Summary
Autoregulatory loops often provide precise control of the level of expression of specific genes that encode key regulatory proteins. Here we have defined a pathway by which Yra1p, a yeast mRNA export factor, controls its own expression. We show that YRA1 exon1 sequences in cis and Yra1p in trans inhibit YRA1 pre-mRNA splicing and commit the pre-mRNA to nuclear export. Mex67p and Crm1p jointly promote YRA1 pre-mRNA export and, once in the cytoplasm, the pre-mRNA is degraded by a 5’ to 3’ decay mechanism that is dependent on the decapping activator Edc3p and on specific sequences in the YRA1 intron. These results illustrate how common steps in the nuclear processing, export, and degradation of a transcript can be uniquely combined to control the expression of a specific gene and suggest that Edc3p-mediated decay may have additional regulatory functions in eukaryotic cells.
Introduction
mRNA degradation, an integral event in gene expression, provides post-transcriptional titration of transcript levels and ensures the elimination of aberrant mRNAs or those that have exited the translation-competent state (Parker and Song, 2004). Two general and functionally redundant mechanisms, the deadenylation-dependent 5’ to 3’ pathway and the exosome-mediated 3’ to 5’ pathway, are responsible for the decay of most mRNAs in yeast (Coller and Parker, 2004). Subsequent to poly(A) shortening, substrates of the 5’ to 3’ pathway are decapped by the Dcp1p/Dcp2 decapping enzyme complex and then digested exonucleolytically by the 5′ to 3′ exoribonuclease, Xrn1p (Coller and Parker, 2004), whereas those subject to 3’ to 5’ decay are deadenlylated and then degraded by the ten-subunit exosome complex of 3’ to 5’ exonucleases (Mitchell et al., 1997). Components of both pathways are also utilized by several cytoplasmic translation-dependent mRNA surveillance mechanisms that target mRNAs containing premature termination codons (nonsense-mediated mRNA decay [NMD]), mRNAs lacking translational termination codons (non-stop decay [NSD]), and mRNAs impaired in translational elongation (no-go decay [NGD]) (Doma and Parker, 2006; Frischmeyer et al., 2002; Jacobson and Peltz, 1996; van Hoof et al., 2002).
Almost all mRNAs subject to 5′ to 3′ decay, whether normal or aberrant, are shunted into the pathway as a consequence of specific decapping activators (Coller and Parker, 2004). For example, the decapping of conventional mRNAs is promoted by Pat1p, Dhh1p, and the Lsm1p-7p complex (Coller and Parker, 2004), and Upf1p, Nmd2p/Upf2p, and Upf3p serve a similar function for NMD substrates (He and Jacobson, 2001). The precise mechanism of action of these decapping activators has yet to be elucidated, but it is thought that they promote both departure from the translation pathway and recruitment of the Dcp1p/Dcp2p complex (Coller and Parker, 2004; Coller and Parker, 2005). Additional potential regulators of decapping have been identified either as high-copy suppressors of in vivo decapping defects caused by mutations in the DCP1 and DCP2 genes or as factors that interact with the decapping enzyme (Dunckley et al., 2001; Kshirsagar and Parker, 2004). These regulators include Edc1p, Edc2p, and Edc3p. Interestingly, depletion of each these factors does not affect general decapping rates of wild-type mRNAs or the decapping rates of nonsense-containing mRNAs (Dunckley et al., 2001; Kshirsagar and Parker, 2004), suggesting that these factors may be required for the decapping of specific mRNAs.
Cytoplasmic mRNA decay pathways can also play a role in the degradation of intron-containing pre-mRNAs. Normally, such pre-mRNAs are retained in the nucleus to be processed to mRNAs and are rarely exported to the cytoplasm. Pre-mRNAs that fail to complete splicing or 3’-end formation are usually degraded by the nuclear exosome (Bousquet-Antonelli et al., 2000; Torchet et al., 2002). However, cis-mutations in the 5’ splice site or the branchpoint region or trans-mutations in several splicing factors (including Prp6p, Prp9p, and the U1 snRNA), or the Mlp1p nuclear retention factor, result in the export of pre-mRNAs to the cytoplasm (Galy et al., 2004; Hilleren and Parker, 2003; Legrain and Rosbash, 1989; Rain and Legrain, 1997). As shown by our previous studies, pre-mRNAs that escape the nuclear retention system and enter the cytoplasm are usually recognized by ribosomes and degraded in a translation-dependent manner by the NMD pathway (He et al., 1993; Vilardell et al., 2000).
We have been using microarray analysis to better understand the function of the cytoplasmic mRNA decay pathways (He et al., 2003). Application of this methodology to the NMD and 5′ to 3′ decay pathways identified endogenous substrates of the respective pathways and provided insights into the determinants of substrate status (He et al., 2003). Here, we report that comparable delineation of the substrates of Edc3p identified a specific cytoplasmic decay pathway involved in the degradation of intron-containing YRA1 pre-mRNA. Subsequent investigation of the means by which YRA1 pre-mRNA is actively exported to the cytoplasm revealed a complex autoregulatory network that involves splicing inhibition, Crm1p-mediated nuclear export, and Edc3p-mediated decapping of the pre-mRNA.
Results
Deletion of EDC3 Selectively Stabilizes YRA1 Pre-mRNA
To assess the role of Edc3p in mRNA decay, we utilized high-density oligonucleotide microarrays to analyze the effect of EDC3 deletion on global RNA accumulation. Five independent expression profiling experiments with EDC3 and edc3Δ strains indicated that, of 7839 potential transcripts analyzed, only two were differentially expressed (see Supplementary Data) and both showed increased levels in the edc3Δ strain. One transcript, from the RPS28B gene, codes for a 40S ribosomal protein (Lecompte et al., 2002) and the other, from the YRA1 gene, codes for an hnRNP-like protein (Yra1p) involved in an early stage of mRNA export (Portman et al., 1997; Strasser and Hurt, 2000). Significantly, the RPS28B transcript, identified in a similar screen as the sole differentially expressed mRNA in edc3Δ cells, has been shown to be degraded through an Ecd3p-mediated mRNA decay pathway (Badis et al., 2004). Thus, in this study, we focused our analysis on the YRA1 transcript(s).
The YRA1 gene contains an intron in the middle of its coding region and has the potential to produce an intron-containing pre-mRNA and a mature mRNA. To validate our microarray data and to identify the RNA species affected by deleting EDC3, we examined the steady-state levels of the YRA1 transcripts in EDC3 and edc3Δ strains. As shown in Figure 1A, deletion of EDC3 had no effect on the level of YRA1 mRNA, but resulted in a five-fold increase in YRA1 pre-mRNA. As controls, we found that deletion of EDC3 did not affect the levels of the intron-lacking CYH2 and PGK1 mRNAs or the intron-containing CYH2 and DBP2 pre-mRNAs (data not shown).
Figure 1. YRA1 Pre-mRNA is Selectively Stabilized in edc3Δ Cells.
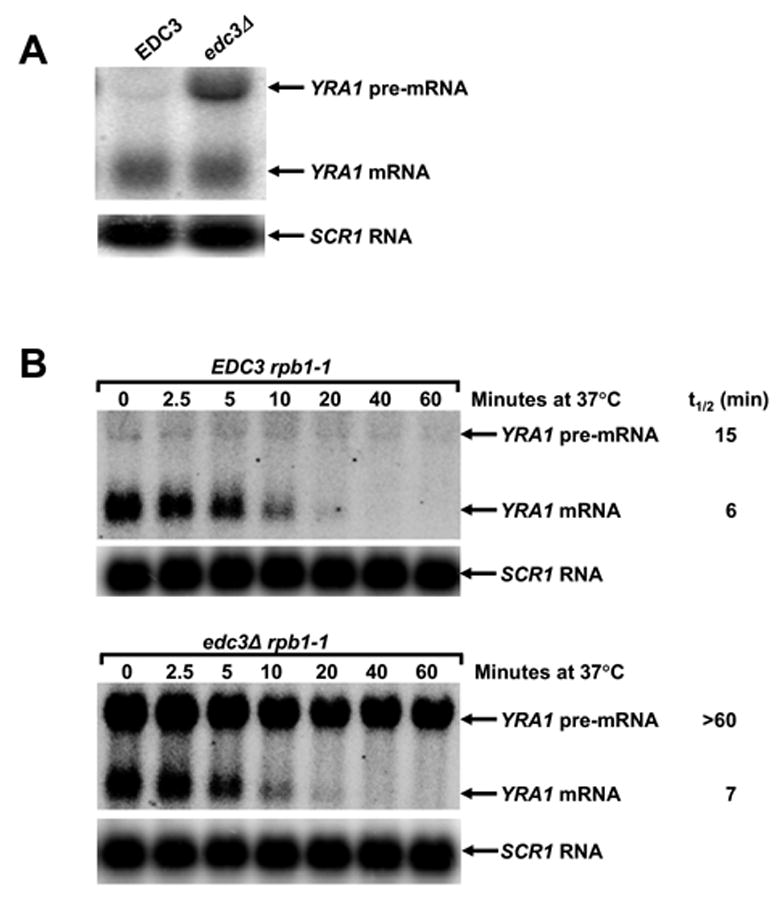
(A) Northern analysis of YRA1 pre-mRNA and mRNA levels in EDC3 and edc3Δ cells.
(B) Determination of YRA1 pre-mRNA and mRNA half-lives. Cultures of wild-type and edc3Δ cells harboring the rpb1-1 allele were shifted from 25°C to 37°C to inactivate transcription by RNA polymerase II and samples were taken for northern analysis at different times after the shift.
In panels A and B blots were hybridized with probes complementary to YRA1 and SCR1 transcripts, with the latter serving as a loading control.
To determine whether Edc3p plays a direct role in YRA1 pre-mRNA degradation, we monitored YRA1 pre-mRNA decay kinetics subsequent to inhibiting transcription. This analysis revealed that the YRA1 pre-mRNA has a half-life >60 min in edc3Δ cells and ~15 min in EDC3 cells (Figure 1B). In contrast, deletion of EDC3 did not alter the decay rate of the YRA1 mRNA (t1/2~6 min). Deletion of EDC3 also did not alter the decay rates of the CYH2, PGK1, and RPS28A mRNAs (data not shown). Taken together, these results indicate that Edc3p directly controls YRA1 pre-mRNA degradation.
