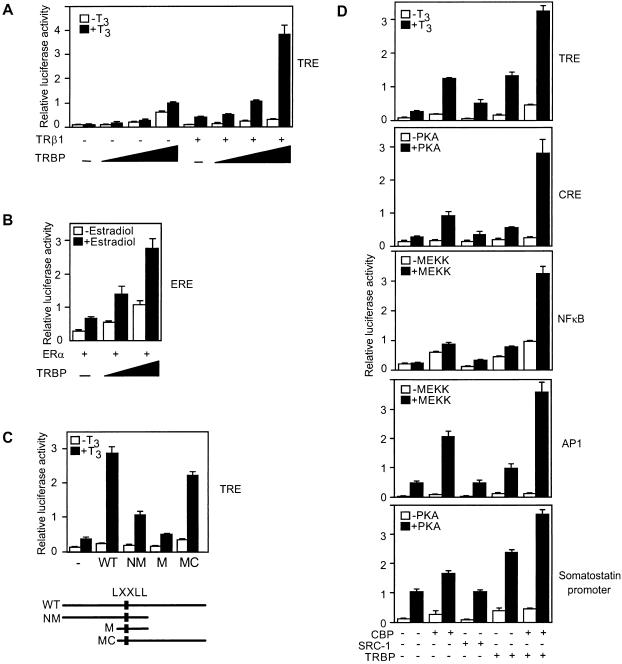
To investigate whether TRBP functions as a coactivator, transient transfection studies were performed with luciferase reporters driven by several different enhancer elements. TRBP potently coactivates a thyroid hormone-response element (TRE) and an estrogen-response element (ERE) in a dose-dependent manner (Fig. 3 A and B). Because the stimulation was also observed with liganded TR-mediated as well as basal transcription, this effect was both ligand dependent and independent. A transfection study with different regions of TRBP revealed that the major activation domain is located at the C-terminal region of TRBP, although the N-terminal region also showed moderate activity (Fig. 3C). The expression level of each TRBP region was monitored by immunoblotting to ensure that the effect on transcription was not caused by altered protein expression. To assess whether TRBP may also serve as a general coactivator, it was compared with CBP and SRC-1 for activity on several enhancer elements, including TRE, CRE, AP-1, and NFκB, and on the human somatostatin gene promoter. Unlike SRC-1, which largely coactivates through nuclear receptors, TRBP and CBP both function on all elements tested (Fig. 3D). This result indicated that TRBP is not enhancer element-specific but rather functions as a general coactivator similar to CBP. Furthermore, the effects of TRBP and CBP were strongly synergistic, suggesting that these two general coactivators may act cooperatively to activate the transcription of a variety of promoters.
Figure 3.
TRBP is a general coactivator. (A) TRBP dose-dependent coactivator activity on TRE (F2). CV-1 cells were cotransfected, with or without TRβ1 (0.1 μg), and with increasing amounts of cytomegalovirus (CMV) promoter-driven TRBP (0, 0.1, 0.5, and 2 μg). Cells were grown in the presence or absence of 10 nM T3. (B) TRBP dose-dependent coactivation of 2XERE luciferase reporter. CV-1 cells were cotransfected with ER (0.1 μg) and TRBP (0, 0.5, and 2 μg), in the presence or absence of 1 μM estradiol. (C) A comparison of the coactivator activities of TRBP deletion fragments on the F2 TRE in CV-1 cells. Schematic below depicts the nature of the TRBP fragments. (D) TRBP functions as a general coactivator. TRBP, CBP, and SRC-1 were cotransfected with luciferase reporters containing different minimal enhancer elements (TRE, 4XCRE, 7XAP-1, 5XNFκB) in CV-1 cells or somatostatin gene promoter (nucleotides −360 to +91) in 293 cells, with or without PKA (0.5 μg) or MEKK (0.1 μg).
For further insight into the structure and function of TRBP, a secondary structure analysis using the Jpred/PHD program was performed. Study of CBP/p300, SRC-1, and pCAF revealed the presence of structured α-helices and β-strands among their histone acetyltransferase (HAT) and transcription factor-binding domains, whereas the C-terminal activation domain of CBP/p300 is predicted to be unstructured, or to display disordered structure in solution. Strikingly, TRBP, with a high percentage of proline, glutamine, serine, and hydrophobic residues, a common feature seen in transcriptional activation domains (23–25), is predicted to be largely unstructured. Of particular note, the only predicted structured region of TRBP is present at its extreme C terminus with β-strands (data not shown). This observation suggests that the C terminus of TRBP may serve as a structured domain that interacts with promoter-bound components to confer coactivator function to TRBP.
TRBP Associates with DNA-PK Complex, and Phosphorylation of TRBP C Terminus by DNA-PK Is DNA Independent.
