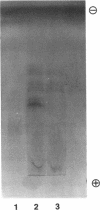
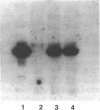
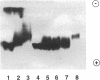
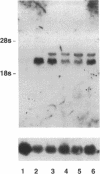
Abstract
Individuals heterozygous or homozygous for the variant aldehyde dehydrogenase (ALDH2) allele (ALDH2*2), which encodes a protein differing only at residue 487 from the normal protein, have decreased ALDH2 activity in liver extracts and experience cutaneous flushing when they drink alcohol. The mechanisms by which this allele exerts its dominant effect is unknown. To study this effect, the human ALDH2*1 cDNA was cloned and the ALDH2*2 allele was generated by site-directed mutagenesis. These cDNAs were transduced using retroviral vectors into HeLa and CV1 cells, which do not express ALDH2. The normal allele directed synthesis of immunoreactive ALDH2 protein (ALDH2E) with the expected isoelectric point. Extracts of these cells contained increased aldehyde dehydrogenase activity with low Km for the aldehyde substrate. The ALDH2*2 allele directed synthesis of mRNA and immunoreactive protein (ALDH2K), but the protein lacked enzymatic activity. When ALDH2*1-expressing cells were transduced with ALDH2*2 vectors, both mRNAs were expressed and immunoreactive proteins with isoelectric points ranging between those of ALDH2E and ALDH2K were present, indicating that the subunits formed heteromers. ALDH2 activity in these cells was reduced below that of the parental ALDH2*1-expressing cells. Thus, the ALDH2*2 allele is sufficient to cause ALDH2 deficiency in vitro.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borén J., Graham L., Wettesten M., Scott J., White A., Olofsson S. O. The assembly and secretion of ApoB 100-containing lipoproteins in Hep G2 cells. ApoB 100 is cotranslationally integrated into lipoproteins. J Biol Chem. 1992 May 15;267(14):9858–9867. [PubMed] [Google Scholar]
- Cao Q. N., Tu G. C., Weiner H. Mitochondria as the primary site of acetaldehyde metabolism in beef and pig liver slices. Alcohol Clin Exp Res. 1988 Oct;12(5):720–724. doi: 10.1111/j.1530-0277.1988.tb00271.x. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Crabb D. W., Edenberg H. J., Bosron W. F., Li T. K. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest. 1989 Jan;83(1):314–316. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto N., Takase S., Yasuhara M., Takada A. Acetaldehyde metabolism in different aldehyde dehydrogenase-2 genotypes. Alcohol Clin Exp Res. 1991 Feb;15(1):141–144. doi: 10.1111/j.1530-0277.1991.tb00532.x. [DOI] [PubMed] [Google Scholar]
- Farrés J., Guan K. L., Weiner H. Primary structures of rat and bovine liver mitochondrial aldehyde dehydrogenases deduced from cDNA sequences. Eur J Biochem. 1989 Mar 1;180(1):67–74. doi: 10.1111/j.1432-1033.1989.tb14616.x. [DOI] [PubMed] [Google Scholar]
- Farrés J., Wang X., Takahashi K., Cunningham S. J., Wang T. T., Weiner H. Effects of changing glutamate 487 to lysine in rat and human liver mitochondrial aldehyde dehydrogenase. A model to study human (Oriental type) class 2 aldehyde dehydrogenase. J Biol Chem. 1994 May 13;269(19):13854–13860. [PubMed] [Google Scholar]
- Ferencz-Biro K., Pietruszko R. Human aldehyde dehydrogenase: catalytic activity in oriental liver. Biochem Biophys Res Commun. 1984 Jan 13;118(1):97–102. doi: 10.1016/0006-291x(84)91072-6. [DOI] [PubMed] [Google Scholar]
- Goedde H. W., Agarwal D. P., Harada S., Meier-Tackmann D., Ruofu D., Bienzle U., Kroeger A., Hussein L. Population genetic studies on aldehyde dehydrogenase isozyme deficiency and alcohol sensitivity. Am J Hum Genet. 1983 Jul;35(4):769–772. [PMC free article] [PubMed] [Google Scholar]
- Goedde H. W., Singh S., Agarwal D. P., Fritze G., Stapel K., Paik Y. K. Genotyping of mitochondrial aldehyde dehydrogenase in blood samples using allele-specific oligonucleotides: comparison with phenotyping in hair roots. Hum Genet. 1989 Mar;81(4):305–307. doi: 10.1007/BF00283679. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Gritz L., Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983 Nov;25(2-3):179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- Harada S., Agarwal D. P., Goedde H. W. Aldehyde dehydrogenase deficiency as cause of facial flushing reaction to alcohol in Japanese. Lancet. 1981 Oct 31;2(8253):982–982. doi: 10.1016/s0140-6736(81)91172-7. [DOI] [PubMed] [Google Scholar]
- Hempel J., Kaiser R., Jörnvall H. Human liver mitochondrial aldehyde dehydrogenase: a C-terminal segment positions and defines the structure corresponding to the one reported to differ in the Oriental enzyme variant. FEBS Lett. 1984 Aug 6;173(2):367–373. doi: 10.1016/0014-5793(84)80807-8. [DOI] [PubMed] [Google Scholar]
- Hsu L. C., Bendel R. E., Yoshida A. Genomic structure of the human mitochondrial aldehyde dehydrogenase gene. Genomics. 1988 Jan;2(1):57–65. doi: 10.1016/0888-7543(88)90109-7. [DOI] [PubMed] [Google Scholar]
- Johnson C. T., Bosron W. F., Harden C. A., Li T. K. Purification of human liver aldehyde dehydrogenase by high-performance liquid chromatography and identification of isoenzymes by immunoblotting. Alcohol Clin Exp Res. 1987 Feb;11(1):60–65. doi: 10.1111/j.1530-0277.1987.tb01264.x. [DOI] [PubMed] [Google Scholar]
- Miller A. D., Rosman G. J. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989 Oct;7(9):980-2, 984-6, 989-90. [PMC free article] [PubMed] [Google Scholar]
- Poole R. C., Halestrap A. P. Purification of aldehyde dehydrogenase from rat liver mitochondria by alpha-cyanocinnamate affinity chromatography. Biochem J. 1989 Apr 1;259(1):105–110. doi: 10.1042/bj2590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant C., Houdebine L. M. An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Biotechniques. 1990 Feb;8(2):148–149. [PubMed] [Google Scholar]
- Schwitters S. Y., Johnson R. C., Johnson S. B., Ahern F. M. Familial resemblances in flushing following alcohol use. Behav Genet. 1982 May;12(3):349–352. doi: 10.1007/BF01067854. [DOI] [PubMed] [Google Scholar]
- Svanas G. W., Weiner H. Aldehyde dehydrogenase activity as the rate-limiting factor for acetaldehyde metabolism in rat liver. Arch Biochem Biophys. 1985 Jan;236(1):36–46. doi: 10.1016/0003-9861(85)90603-4. [DOI] [PubMed] [Google Scholar]
- Yoshida A., Huang I. Y., Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci U S A. 1984 Jan;81(1):258–261. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]