YRA1 Pre-mRNA is Degraded Through a 5’ to 3’ Decay Mechanism
To elucidate the mechanism of YRA1 pre-mRNA decay, we analyzed its level in strains containing deletions of genes encoding well characterized factors involved in deadenylation, the general 5’ to 3’ or 3’ to 5’ decay pathways, or the NMD pathway. Among these factors, only deletion of the genes encoding Dcp1p, a component of the decapping enzyme, and Xrn1p, the cytoplasmic 5’ to 3’ exoribonuclease, affected YRA1 pre-mRNA levels (Figure 2A). Compared to the level in wild-type cells, deletion of these two genes resulted in 5- to 10-fold increases in YRA1 pre-mRNA levels. In contrast, deletion of the genes encoding all the other factors, including the major cytoplasmic deadenylase, Ccr4p (Tucker et al., 2002), the exosome components, Ski2p and Ski7p, the decapping activators, Pat1p, Dhh1p, Lsm1p, and Lsm7p, and the NMD factors, Upf1p, Nmd2p, and Upf3p, had no effect on YRA1 pre-mRNA accumulation (Figures 2A, 2B, and 2C). These results indicate that YRA1 pre-mRNA is degraded by a 5’ to 3’ mechanism that requires decapping by Dcp1p and Dcp2p, and 5’ to 3’ exonucleolytic digestion by Xrn1p.
Figure 2. YRA1 Pre-mRNA is Degraded by Deadenylation-independent Decapping and 5′ to 3′ Exonucleolytic Digestion.
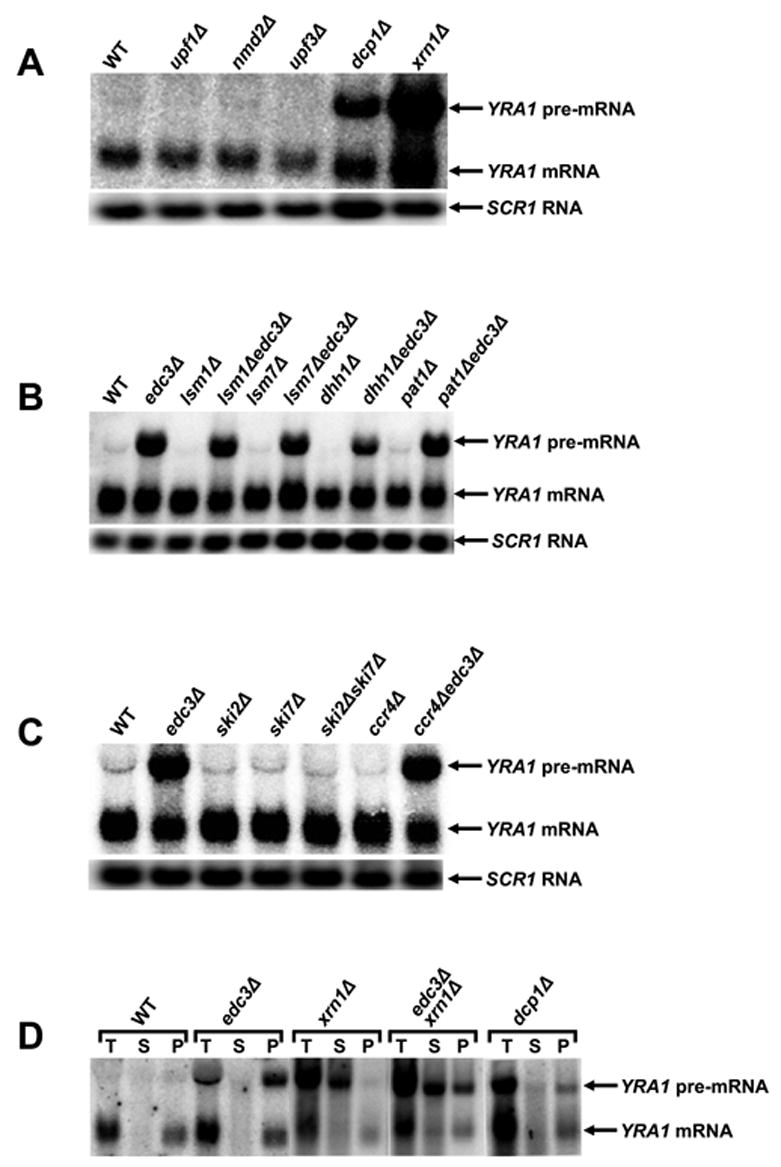
(A–C) Northern analysis of YRA1 pre-mRNA and mRNA in yeast strains containing deletions of genes encoding: (A) the principal NMD factors, a subunit of the decapping enzyme, or the 5’ to 3’ exoribonuclease; (B) decapping activators of the general 5’ to 3’ mRNA decay pathway; or (C) components of the cytoplasmic exosome or the major cytoplasmic deadenylase complex. Blots were hybridized to a SCR1 probe to serve as a loading control.
(D) Analysis of the 5’ cap status of the YRA1 pre-mRNA by anti-m7G immunoprecipitation and northern blotting. T: total input RNA, S: supernatant (uncapped), P: pellet (capped).
Edc3p-mediated YRA1 Pre-mRNA Degradation Occurs in the Cytoplasm
The observation that YRA1 pre-mRNA degradation requires Dcp1p and Xrn1p, two cytoplasmic factors, strongly suggests that Edc3p-mediated degradation of YRA1 pre-mRNA occurs in the cytoplasm. To test this hypothesis, we assessed the subcellular localization and levels of YRA1 pre-mRNA in wild-type, edc3Δ, dcp1Δ, and xrn1Δ strains by fluorescent in situ hybridization (FISH) analysis. We utilized sets of Cy3- and Cy5-labelled oligonucleotide probes to respectively detect YRA1 intron and exon sequences. In wild-type cells, the exon signal was detected in the nucleus and cytoplasm, whereas the intron signal was mainly detected in the nucleus, co-localizing with the exon signal and likely reflecting nascent YRA1 transcripts (Figure 3, panels a,b,c). Compared to the YRA1 exon and intron signals in wild-type cells, edc3Δ, xrn1Δ, dcp1Δ, and xrn1Δedc3Δ cells all showed significant increase in both the exon and intron signals in the cytoplasm. In addition, the cytoplasmic intron signals in these cells co-localized largely with the exon signals (Figure 3, panels e-t). Interestingly, dcp1Δ cells displayed a local enrichment of YRA1 pre-mRNA in cytoplasmic dots, suggesting that YRA1 pre-mRNA may enter P-bodies but fail to be degraded due to the lack of Dcp1p (Figure 3, panels q–t). These data demonstrate that YRA1 pre-mRNA degradation occurs after the transcript has been exported to the cytoplasm and that when degradation is inhibited by deleting EDC3, DCP1, or XRN1, YRA1 pre-mRNA accumulates in the cytoplasm.
Figure 3. Edc3p-mediated YRA1 Pre-mRNA Degradation Occurs in the Cytoplasm.
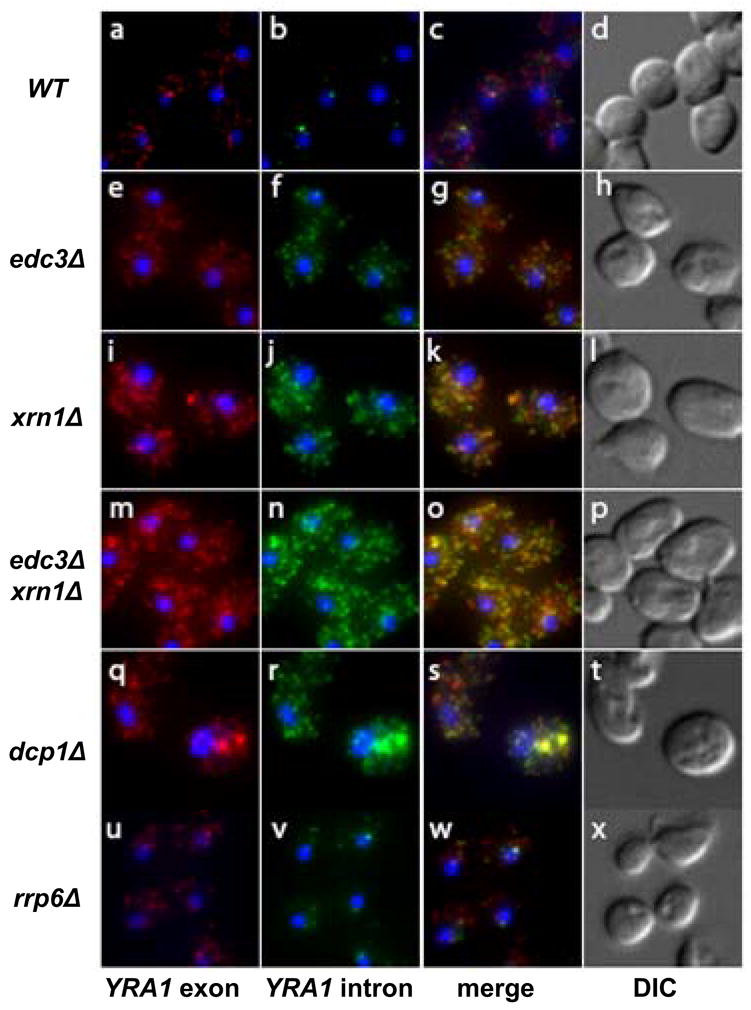
Localization of YRA1 pre-mRNA in wild-type, edc3Δ, xrn1Δ, xrn1Δedc3Δ, dcp1Δ, and rrp6Δ strains by in situ hybridization (FISH). Intron probes were labeled with Cy3 (green) and exon probes were labeled with Cy5 (red). DAPI labeling was used to identify the position of the nucleus. The corresponding merged and phase-contrast images are shown on the right side of the figure.
We also analyzed the effects of inactivation or deletion of nuclear decay factors on YRA1 pre-mRNA accumulation. We found that neither inactivation of the 5’ to 3’ exoribonuclease, Rat1p, an essential component of the nuclear 5’ to 3’ decay pathway (Bousquet-Antonelli et al., 2000), nor deletion of the gene encoding Rrp6p, a component of the nuclear exosome involved in the nuclear 3’ to 5’ decay pathway (Bousquet-Antonelli et al., 2000), affected YRA1 pre-mRNA levels (Figure 1S, Supplementary Data). Taken together, these results indicate that Edc3p-mediated YRA1 pre-mRNA degradation occurs in the cytoplasm.