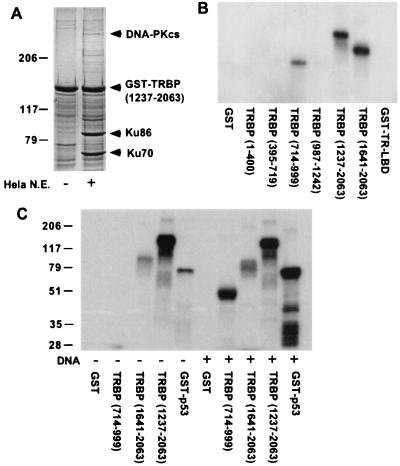
The transfection results and the predicted secondary structure suggested that TRBP might associate with nuclear proteins involved in transcription regulation. Because TRBP is present in very low abundance in vivo, recombinant TRBP fragments were used to augment the detection of interacting proteins in nuclear extracts. A distinct set of TRBP-associated proteins, specifically bound to the C terminus but not the other regions of TRBP, was detected. Similar patterns of binding proteins were observed in both HeLa and GH3 nuclear extracts (data not shown). Microsequencing of five bound proteins from HeLa nuclear extract yielded their identities, with each having more than 20 proteolytic peptide fragments matched to a single known protein. These five proteins are human DNA-PK catalytic subunit (DNA-PKcs), DNA-PK regulatory subunits (Ku70 and Ku86), poly(ADP-ribose) polymerase (PARP), and DNA topoisomerase I. DNA-PK regulatory subunits were particularly abundant (Fig. 4A). PARP is the only nuclear chromatin-associated ADP-ribosylating enzyme, and its targets include histone and DNA topoisomerase. PARP stimulates DNA-PK kinase activity and can be directly phosphorylated by DNA-PK (26–28). DNA-PK was originally characterized as a DNA-dependent protein kinase requiring double-strand DNA ends as cofactor. Recently, DNA-independent activation of DNA-PK by a protein factor has also been reported (29), and the proposed roles of DNA-PK in transcriptional regulation besides recombination and double-stranded DNA repair are increasing. The high avidity of the TRBP C terminus for the multiple DNA-PK/PARP complex components from nuclear extract suggests that TRBP may associate with the entire complex in vivo. In additional to its interaction with DNA-PK complex, TRBP was found to be a potent substrate of purified DNA-PK in vitro. When each TRBP region was tested, the major phosphorylation site(s) was located at both the LXXLL-containing middle region (residues 714–999) and the C terminus of TRBP (residues 1237–2063 and 1641–2063) (Fig. 4B). GST alone was not phosphorylated by DNA-PK, suggesting the phosphorylation occurred only at TRBP. Notably, the phosphorylation of the middle region is DNA dependent and C-terminal phosphorylation is DNA independent compared with p53, a known DNA-dependent substrate (Fig. 4C). These data suggest that the association of TRBP C terminus with the DNA-PK complex can induce kinase activity without DNA. The in vivo physiological substrate for this DNA-independent kinase activity is not yet known. Nevertheless, DNA-PK associates with and direct phosphorylates coactivator TRBP and is potentially activated by TRBP, indicating that DNA-PK might be a the key nuclear protein kinase involved in transcriptional regulation.
Figure 4.
DNA-PK complex interacts with TRBP and phosphorylates TRBP C terminus in the absence of DNA. (A) HeLa nuclear extract was incubated with GST- TRBP-(1237–2063), and bound nuclear proteins were analyzed by SDS/PAGE with Coomassie blue staining. Protein bands of interest were excised from the gel and subjected to protein microsequencing. DNA-PK catalytic subunit (cs) and regulatory subunits, Ku70 and Ku86, identified by protein sequencing are indicated. (B) DNA-PK phosphorylates TRBP in vitro. GST-TRBP fusion proteins indicated were used as substrates in DNA-PK kinase assay in the presence of linear double-stranded DNA as DNA-PK activator. Phosphorylated TRBP fusion proteins ([γ-32P]ATP labeled) were detected by autoradiography. GST alone and GST-TR-LBD were used as negative controls. (C) DNA-PK phosphorylation of TRBP C terminus is DNA independent. DNA-PK kinase assay was performed with (+) and without (−) linear double-stranded DNA by using TRBP C-terminal (residues 1237–2063 and 1641–2063) or TRBP middle region (residues 714–999) GST fusion proteins as substrates. GST alone and GST-p53 are negative and positive controls, respectively.
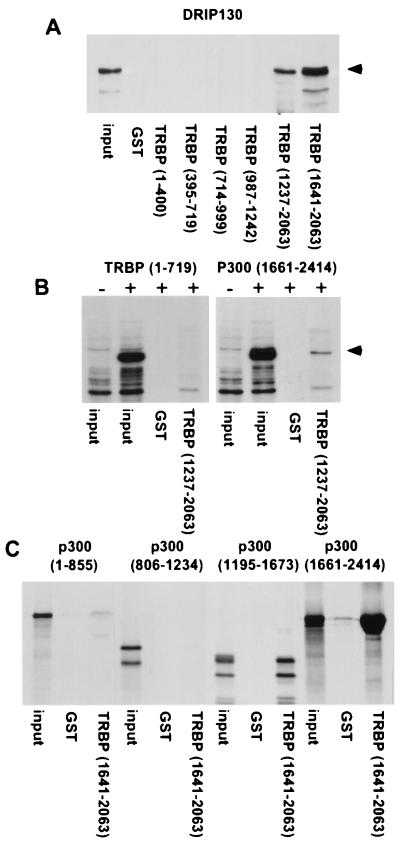
TRBP Associates with DRIP130 and p300.
To detect other proteins from a coactivator complex that is bound to TRBP, Western blotting was performed with a DRIP130 antibody. DRIP130, a component in DRIP complex associated with the liganded nuclear receptors, is shown to interact with high affinity to TRBP by means of its C terminus. This finding suggests that TRBP interacts with the DRIP complex (Fig. 5A). Moreover, the TRBP C terminus binds to p300 (Fig. 5B), in a likely direct interaction (Fig. 5C). This feature might explain the synergistic effect of wild-type TRBP and CBP in transient transfection experiments (Fig. 3D). As TRBP does not possess inherent HAT activity (data not shown), and lacks the typical HAT secondary structure (30), the association with CBP/p300 may permit TRBP to recruit HAT activity during activation. Taken together, these results indicate that TRBP may exert its general coactivator function by participating in coactivator and chromatin remodeling complexes.
Figure 5.
TRBP associates with DRIP130 and p300. (A) TRBP C terminus interacts with endogenous DRIP130 from GH3 nuclear extract. GST-TRBP fusion proteins were incubated with GH3 nuclear extract, and TRBP-bound proteins were subjected to Western blot analysis. Arrow indicates DRIP130 detected by anti-DRIP130. Input was 5%. (B) 293 cells were transfected with (+) or without (−) indicated Flag-p300-(1661–2414) (5) or Flag-TRBP-(1–719) as control. Nuclear extracts from transfected cells were incubated with GST or GST-TRBP. Washed GST beads were subjected to Western blot analysis. Arrow indicates TRBP-bound p300 detected by anti-Flag antibody. Input was 10%. (C) In vitro translated, 35S-labeled p300 fragments were tested for binding to GST and GST-TRBP-(1641-2063). Bound p300 was detected by SDS/PAGE and autoradiography. Input was 20%.
Discussion
Coactivators act as members of large complexes rather than as individual factors (1, 2). The collaborative action of coactivators with overlapping and distinct properties results in the targeting to common transcription machinery. TRBP, although deficient in HAT activity, may still serve as a general coactivator through its interaction with the coactivator complex. Secondary structure analysis of TRBP suggests a high degree of activation domain character that is distinguishable from CBP/p300 and SRC-1, indicating its nature as a unique coactivator. After we submitted this manuscript, we noted that there are two additional identical TRBP human clones (ASC-2 and RAP250) (31, 32). ACS-2 was isolated as a previously identified gene amplified in human breast cancer (AIB3) (31). The involvement of CBP, SRC-1 family members, and TRBP/ASC-2 in gene rearrangement or amplification in cancer suggests that TRBP is also a bona fide coactivator that profoundly affects transcription when altered.
Chromatin remodeling is essential and integral to DNA replication, double-strand DNA repair, and transcriptional control (33). DNA-PK, a nuclear serine/threonine kinase required for double-strand DNA repair, has been implicated as a factor in chromatin remodeling and transcriptional activation. DNA-PK complexes have been shown to phosphorylate polymerase II CTD and other transcriptional factors (34). Recent data suggest that protein phosphorylation by DNA-PK may produce multiple effects, such as silencing heterochromatin in yeast, stimulating general transcription after initiation (35), and regulating chromatin integrity and architecture (36). TRBP provides the evidence for an association between a general coactivator and the DNA-PK complex.
In sum, TRBP (i) is a ubiquitous nuclear protein with a functional LXXLL motif; (ii) has homology with the activation domain of CBP/p300; (iii) is a transcriptional coactivator for a broad spectrum of enhancer elements including TRE, ERE, CRE, AP-1, and NFκB; (iv) synergizes with CBP; (v) lacks inherent HAT activity; and (vi) possesses a C terminus with strong capacity for interaction with the DNA-PK complex, CBP/p300, and DRIP130. Thus, TRBP is a general coactivator, which may serve as an important component in the general transcriptional machinery.