Edc3p Activates, But Does Not Catalyze Decapping of the YRA1 Pre-mRNA
To define the functional role of Edc3p inYRA1 pre-mRNA decay, we analyzed YRA1 pre-mRNA cap status. As shown in Figure 2D, YRA1 pre-mRNAs that accumulate in edc3Δ and dcp1Δ cells are essentially all in the capped fraction. In contrast, pre-mRNA transcripts that accumulate in xrn1Δ cells are essentially all in the uncapped fraction. However, the pre-mRNAs that accumulate in edc3Δxrn1Δ cells contain both capped (~40%) and uncapped (~60%) species, demonstrating that deletion of EDC3 inhibits but does not eliminate decapping and that Edc3p activates but does not catalyze decapping of YRA1 pre-mRNA.
Edc3p-mediated Degradation of YRA1 Pre-mRNA is a Component of an Autoregulatory Negative Feedback Loop
Previous studies indicated that YRA1 regulates its own expression through a negative feedback loop and that this autoregulation requires the YRA1 intron (Preker et al., 2002; Rodriguez-Navarro et al., 2002). Our observation that Edc3p is required for YRA1 pre-mRNA degradation raised the possibility that Edc3p plays a role in YRA1 autoregulation. To test this notion, we examined the effects of increasing YRA1 gene copy number on the levels of its pre-mRNA, mRNA, and protein in wild-type and edc3Δ strains. As shown in Figure 4A, introduction of extra copies of the intron-containing YRA1 allele altered neither YRA1 mRNA levels nor Yra1p levels in EDC3 and edc3Δ strains. Interestingly, introduction of extra copies of YRA1 differentially affected YRA1 pre-mRNA levels, resulting in only a 2-fold increase in the EDC3 strain but a 24-fold increase in the edc3Δ strain. In contrast to the intron-containing YRA1 allele, introduction of extra copies of an intron-lacking YRA1 allele increased the levels of YRA1 mRNA 4-fold and Yra1p 3-fold in both the EDC3 and edc3Δ strains. These results confirm that Yra1p negatively regulates its own level of expression (Preker et al., 2002; Rodriguez-Navarro et al., 2002), and further indicate that Edc3p-mediated YRA1 pre-mRNA degradation is a component of this negative feedback loop. Since Edc3p-mediated YRA1 pre-mRNA degradation occurs in the cytoplasm (see above), this result also suggests that Yra1p regulates its expression by inhibiting YRA1 pre-mRNA splicing and effecting or promoting pre-mRNA nuclear export.
Figure 4. YRA1 Autoregulation Requires Edc3p and Two Distinct Regulatory Elements Localized in Exon1 and the Intron of YRA1 Pre-mRNA.
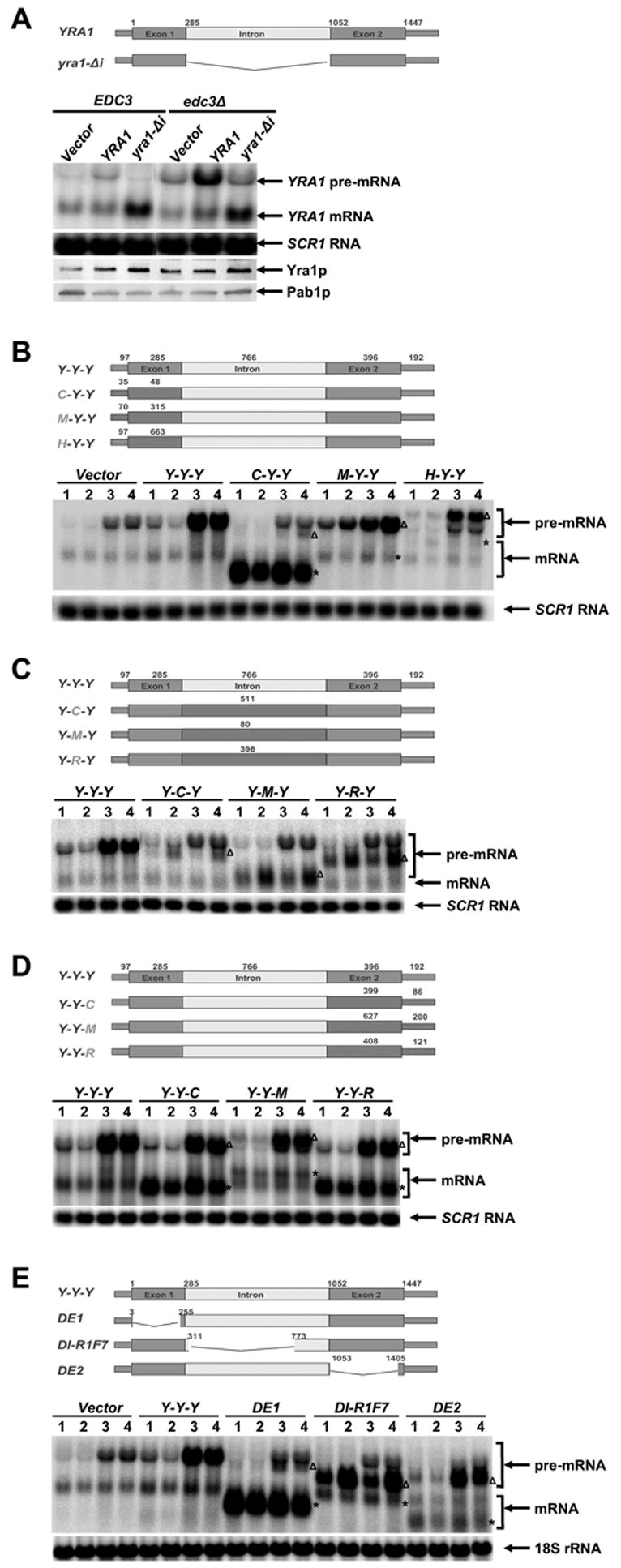
(A) The effects of overexpressing intron-containing or intron-lacking YRA1 alleles on the levels of YRA1 pre-mRNA, mRNA, and Yra1p in EDC3 and edc3Δ strains.
(B–D) Effects of replacing YRA1 exon1 (B), intron (C), and exon2 (D) with different sequences on YRA1 pre-mRNA and mRNA expression. A three-letter code was used to denote the order of exon1, intron, and exon2 of each chimeric construct, with the first letter of each of these genes specifying its origin. Y: YRA1, C: CYH2, M: MER2, R: RPS51A, and H: HIS3.
(E) Effects of internal deletions in YRA1 exon1, intron, or exon2 on YRA1 pre-mRNA and mRNA expression.
In panels B, C, D, and E YCp low-copy plasmids harboring the wild-type YRA1 gene, or a chimeric or deletion allele (depicted above the corresponding blots), were introduced into wild-type (1), upf1Δ (2), edc3Δ (3), or upf1Δ edc3Δ (4) strains and the levels of the respective pre-mRNAs and mRNAs encoded by these alleles were analyzed by northern blotting. The positions of chimeric or mutant pre-mRNAs and mRNAs are marked by Δ and *, respectively. Northern blots were hybridized to a SCR1 or an 18S rRNA probe to serve as loading controls. Pab1p served as a loading control for the western blot in panel A.
Yra1p Autoregulation Requires Two Functionally Distinct cis-regulatory Elements
As described above, YRA1 autoregulation likely involves splicing inhibition, nuclear export, and cytoplasmic degradation of YRA1 pre-mRNA. To identify the cis-regulatory elements involved in these functions, we constructed chimeric RNAs encompassing segments of the YRA1 transcript and other non-Edc3p substrate RNAs and examined their decay phenotypes in wild-type, upf1Δ, edc3Δ, and upf1Δedc3Δ strains. We included the upf1Δ strains in this analysis because we speculated that chimeric RNAs that fail to autoregulate are likely to be degraded by the NMD pathway. YRA1 exon1, intron, and exon2 sequences were replaced with the corresponding parts of the CYH2, MER2, and RPS51A pre-mRNAs, three transcripts that differ in splicing efficiency (during vegetative growth, the CYH2 and MER2 pre-mRNAs are inefficiently spliced, but the RPS51A pre-mRNA is efficiently spliced; (He et al., 1993). In addition, we also replaced YRA1 exon1 with the HIS3 coding sequence. Analyses of the steady-state levels of these chimeric pre-mRNAs and their spliced products in wild-type, upf1Δ, edc3Δ, and upf1Δedc3Δ strains led to several important observations. First, replacement of YRA1 exon1 with the CYH2 or MER2 exon1, or the HIS3 coding sequence differentially affected pre-mRNA splicing efficiency. Substitution of YRA1 exon1 with the shorter CYH2 exon1 (285 nt vs. 48 nt) dramatically increased the splicing efficiency of the pre-mRNA, as negligible C-Y-Y pre-mRNA signals were detected in the wild-type, upf1Δ, edc3Δ, and upf1Δedc3Δ strains but high levels of mRNA accumulated in these strains (Figure 4B). In contrast, substitution of YRA1 exon1 by the comparably sized MER2 exon1 (315 nt), or by a longer HIS3 coding region (633 nt), did not improve the splicing efficiency of the pre-mRNA. Interestingly, like wild-type YRA1 pre-mRNA, both the M-Y-Y and H-Y-Y pre-mRNAs accumulated to high levels in edc3Δ strains (Figure 4B and Figure 4S, Supplementary Data). Second, replacement of the YRA1 intron with the CYH2, MER2, or RPS51A intron did not alter pre-mRNA splicing efficiency but, in each case, resulted in a pre-mRNA that is insensitive to Edc3p, but sensitive to Upf1p (Figure 4C and Figure 4S, Supplementary Data). Third, replacement of YRA1 exon2 with the CYH2, MER2 or RPS51A exon2 did not alter pre-mRNA splicing efficiency and, in each case, resulted in a pre-mRNA that behaved the same as the wild-type YRA1 pre-mRNA (Figure 4D and Figure 4S, Supplementary Data). We also noted that the levels of mRNAs generated from the Y-Y-C and Y-Y-R pre-mRNAs were greatly increased in all four yeast strains (Figure 4D). This is most likely due to increased stability of these chimeric mRNAs. These results indicate that sequences in exon1 and the intron of the YRA1 pre-mRNA are required for Yra1p autoregulation and that these sequences have distinct functions. Sequences in exon1 inhibit YRA1 pre-mRNA splicing and effect or promote nuclear export of the pre-mRNA while sequences in the intron dictate the substrate specificity for Edc3p-mediated decay. Consistent with these conclusions, deletion analysis of YRA1 pre-mRNA showed that: a) a 252-nt internal deletion in exon1 promoted more efficient splicing of the YRA1 pre-mRNA; b) a 462-nt internal deletion in the intron did not significantly improve the splicing efficiency but resulted in a pre-mRNA that is degraded by NMD; and c) a 352-nt internal deletion in exon 2 did not affect YRA1 autoregulation and resulted in a pre-mRNA that behaved the same as wild-type YRA1 pre-mRNA (Figure 4E).