Acknowledgments
We thank A. Aronheim and M. Karin for the Sos-Ras yeast two-hybrid system; T. Nagase from Kazuka DNA Research Institute for the KIAA0181 clone; W. S. Lane (Harvard Microchemistry Facility) for microsequencing; and L. P. Freedman for the anti-DRIP130. This work was supported in part by grants from the National Institutes of Health.
Abbreviations
- LBD
ligand-binding domain
- DBD
DNA-binding domain
- TR
thyroid hormone receptor
- TRBP
TR-binding protein
- TRE
thyroid hormone-response element
- DNA-PK
DNA-dependent protein kinase
- DRIP
vitamin D receptor-interacting protein
- SRC-1
steroid receptor coactivator 1
- CBP
CREB-binding protein
- PKA
protein kinase A
- MEKK
MEK kinase
- Triac
3,3′,5-triiodothyroacetic acid
- T3
3,3′,5-triiodo-l-thyronine
- GST
glutathione S-transferase
- HAT
histone acetyltransferase
Footnotes
References
- 1.Freedman L P. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 2.Xu L, Glass C K, Rosenfeld M G. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 3.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 4.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 5.Arany Z, Sellers W R, Livingston D M, Eckner R. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 6.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Nature (London) 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 7.Yong X, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 8.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 9.Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 10.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Nature (London) 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 11.Lanz R B, McKenna N J, Onate S A, Albrecht U, Wong J, Tsai S Y, Tai M J, O'Malley B W. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 12.Chen D, Ma H, Hong H, Koh S S, Huang S M, Schurter B T, Aswad D W, Stallcup M R. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 13.Naar A M, Breaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Nature (London) 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 14.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Nature (London) 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 15.Ito M, Yuan C X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z Y, Zhang X, Qin J, Roeder R G. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 16.Ryu S, Zhou S, Ladurner A G, Tjian R. Nature (London) 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 17.Smith G C M, Jackson S P. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 18.Aronheim A, Zandi E, Hennemann H, Elledge S J, Karin M. Mol Cell Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagase T, Seki N, Ishikawa K, Tanaka A, Nomura N. DNA Res. 1996;3:17–24. doi: 10.1093/dnares/3.1.17. [DOI] [PubMed] [Google Scholar]
- 20.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y S, Yen P M, Chin W W, Pfaff D W. Proc Natl Acad Sci USA. 1996;93:12587–12592. doi: 10.1073/pnas.93.22.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J W, Ryan F, Swaffield J C, Johnston S A, Moore D D. Nature (London) 1995;374:91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- 23.Triezenberg S J. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 24.Schmitz M L, Silva M A S, Altmann H, Czisch M, Holak T A, Baeuerle P A. J Biol Chem. 1994;269:25613–25620. [PubMed] [Google Scholar]
- 25.Hoy M V, Leuther K K, Kodadek T, Johnston S A. Cell. 1993;72:587–594. doi: 10.1016/0092-8674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- 26.Galande S, Kohwi-Shigematsu T. J Biol Chem. 1999;274:20521–20528. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- 27.Ruscetti T, Lehnert B E, Halbrook J, Trong H L, Hoekstra M F, Chen D J, Peterson S R. J Biol Chem. 1998;273:14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- 28.Meisterernst M, Stelzer G, Roeder R G. Proc Natl Acad Sci USA. 1997;94:2261–2265. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yavuzer U, Smith G C M, Bliss T, Werner D, Jackson S P. Genes Dev. 1998;12:2188–2199. doi: 10.1101/gad.12.14.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutnall R N, Tafrov S T, Sternglanz R, Ramakrishnan V. Cell. 1998;94:427–438. doi: 10.1016/s0092-8674(00)81584-6. [DOI] [PubMed] [Google Scholar]
- 31.Lee S K, Anzick S L, Choi J E, Bubendorf L, Guan X Y, Jung Y K, Kallioniemi O P, Kononen J, Trent J M, Azorsa D, et al. J Biol Chem. 1999;274:34283–34293. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- 32.Caira F, Antonson P, Pelto-Huikko M, Treuter E, Gustafsson J A. J Biol Chem. 2000;275:5308–5317. doi: 10.1074/jbc.275.8.5308. [DOI] [PubMed] [Google Scholar]
- 33.Tyler J K, Adams C R, Chen S R, Kobayashi R, Kamakaka R T, Kadonaga J T. Nature (London) 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 34.Lees-Miller S P. Biochem Cell Biol. 1996;74:503–512. doi: 10.1139/o96-054. [DOI] [PubMed] [Google Scholar]
- 35.Woodard R L, Anderson M G, Dynan W S. J Biol Chem. 1999;274:478–485. doi: 10.1074/jbc.274.1.478. [DOI] [PubMed] [Google Scholar]
- 36.Featherstone C, Jackson S P. Br J Cancer. 1999;80,(Suppl. 1):14–19. [PubMed] [Google Scholar]