YRA1 Exon1 Sequences Inhibit its Pre-mRNA Splicing in a Size-dependent But Sequence-independent Manner
Since YRA1 exon1 appeared to regulate its pre-mRNA splicing through a size-dependent but sequence-independent mechanism (see above) we analyzed the effects of shortening YRA1 exon1 or replacing exon1 coding sequences with their complementary sequences. This analysis revealed that incremental deletions of exon1 resulted in incremental increases in YRA1 mRNA levels in EDC3 and edc3Δ strains (Figure 5A). Importantly, the incremental increases in YRA1 mRNA levels in both strains were all accompanied by corresponding decreases in YRA1 pre-mRNA levels in the edc3Δ strain (Figure 5A). These data show that YRA1 exon1 functions to inhibit pre-mRNA splicing in cis.
Figure 5. YRA1 Exon1 and Yra1p are cis- and trans-acting Negative Regulators of YRA1 Pre-mRNA Splicing.
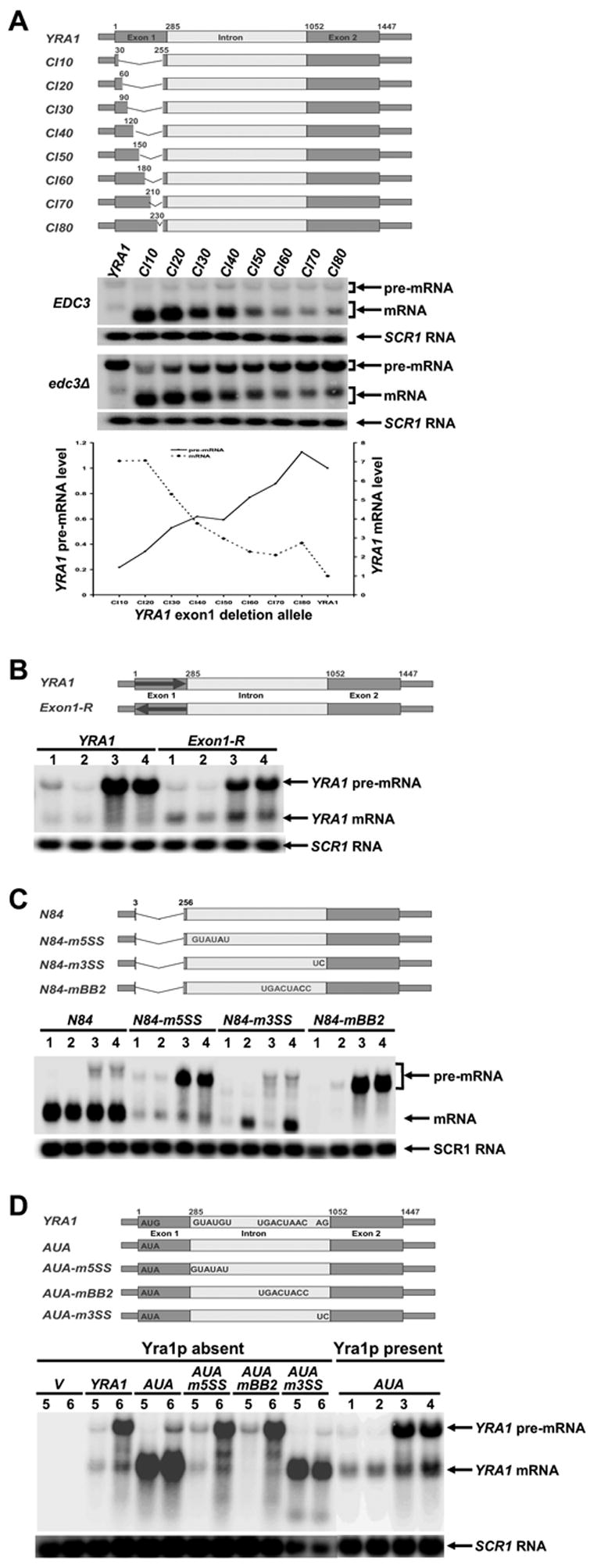
(A-D) Effects on the levels of YRA1 pre-mRNA and mRNA engendered by: (A) internal deletions in YRA1 exon1; (B) substituting YRA1 exon1 coding sequences with their complementary sequences; (C) mutations in the 5’ splice site, the 3’ splice site, or the branchpoint region of the YRA1 intron; and (D) mutations in the translation initiation codon, 5’ splice site, 3’ splice site, and branchpoint region of the YRA1 intron in the presence or absence of Yra1p. Note that, in panel B, the replaced complementary sequence does not contain in-frame premature stop codons.
In panels A, B, C, and D plasmids carrying the wild-type YRA1 gene, or one of the mutant YRA1 alleles (depicted above the corresponding northern blots), were introduced into different yeast strains and the levels of the respective pre-mRNAs and mRNAs encoded by these alleles were analyzed by northern blotting. The relevant genotypes of yeast strains used in these experiments are indicated (panel A) or denoted by numbers in panels B, C, and D: 1: wild-type, 2: upf1Δ, 3: edc3Δ, 4: upf1Δ edc3Δ, 5:EDC3 yra1Δ yra2Δ (YEplac112-YRA2), and 6: edc3Δ yra1Δ yra2Δ (YEplac112-YRA2). In panel A, the levels of mutant YRA1 pre-mRNAs and mRNAs in edc3Δ cells were quantified, normalized to the corresponding wild-type pre-mRNA and mRNA levels, and graphed below the northern blot. Blots were hybridized to a SCR1 probe to serve as a loading control.
Figure 5A also reveals that the splicing efficiency of mutant pre-mRNAs correlates positively with the size of deletions and thus negatively with the size of the remaining exon1, suggesting that the inhibitory function of exon1 on YRA1 pre-mRNA splicing is dictated by its length. Replacing the YRA1 exon1 coding sequence with its complementary sequence resulted in a pre-mRNA that behaved very similarly to the wild-type pre-mRNA, i.e., it was inefficiently spliced and degraded by the Edc3p-mediated decay pathway (Figure 5B). Since the complementary sequences of exon1 function as well as the coding sequences the ability of YRA1 exon1 to inhibit its pre-mRNA splicing must be dictated by its size, but not its primary sequence.
Our analysis of exon1 deletion mutants in the edc3Δ strain also revealed an inverse correlation between the levels of YRA1 pre-mRNA and mRNA (Figure 5A). This observation, combined with the fact that Edc3p-mediated YRA1 pre-mRNA degradation occurs in the cytoplasm, suggests that YRA1 pre-mRNA splicing and nuclear export are functionally linked and compete for common substrates. One explanation for this effect is that exon1 functions primarily to inhibit pre-mRNA splicing in cis and thus results in nuclear export of the pre-mRNA. Alternatively, YRA1 exon1 could primarily promote nuclear export of its pre-mRNA and, as a consequence, inhibit pre-mRNA splicing. To distinguish these possibilities, we tested whether cis-mutations that inhibit splicing can bypass the regulatory function of YRA1 exon1. We used yra1-N84, a complete loss-of-regulation allele that contains a 252-nt deletion in the coding region of exon1 and encodes a pre-mRNA that is efficiently spliced, as evidenced by high levels mRNA but almost no pre-mRNA in wild-type, upf1Δ, edc3Δ, and upf1Δedc3Δ strains (Figure 5C). We found that mutations in the 5’ splice site (m5SS) or the branch point region (mBB2), which block splicing prior to the first step of splicing (Jacquier et al., 1985; Parker and Guthrie, 1985), greatly reduced or eliminated splicing of the yra1-N84 pre-mRNA in wild-type, upf1Δ, edc3Δ, and upf1Δedc3Δ strains (Figure 5C). Remarkably, the m5SS and mBB2 mutations also restored yra1-N84 pre-mRNA to wild-type pre-mRNA regulation, i.e., the yra1-N84-m5SS and yra1-N84-mBB2 pre-mRNAs accumulated to high levels in edc3Δ strains (Figure 5C). In contrast, mutation of the 3’ splice site (m3SS), which inhibits the second step of splicing (Rymond et al., 1987), reduced yra1-N84 pre-mRNA splicing only modestly, as significant levels of yra1-N84 mRNA accumulated in upf1Δ cells (Figure 5C). The modest effect of the m3SS mutation on yra1-N84 pre-mRNA splicing is likely due to the use of alternative 3’ splicing signals in exon2 and the sensitivity of yra1-N84 mRNA to UPF1 probably reflects the creation of a premature stop codon as a consequence of alternative 3’ splicing. Significantly, the m3SS mutation did not restore the yra1-N84 pre-mRNA to wild-type YRA1 pre-mRNA regulation. These data show that cis-mutations that inhibit the first but not the second step of YRA1 pre-mRNA splicing completely suppress the defect caused by the N84 deletion and thus result in a bypass of the regulatory function of YRA1 exon1. These results indicate that: a) the primary function of YRA1 exon1 is to inhibit YRA1 pre-mRNA splicing, not to promote YRA1 pre-mRNA nuclear export and b) YRA1 exon1 most likely exerts its inhibitory function at or before the first step of the splicing pathway.
Yra1p Autoregulates its Level of Expression by Inhibiting YRA1 Pre-mRNA Splicing and Committing the Pre-mRNA to Nuclear Export
To further understand the role of Yra1p in its autoregulation, we used the ts yra1-1 allele to analyze the effect of Yra1p inactivation on YRA1 pre-mRNA and mRNA levels in EDC3 and edc3Δ cells. Inactivation of Yra1p resulted in decreased levels of YRA1 pre-mRNA, but increased levels of YRA1 mRNA in edc3Δ cells (Figure 2S, Supplementary Data), a result suggesting that Yra1p regulates its expression by inhibiting YRA1 pre-mRNA splicing. To test this idea further, we analyzed the effects of eliminating Yra1p. In this experiment, we generated a yra1-AUA allele that encodes a G→A substitution (AUG to AUA) in the translation initiation codon. This mutation eliminated Yra1p production and resulted in a complete loss of YRA1 function, as assessed by western blotting analysis and a genetic complementation assay (data not shown). We cloned the yra1-AUA allele, as well as the wild-type YRA1 gene, into low-copy plasmids and introduced these plasmids into EDC3 and edc3Δ strains that contain chromosomal deletions of both YRA1 and YRA2 but harbor YRA2 on a high-copy plasmid. YRA2 codes for Yra2p, a yeast homolog of Yra1p that has been previously shown to suppress the lethality of YRA1 deletion when overexpressed (Zenklusen et al., 2001). Wild-type YRA1 generated low levels of YRA1 mRNA in both EDC3 and edc3Δ cells and high levels of YRA1 pre-mRNA in an edc3Δ background (Figure 5D). These data show that introduction of wild-type YRA1 into yra1Δ strains recapitulated YRA1 autoregulation, and suggest that overexpression of YRA2 has little or no effect on YRA1 autoregulation. Compared to wild-type YRA1, the yra1-AUA allele generated 6-fold higher levels of YRA1 mRNA in both EDC3 and edc3Δ cells, and generated a 3-fold lower level of YRA1 pre-mRNA in edc3Δ cells (Figure 5D). These data indicate that YRA1-AUA pre-mRNA is efficiently spliced in both yra1Δ strains, an effect attributable to the absence of Yra1p, not the AUA mutation. We justify the latter conclusion because, when the yra1-AUA allele was introduced into EDC3 and edc3Δ strains that contain the endogenous YRA1 gene, the yra1-AUA allele behaved the same as wild-type YRA1, in that yra1-AUA pre-mRNA was inefficiently spliced and a high level of this pre-mRNA accumulated in an edc3Δ background (Figure 5D). Taken together, these results indicate that, in the absence of Yra1p, YRA1 pre-mRNA is efficiently spliced, suggesting that Yra1p regulates its level of expression by inhibiting splicing of its pre-mRNA.
A role for Yra1p in the inhibition of its pre-mRNA splicing raised the question of its primary function in YRA1 autoregulation. One possibility is that Yra1p primarily promotes YRA1 pre-mRNA export and, as a consequence, inhibits YRA1 pre-mRNA splicing. A second possibility is that Yra1p primarily inhibits YRA1 splicing and, as a consequence, promotes or commits the pre-mRNA to nuclear export. A third possibility is that Yra1p is required for both splicing inhibition and active nuclear export. To distinguish these possibilities, we tested whether cis-mutations that inhibit YRA1 pre-mRNA splicing can bypass the regulatory function of Yra1p. Accordingly, we introduced the same m5SS, mBB2, and m3SS mutations described above into the 5’ splice site, the branch-point region, and the 3’ splice site of the yra1-AUA intron. We cloned the resulting alleles into low-copy plasmids that were then introduced into the EDC3 and edc3Δ strains that contain chromosomal deletions of both YRA1 and YRA2 but harbor YRA2 on a high-copy plasmid. Northern analysis revealed that the m5SS and mBB2 mutations fully restored yra1-AUA pre-mRNA to wild-type regulation but the m3SS mutation did not (Figure 5D). These data show that cis mutations (m5SS and mBB2) that inhibit the first but not the second step of YRA1 pre-mRNA splicing completely suppress the YRA1 autoregulation defect caused by Yra1p elimination, thus bypassing the regulatory function of Yra1p. These results indicate that the primary autoregulatory function of Yra1p is to inhibit YRA1 pre-mRNA splicing, thus committing the pre-mRNA to nuclear export, and that Yra1p likely inhibits at or before the first step of YRA1 pre-mRNA splicing.
Autoregulation of YRA1 Expression Involves Mex67p
Mex67p, a general mRNA export factor in yeast, interacts genetically and physically with Yra1p (Strasser and Hurt, 2000; Zenklusen et al., 2001). To assess whether Mex67p plays a role in YRA1 autoregulation, we utilized the ts mex67-5 allele and analyzed the effect of its inactivation on the accumulation of YRA1 pre-mRNA and mRNA in edc3Δ cells. mex67-5edc3Δ and MEX67edc3Δ cells grown at 25°C accumulated comparable levels of YRA1 pre-mRNA and mRNA (Figure 6A, compare t0 samples). At 37°C, the same strains exhibited dramatically different YRA1 expression patterns (Figure 6A), including: a) MEX67edc3Δ cells had slightly decreased but sustained levels of YRA1 pre-mRNA during a 24 min time course, but the same transcript decreased rapidly in mex67-5edc3Δ cells; b) MEX67edc3Δ cells had significantly lower levels of YRA1 mRNA at 24 min than at 0 min whereas mex67-5edc3Δ cells accumulated similar mRNA levels at both time points; and c) at the 12 and 24 min time points mex67-5edc3Δ cells accumulated a novel YRA1 transcript migrating slightly slower than normal YRA1 mRNA. Since this RNA species hybridized with an mRNA-specific oligonucleotide that spanned the junction of YRA1 exons 1 and 2 and also to an oligonuclotide complementary to sequences 92-nt downstream of the mapped canonical poly(A) site (Figure 6B), we conclud that it was comprised of YRA1 mRNA with an extended 3’-UTR. MEX67edc3Δ cells also accumulated a YRA1 RNA species of similar size at 12 and 24 min. However, this RNA hybridized to an intron-specific oligonucleotide, but not to the mRNA-specific oligonucleotide and the 3’-UTR oligonucleotide (data not shown), suggesting that it is a 3’ to 5’ decay intermediate of YRA1 pre-mRNA.
Figure 6. YRA1 Autoregulation Requires the Function of Mex67p.
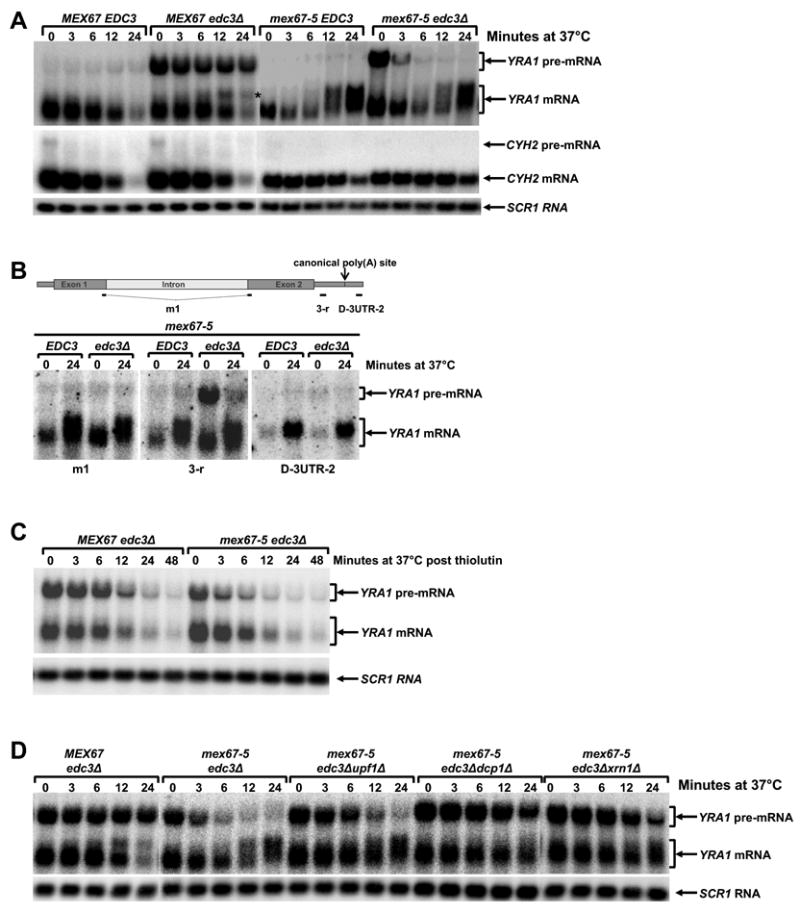
(A) Inactivation of Mex67p alters the levels of the YRA1, but not the CYH2 pre-mRNA and mRNA and also results in the accumulation of an atypical YRA1 RNA species in edc3Δ cells.
(B) Inactivation of Mex67p promotes accumulation of YRA1 mRNA with an extended 3’-UTR. RNA samples from time points 0 and 24 min of the Mex67-5 EDC3 and Mex67-5 edc3Δ strains shown in panel A were analyzed with a YRA1 oligonucleotide probe that spans exon1 and exon2 (Left), an oligonucleotide probe downstream of the YRA1 translation termination codon (Middle), or an oligonucleotide probe downstream of the canonical poly(A) addition site (Right).
(C) The effects of Mex67p inactivation on YRA1 mRNA level and the accumulation of YRA1 mRNA with an extended 3’-UTR requires ongoing transcription.
(D) Deletion of UPF1, DCP1 or XRN1 restores YRA1 pre-mRNA levels at 37°C in mex67-5 edc3Δ cells.
In panels A, C, and D, yeast strains of the indicated genotypes were grown at 25°C and then shifted to 37°C and the levels of YRA1 or CYH2 transcripts were analyzed by northern blotting. In panel C, thiolutin was added to cell cultures just before the temperature shift. A putative 3’ to 5’ decay intermediate of YRA1 pre-mRNA detected in the MEX67edc3Δ strain in panel A is marked by *. Blots were hybridized to a SCR1 probe to serve as a loading control.
Our observation that, at 37°C, mex67-5edc3Δ cells accumulated lower levels of YRA1 pre-mRNA but higher levels of YRA1 mRNA than the MEX67edc3Δ strain, and that mex67-5edc3Δ cells also accumulated YRA1 mRNA with an extended 3’-UTR, suggests that inactivation of Mex67p alters nuclear YRA1 pre-mRNA metabolism. For example, Mex67p might inhibit YRA1 pre-mRNA splicing and commit the pre-mRNA to nuclear export such that inactivation of Mex67p would allow a fraction of newly synthesized YRA1 pre-mRNA normally committed to nuclear export to adopt an alternative fate and proceed to the splicing pathway. Consistent with this interpretation, we found that the effects of inactivating Mex67p on YRA1 mRNA expression were dependent on ongoing transcription since simultaneous inhibition of transcription with thiolutin and thermal inactivation of Mex67p function resulted in decreased expression of YRA1 mRNA and eliminated the formation of YRA1 mRNA with extended 3’-UTRs (Figure 6C). Additional control experiments revealed that inactivation of Mex67p did not affect the level of the CYH2 pre-mRNA, an NMD substrate. At 37°C, mex67-5edc3Δ and MEX67edc3Δ strains accumulated similar levels of this pre-mRNA as well as its mRNA product (Figure 6A). These results indicate that the effect of Mex67p inactivation on pre-mRNA splicing is specific for YRA1 pre-mRNA.
Thermal inactivation of Mex67p resulted in almost complete disappearance of YRA1 pre-mRNA in mex67-5edc3Δ cells in 24 min (Figure 6A). While this rapid disappearance is at least partially attributable to a loss of Mex67p inhibition of YRA1 pre-mRNA splicing, other mechanisms must be operative. Since, in steady-state, YRA1 pre-mRNA is predominantly cytoplasmic in edc3Δ cells (Figure 3A) and since, in a MEX67edc3Δ background, YRA1 pre-mRNA has a half-life of ≥60 min (Figure 1B), the rapid disappearance of YRA1 pre-mRNA in mex67-5edc3Δ cells must be due principally to accelerated cytoplasmic decay of the pre-mRNA. In turn, this suggests that Mex67p is a component of the cytoplasmic YRA1 pre-mRNP and functions to repress YRA1 pre-mRNA translation and thus inhibit NMD. To test this hypothesis, we assessed whether deletion of UPF1, DCP1, or XRN1 could restore the steady-state levels of YRA1 pre-mRNA in mex67-5edc3Δ cells at 37°C. Indeed, Figure 6D shows that deletion of UPF1 restored YRA1 pre-mRNA levels at early time points and deletion of DCP1 or XRN1 restored YRA1 pre-mRNA levels at almost all time points. These results indicate that inactivation of Mex67p also triggers rapid cytoplasmic degradation of YRA1 pre-mRNA by NMD. Our observation that deletion of UPF1 resulted in a partial restoration and deletion of DCP1 or XRN1 results in a complete restoration of YRA1 pre-mRNA levels at 37°C in mex67-5 edc3Δ cells suggests that, when NMD is inactivated, the YRA1 pre-mRNA is degraded by an alternative mechanism, most likely the general 5’ to 3’ decay pathway, as we have observed previously for other nonsense-containing mRNAs (He et al., 2003). Taken together, the data of Figure 6 show that inactivation of Mex67p promotes YRA1 pre-mRNA splicing and triggers rapid cytoplasmic degradation of the pre-mRNA by NMD.
YRA1 Pre-mRNPs are Exported to the Cytoplasm by the Crm1p-mediated Export Pathway
Genome-wide two-hybrid analyses have identified an interaction between Edc3p and Crm1p (Ito et al., 2001), a result that we have confirmed (Figure 3S, Supplementary Data). Since the human Crm1p homolog is implicated in the nuclear export of unspliced or incompletely spliced HIV RNAs in mammalian cells (Cullen, 2003), the yeast Edc3p:Crm1p interaction suggested the possibility that Crm1p is involved in YRA1 pre-mRNA nuclear export. To assess the physiological relevance of the Edc3p:Crm1p interaction, we used yeast strains harboring the leptomycin-sensitive CRM1-T539C allele and analyzed the effect of inhibition of Crm1p function by leptomycin treatment. In both EDC3 and edc3Δ cells, leptomycin treatment resulted in increased accumulation of YRA1 mRNA and the appearance of a novel YRA1 mRNA with an extended 3’-UTR (Figure 7A and B). The increased accumulation of YRA1 mRNA and the appearance of YRA1 mRNA with an extended 3’-UTR in both EDC3 and edc3Δ cells after leptomycin treatment are likely to reflect one of the consequences of inhibition of Crm1p-mediated YRA1 pre-mRNA nuclear export. A simple interpretation of these results is that, when Crm1p function is inhibited, a fraction of newly synthesized YRA1 pre-mRNA that is normally committed to nuclear export adopts an alternative fate and proceeds to the splicing pathway. Consistent with this interpretation, we found that the effect of leptomycin treatment on YRA1 mRNA expression is dependent on both ongoing transcription and splicing of YRA1 pre-mRNA (Figure 7C, D and E).
Figure 7. Crm1p Mediates YRA1 Pre-mRNP Export to the Cytoplasm.
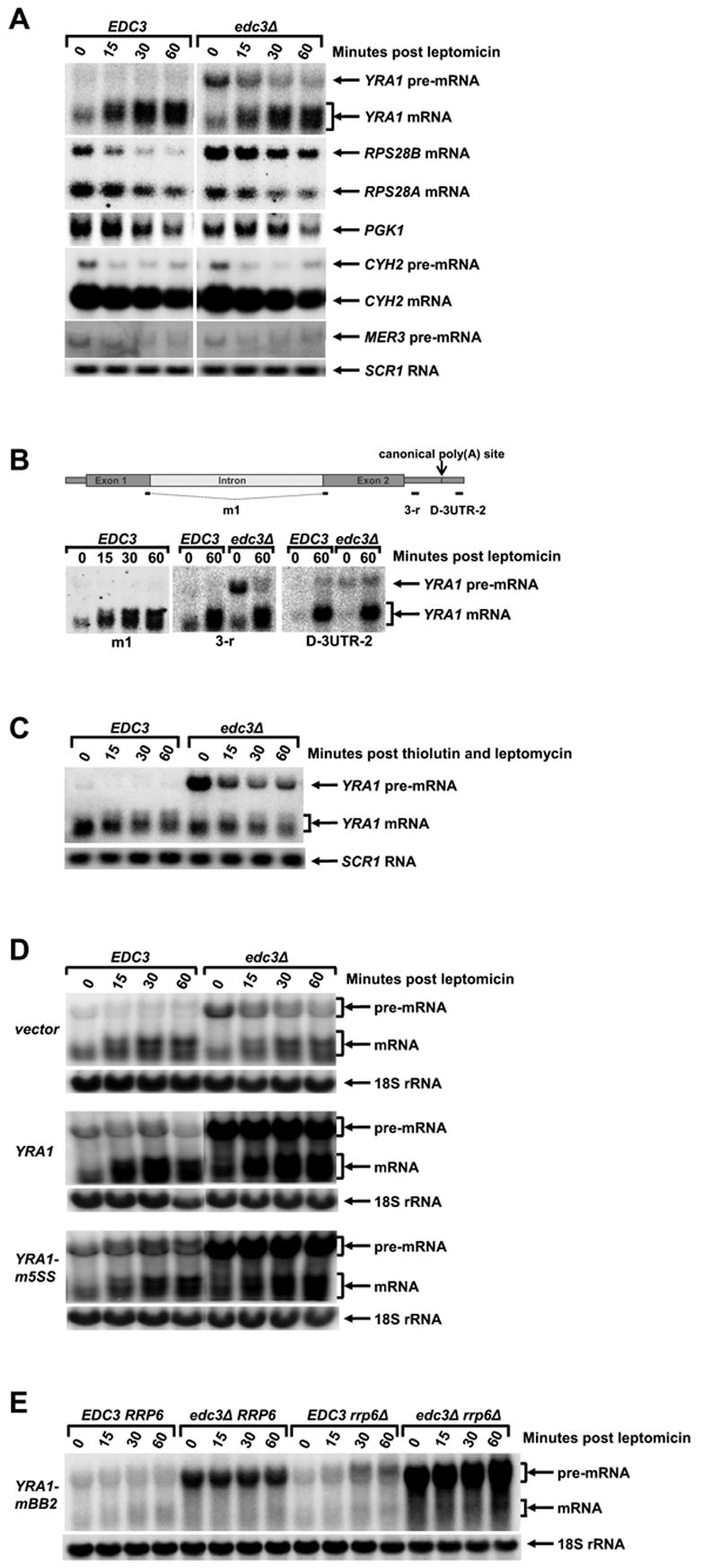
(A) Inhibition of Crm1p function increases expression of YRA1 mRNA, but slightly decreases expression of other control RNAs including the CYH2 and MER3 pre-mRNAs, and the RPS28A, RPS28B, and PGK1 mRNAs
(B) Inhibition of Crm1p promotes accumulation of YRA1 pre-mRNA and mRNA with extended 3’-UTRs. RNA samples from the indicated time points of the EDC3 and edc3Δ strains shown in panel A were analyzed with the same set of oligonucleotide probes used in Figure 6B.
(C–D) Leptomycin-promoted increases in YRA1 mRNA levels and the accumulation of YRA1 mRNA with an extended 3’-UTR requires ongoing transcription (C) and YRA1 pre-mRNA splicing (D).
(E) YRA1 pre-mRNA defective in splicing becomes a target of the nuclear exosome when Crm1p function is inhibited.
In panels A, C, D, and E yeast strains of the indicated genotypes (all harboring the CRM1-T539C allele) were treated with leptomycin and the effect on YRA1 pre-mRNA and mRNA were analyzed by northern blotting. In panel C, thiolutin and leptomycin were simultaneously added to EDC3 and edc3Δ cells. In panel D, a YCp plasmid harboring the YRA1 gene or its yra1-m5SS allele, as well as the empty vector, were introduced into EDC3 and edc3Δ strains. In panel E, a YCp plasmid harboring the yra1-mBB2 allele was introduced into yeast strains of the indicated genotypes. Blots were hybridized to a SCR1 or an 18S rRNA probe to serve as loading controls.
Leptomycin treatment also resulted in the accumulation of longer YRA1 pre-mRNA transcripts in both EDC3 and edc3Δ cells (Figure 7E). These longer pre-mRNA species hybridize with an oligonuclotide probe downstream of the mapped canonical poly(A) site (Figure 7B), indicating that these transcripts have an extended 3’-UTR. The accumulation of mRNAs with extended 3’-UTRs has been observed in many yeast mRNA export mutant strains and is a general characteristic of mRNA nuclear export defects (Forrester et al., 1992; Hammell et al., 2002). Interestingly, as observed for other mRNAs, YRA1 pre-mRNAs with extended 3’-UTRs are also the target of the nuclear exosome surveillance system (Torchet et al., 2002). When Crm1p is functional, elimination of the exosome component Rrp6p had no effect on levels of the pre-mRNA transcripts encoded by either endogenous wild-type YRA1 or by the exogenous yra1-mBB2 allele in both EDC3 and edc3Δ backgrounds (Figure 1S, Supplementary Data and Figure 7E-t0). In contrast, when Crm1p function is inhibited by leptomycin, deletion of RRP6 resulted in increased accumulation of YRA1 and yra1-mBB2 pre-mRNA transcripts with an extended 3’-UTR in both EDC3 and edc3Δ backgrounds (Figure 7E). These results show that inhibition of Crm1p function leads a fraction of newly synthesized YRA1 pre-mRNAs that normally commit to nuclear export to adopt yet another alternative fate and be degraded by the nuclear exosome.
In both EDC3 and edc3Δ backgrounds, inhibition of Crm1p function by leptomycin treatment did not alter the metabolism of several other intron-containing pre-mRNAs and intron-lacking mRNAs (Figure 7A). These include the intron-containing CYH2 and MER3 pre-mRNAs that are substrates of the NMD pathway; the RPS28B mRNA, a second substrate of the Edc3p-mediated decay pathway; and the PGK1 and RPS28A mRNAs, substrates of the general 5’ to 3’ and 3’ to 5’ decay pathways. We thus conclude that YRA1 pre-mRNPs are transported to the cytoplasm by the Crm1p-mediated export pathway and that, when this pathway is inhibited, the pre-mRNA transcripts are trapped in the nucleus and either spliced to generate mRNA or degraded by the nuclear exosome. In contrast to inactivation of Mex67p, inhibition of Crm1p function by leptomycin did not alter the YRA1 pre-mRNA decay rate in edc3Δ cells (compare Figures 7C and 1B), indicating that Crm1p function is not required for the cytoplasmic degradation of YRA1 pre-mRNA.
Discussion
YRA1 Pre-mRNA is a Cytoplasmic Substrate for Edc3p-mediated 5′ to 3′ Decay
Intron-containing pre-mRNAs are generally processed or degraded within the nucleus and rarely enter the cytoplasm (Bousquet-Antonelli et al., 2000; Legrain and Rosbash, 1989). In the limited instances where such export occurs these transcripts are often degraded by the translation-dependent NMD pathway (He et al., 2003; He et al., 1993; Vilardell et al., 2000). As shown here, the intron-containing YRA1 pre-mRNA is a notable exception to this rule. A sizeable fraction of YRA1 pre-mRNA can be exported to the cytoplasm and subsequently degraded via an NMD-independent 5’ to 3’ mechanism that requires the Dcp1p/Dcp2p decapping enzyme, the Xrn1p exoribonuclease, and a specific regulator, Edc3p.
The atypical localization of this decay pathway is surprising, but is consistent with the results of several independent approaches. Cytoplasmic localization of Edc3p-mediated YRA1 pre-mRNA degradation follows from: a) the involvement of the Dcp1p/Dcp2p complex and Xrn1p, two cytoplasmic ribonucleases (Figure 2), b) the in situ detection of YRA1 pre-mRNA in the cytoplasm of edc3Δ, dcp1Δ, or xrn1Δ cells (Figure 3), c) the resistance of YRA1 pre-mRNA degradation to inactivation or depletion of the nuclear exonucleases, Rat1p or Rrp6p, and d) the unchanged levels of YRA1 mRNA in edc3Δ cells (Figure 1), a result to be contrasted to the increased levels of mature mRNAs that generally follow the inhibition of intron-containing pre-mRNA degradation in the nucleus (Bousquet-Antonelli et al., 2000; Danin-Kreiselman et al., 2003).
Edc3p Activates Decapping of YRA1 Pre-mRNA
Our results indicate that Edc3p activates, but does not catalyze decapping of YRA1 pre-mRNA. In light of the physical interactions between Edc3p and Dcp1p or Dcp2p (Gavin et al., 2002; Ho et al., 2002; Ito et al., 2001; Uetz et al., 2000), this activation may reflect Edc3p recruitment of the decapping enzyme or one of its subunits. In the general 5’ to 3’ decay pathway, decapping requires the functions of Dhh1p, Pat1p, and the Lsm1–7p complex (Coller and Parker, 2004) and, in the NMD pathway, decapping requires the functions of Upf1p, Nmd2p/Upf2p, and Upf3p (He and Jacobson, 2001). Like Edc3p, all of these factors function as decapping activators (Coller and Parker, 2004). Edc3p-mediated decapping has several unique features, of which the most notable is its limited number of substrate transcripts. At present, we cannot exclude the possibility that Edc3p targets a much larger set of mRNAs that are underrepresented in conventional microarray analyses, e.g., transcripts lacking poly(A) tails. Another unique feature of Edc3p-mediated decapping is that it requires specific cis-regulatory elements in its target transcripts. For YRA1 pre-mRNA, we mapped the cis-element to its intron (Figure 4) and, in RPS28B mRNA, the cis-element has been localized to its 3’ UTR (Badis et al., 2004). These sequence requirements indicate that Edc3p substrate specificity is likely dictated by a specific structure and/or composition of the target mRNPs. While specific mRNP structures may also be required for other decapping activators, these structures do not appear to be mRNA-specific (Coller and Parker, 2004).
While our data and those of a previous study (Badis et al., 2004) demonstrate that decapping activation by Edc3p is transcript-specific, several observations also suggest that Edc3p may have a general function in decapping: a) Edc3p appears to exist in a complex with Dcp1p and Dcp2p in vivo (Gavin et al., 2002; Ho et al., 2002), b) Edc3p and Dcp1p have comparable numbers of molecules per cell (Huh et al., 2003), and c) simultaneous inactivation of Edc3p and mutation of DCP1 or DCP2 promotes synthetic or additive effects on both cell growth and decapping of reporter mRNAs (Kshirsagar and Parker, 2004). Given that mRNA decapping is likely to be a multi-step process in vivo (Coller and Parker, 2004), Edc3p may function at a step that is not rate-limiting for general mRNA decapping (Kshirsagar and Parker, 2004) or may have a function that is redundant with another component of the decapping complex.
YRA1 Autoregulation: Yra1p Inhibits Pre-mRNA Splicing and Commits the Transcript to Nuclear Export
Our observation that introduction of extra copies of the intron-containing YRA1 gene into EDC3 or edc3Δ strains altered neither the YRA1 mRNA level nor the Yra1p level in these cells but yielded a >10-fold, higher level of YRA1 pre-mRNA in edc3Δ cells (Figure 4) confirms that YRA1 autoregulates its own expression through a negative feedback loop (Preker et al., 2002; Rodriguez-Navarro et al., 2002) and further indicates that Edc3p-mediated YRA1 pre-mRNA degradation is a component of YRA1 autoregulation.
Further experiments defined the functional role of Yra1p in its autoregulation. Since inactivation or depletion of Yra1p resulted in increased accumulation of YRA1 mRNA in EDC3 or edc3Δ cells, but diminished accumulation of YRA1 pre-mRNA in edc3Δ cells, Yra1p must inhibit YRA1 pre-mRNA splicing (Figures 2S and 5). Moreover, since cis mutations (m5SS and mBB2) that inhibit the first but not the second step (m3SS) of YRA1 pre-mRNA splicing completely suppress the autoregulation defect caused by Yra1p depletion (Figure 5), Yra1p must inhibit its pre-mRNA splicing at or before the first step of the splicing reaction. These observations also indicate that the sole function of Yra1p in its autoregulation is to inhibit YRA1 pre-mRNA splicing and that Yra1p per se is not required for nuclear export of its pre-mRNA. Combined with our observation that yra1-AUA pre-mRNA is efficiently exported to the cytoplasm in the presence of Yra1p, but is efficiently spliced in the nucleus in the absence of Yra1p (Figure 5), these findings also suggest that Yra1p inhibits YRA1 pre-mRNA splicing by committing the transcript to nuclear export. A simple interpretation of these data is that YRA1 pre-mRNA splicing and nuclear export compete for common substrates and that, by enhancing nuclear export of its pre-mRNA, Yra1p inhibits its splicing.
Autoregulatory Roles of the Intron and Exon1
We have identified two functionally distinct cis-regulatory elements that are involved in YRA1 autoregulation: exon1 inhibits YRA1 pre-mRNA splicing and the intron is required for Edc3p-mediated YRA1 pre-mRNA degradation (Figures 4 and 5). Unexpectedly, splicing autoregulation by exon1 was found to be dictated by its length, not by specific sequences, and cis mutations that inhibit the first but not the second step of YRA1 pre-mRNA splicing completely suppress the autoregulation defect caused by the N84 deletion of exon1 (Figure 5). These findings indicate that: exon1 exerts its inhibitory function at or before the first step of the splicing reaction, the sole autoregulatory role of exon1 is to inhibit YRA1 pre-mRNA splicing, and exon1 per se is not required for nuclear export of YRA1 pre-mRNA. Further, the shared autoregulatory roles, as well as the identical patterns of genetic interactions with cis-acting mutations in YRA1 pre-mRNA, strongly suggest that Yra1p’s inhibitory function on YRA1 pre-mRNA splicing is mediated through exon1.
The primacy of size, not sequence, for the splicing inhibition function of YRA1 exon1 also indicates that the position of the YRA1 intron, rather than the intron itself, is the major determinant in YRA1 autoregulation. This contrasts with a previous conclusion that intron size is the major determinant in YRA1 autoregulation (Preker and Guthrie, 2006). Although this discrepancy may reflect differences in experimental designs and assays for splicing efficiency, our conclusion is strongly supported by the following observations. First, internal deletions in exon1 can result in more than 8-fold increases in the level of YRA1 mRNA (Figure 5A), but internal deletions in the intron had little (<2-fold) or no comparable effect (Figure 4E, and data not shown). Second, replacing the YRA1 intron with any of five other introns (from the CYH2, MER2, RPS51A, RPL25, or UBC8 transcripts) differing in size and splicing efficiency had little or no effect on YRA1 mRNA levels (Figures 4B and 4C; (Rodriguez-Navarro et al., 2002). Third, an intron position effect on splicing has also been seen with pre-mRNAs derived from the MER2 gene and from an ACT1 reporter gene (Klinz and Gallwitz, 1985; Nandabalan and Roeder, 1995).
Our observation that the inhibitory function of YRA1 exon 1 on YRA1 pre-mRNA splicing is dependent on its length, but not specific sequences, suggested a kinetic component to YRA1 autoregulation. Given that Yra1p normally functions in an early stage of mRNA export and this function is linked to polymerase II transcription elongation through the THO complex (Strasser et al., 2002), this kinetic component is likely to reflect a time window in which Yra1p is recruited to the polymerase II elongation complex and loaded onto a nascent mRNP. Such loading may be dictated by both the rate of elongation and the nuclear Yra1p concentration and we suggest that, for intron-containing pre-mRNAs, co-transcriptional packaging of Yra1p onto a nascent mRNP likely promotes the recruitment of the mRNA export receptor Mex67p and thus inhibits spliceosome assembly. For YRA1 autoregulation, high Yra1p levels would thus favor the packaging of Yra1p into YRA1 pre-mRNPs, commit the pre-mRNA to nuclear export, and as a consequence, inhibit pre-mRNA splicing. Given that the vast majority of yeast introns are located at the 5’ ends of their genes (Spingola et al., 1999), tight control of nuclear Yra1p levels could be an important mechanism that prevents premature nuclear export of intron-containing pre-mRNAs.
Roles of Mex67p and Crm1p in YRA1 autoregulation
Our data indicate that, in addition to Yra1p and Edc3p, YRA1 autoregulation also requires the functions of two general nuclear export factors, Mex67p and Crm1p. Since the inactivation of Mex67p or Crm1p function both resulted in decreased accumulation of YRA1 pre-mRNA in edc3Δ cells, increased accumulation of normal YRA1 mRNA, and the appearance of YRA1 mRNA with an extended 3’UTR in EDC3 and edc3Δ cells, it appears that Mex67p and Crm1p jointly promote YRA1 pre-mRNA nuclear export. Given that Mex67p is a general mRNA export receptor (Hieronymus and Silver, 2003; Hurt et al., 2000) and that Crm1p function is usually not required for mRNA export (Neville and Rosbash, 1999), the YRA1 pre-mRNA nuclear export requirement for both factors indicates that the YRA1 pre-RNP is exported to the cytoplasm by a mechanism that is distinct from the general Mex67p-mediated mRNA export pathway. Although the precise functions and the functional order of Mex67p and Crm1p in YRA1 pre-mRNA export are currently not clear, the facts that the function of Yra1p is linked to transcription elongation and that Yra1p physically interacts with Mex67p suggest that Mex67p is likely recruited earlier than Crm1p to YRA1 pre-mRNP. One possibility is that Yra1p first recruits Mex67p to nascent YRA1 pre-mRNP. However, the general mRNA export function of Mex67p is subsequently inhibited in this YRA1 pre-mRNP and Mex67p, in turn, serves as a unique adaptor to access the Crm1p-mediated export pathway. Consistent with this possibility is the observation that the Mex67p C-terminal domain contains a Rev-like NES and exhibits NES activity (Strasser et al., 2000).
Our observation that inactivation of Mex67p, but not Crm1p, triggers rapid degradation of YRA1 pre-mRNA by NMD in edc3Δ cells indicates that Mex67p has a second function in YRA1 autoregulation, i.e., it may be a component of the cytoplasmic YRA1 pre-mRNP and impart Edc3p substrate specificity. Since inactivation of Mex67p leads to rapid degradation of YRA1 pre-mRNA by NMD and activation of NMD is dependent on translation, these observations argue that Mex67p functions at least in part by repressing YRA1 pre-mRNA translation. The possibility that this mRNA export factor has a general role in mRNA translation is consistent with observations that. a) Mex67p shuttles between the nucleus and the cytoplasm (Segref et al., 1997), b) at steady-state, a significant fraction of Mex67p is localized to the cytoplasm and associated with polyribosomes (Segref et al., 1997; Windgassen et al., 2004), and c) the mex67-5 mutation causes aberrant accumulation of Mex67p in the cytoplasm (Segref et al., 1997).
A Model for YRA1 Autoregulation
To integrate Yra1p-mediated splicing inhibition, Crm1p-mediated nuclear export, and Edc3p-mediated cytoplasmic degradation of YRA1 pre-mRNA, we propose the following model for YRA1 autoregulation: When nuclear Yra1p levels are low, YRA1 is transcribed into pre-mRNA that, in turn, is spliced to mRNA. The mature YRA1 mRNA is exported to the cytoplasm, translated to generate Yra1p, and the latter imported into the nucleus, recruited to elongating pol II, and then packaged into nascent mRNPs to serve a function in general mRNA export. However, when nuclear Yra1p levels reach a threshold, Yra1p is likely recruited to the YRA1 gene during an early phase of transcription elongation and packaged into nascent YRA1 pre-mRNPs. Yra1p then recruits Mex67p, inhibiting pre-mRNA splicing and commiting the pre-mRNA to export. The pre-mRNA would then be exported to the cytoplasm by the Crm1p-dependent pathway where it would be degraded by the Edc3p-dependent pathway.
The molecular mechanism uncovered here for YRA1 autoregulation is reminiscent of Rev-mediated unspliced pre-mRNA nuclear export that is required for HIV replication (Cullen, 2003). This observation suggests that Rev may target the same cellular component or pathway as Yra1p to promote nuclear pre-mRNA export. The existence of a specific cytoplasmic pathway dedicated to YRA1 pre-mRNA degradation also raises the question of the biological function of this pathway. In contrast to the general decay pathways that control mRNA levels, and the quality control pathways that eliminate aberrant transcripts, the Edc3p-mediated decay pathway may serve a regulatory function, e.g., it may utilize Crm1p-mediated YRA1 pre-mRNA export to regulate the level of a key nuclear factor or coordinate mRNA decay activity with other cellular processes.
Experimental Procedures
Yeast Strains, Plasmids, Oligonucleotides, Cell Growth Conditions, Microarray Analysis, and Yeast Two-hybrid Analysis
Yeast strains, plasmids, oligonucleotides, cell growth conditions, microarray analysis, and yeast two-hybrid analysis are described in Supplemental Data.
RNA Analysis
RNA isolation and northern blotting procedures were as described previously (He and Jacobson, 1995). Transcript-specific signals were quantified with a Storm phosphorimager and normalized to levels of the 18S rRNA or SCR1 RNA, a stable RNA polymerase III transcript. RNA immunoprecipitations with monoclonal anti-2,2,7-trimethylguanosine antibodies (Calbiochem) were performed as described previously (He and Jacobson, 2001)
Protein Analysis
Preparation of whole cell extracts and western blotting procedures were as described previously (Maderazo et al., 2000). Blots were probed with polyclonal antibodies targeted to Yra1p (a generous gift from Dr. Francoise Stutz, University of Geneve) or Pab1p, with the latter polypeptide serving as a loading control.
In situ Hybridization
RNA-FISH analysis was carried out using Cy3 and Cy5 fluorochrome-conjugated oligonucletides as described at http://www.singerlab.org/protocols/insitu_yeast.htm.
Supplementary Material
Acknowledgments
This work was supported by grants GM27757 to A.J. and GM57071 to R.H.S. from the National Institutes of Health. We thank Drs. Christine Guthrie, Ed Hurt, David Mangus, Michael Rosbash, Francoise Stutz, and Karsten Weis for yeast strains and plasmids and the members of the Jacobson lab for their helpful advice and editorial comments.
Footnotes
Accession Numbers
Microarray data described in this study have been deposited into the Gene Expression Omnibus public database under the accession number GSE6647.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA Degradation by Deadenylation-Independent Decapping. Mol Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R. General Translational Repression by Activators of mRNA Decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Nuclear mRNA export: insights from virology. Trends Biochem Sci. 2003;28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- Danin-Kreiselman M, Lee CY, Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Tucker M, Parker R. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics. 2001;157:27–37. doi: 10.1093/genetics/157.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W, Stutz F, Rosbash M, Wickens M. Defects in mRNA 3′-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- Frischmeyer PA, van Hoof A, O’Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN. Coupling of termination, 3′ processing, and mRNA export. Mol Cell Biol. 2002;22:6441–6457. doi: 10.1128/MCB.22.18.6441-6457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol. 2001;21:1515–1530. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc Natl Acad Sci USA. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Silver PA. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat Genet. 2003;33:155–161. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- Hilleren PJ, Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hurt E, Strasser K, Segref A, Bailer S, Schlaich N, Presutti C, Tollervey D, Jansen R. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J Biol Chem. 2000;275:8361–8368. doi: 10.1074/jbc.275.12.8361. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Jacquier A, Rodriguez JR, Rosbash M. A quantitative analysis of the effects of 5′ junction and TACTAAC box mutants and mutant combinations on yeast mRNA splicing. Cell. 1985;43:423–430. doi: 10.1016/0092-8674(85)90172-2. [DOI] [PubMed] [Google Scholar]
- Klinz FJ, Gallwitz D. Size and position of intervening sequences are critical for the splicing efficiency of pre-mRNA in the yeast Saccharomyces cerevisiae. Nucl Acids Res. 1985;13:3791–3804. doi: 10.1093/nar/13.11.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar M, Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecompte O, Ripp R, Thierry JC, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucl Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- Maderazo AB, He F, Mangus DA, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol. 2000;20:4591–4603. doi: 10.1128/mcb.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Nandabalan K, Roeder G. Binding of a cell-type-specific RNA splicing factor to its target regulatory sequence. Mol Cell Biol. 1995;15:1953–1960. doi: 10.1128/mcb.15.4.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M, Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Guthrie C. A point mutation in the conserved hexanucleotide at a yeast 5′ splice junction uncouples recognition, cleavage, and ligation. Cell. 1985;41:107–118. doi: 10.1016/0092-8674(85)90065-0. [DOI] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nature Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Portman DS, O’Connor JP, Dreyfuss G. YRA1, an essential Saccharomyces cerevisiae gene, encodes a novel nuclear protein with RNA annealing activity. RNA. 1997;3:527–537. [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Guthrie C. Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA. 2006;12:994–1006. doi: 10.1261/rna.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Kim KS, Guthrie C. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA. 2002;8:969–980. doi: 10.1017/s1355838202020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain J-C, Legrain P. In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J. 1997;16:1759–1771. doi: 10.1093/emboj/16.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Strasser K, Hurt E. An intron in the YRA1 gene is required to control Yra1 protein expression and mRNA export in yeast. EMBO Rep. 2002;3:438–442. doi: 10.1093/embo-reports/kvf091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spingola M, Grate L, Haussler D, Ares M., Jr Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Bassler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol Cell. 2002;9:1285–1296. doi: 10.1016/s1097-2765(02)00544-0. [DOI] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Vilardell J, Chartrand P, Singer RH, Warner JR. The odyssey of a regulated transcript. RNA. 2000;6:1773–1780. doi: 10.1017/s135583820000145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol. 2004;24:10479–10491. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Strahm Y, Stutz F. The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol. 2001;21:4219–4232. doi: 10.1128/MCB.21.13.4219-4232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


