Abstract
Chlorinated solvents, especially trichloroethylene (TCE), are the most widespread groundwater contaminants in the United States. Existing methods of pumping and treating are expensive and laborious. Phytoremediation, the use of plants for remediation of soil and groundwater pollution, is less expensive and has low maintenance; however, it requires large land areas and there are a limited number of suitable plants that are known to combine adaptation to a particular environment with efficient metabolism of the contaminant. In this work, we have engineered plants with a profound increase in metabolism of the most common contaminant, TCE, by introducing the mammalian cytochrome P450 2E1. This enzyme oxidizes a wide range of important pollutants, including TCE, ethylene dibromide, carbon tetrachloride, chloroform, and vinyl chloride. The transgenic plants had a dramatic enhancement in metabolism of TCE of up to 640-fold as compared with null vector control plants. The transgenic plants also showed an increased uptake and debromination of ethylene dibromide. Therefore, transgenic plants with this enzyme could be used for more efficient remediation of many sites contaminated with halogenated hydrocarbons.
Trichloroethylene (TCE) is a widespread environmental contaminant throughout the industrialized world. Forty percent of all Superfund sites are contaminated with TCE. It is an Environmental Protection Agency priority pollutant and a suspected human carcinogen. It was commonly used as a metal degreasing agent and a dry cleaning solvent. It is now known that exposure can result in central nervous system depression, hepatotoxicity, and nephrotoxicity (1). TCE persists in the environment for decades, even though it can be metabolized by bacteria through anaerobic, reductive dehalogenation or through cometabolic oxidation (2). Bioremediation of TCE using bacteria has several disadvantages. Cometabolism of TCE is accomplished by bacteria that require the addition of inducing substrates such as methane, phenol, or toluene. The potent carcinogen, vinyl chloride, is a persistent intermediate of the anaerobic pathway (3). Another halogenated hydrocarbon, ethylene dibromide (EDB; dibromoethane), was used as a soil fumigant to kill nematodes and as a gasoline additive. It is a toxic, volatile compound that is suspected to be a human carcinogen (4, 5). Like TCE, EDB is a soil and groundwater contaminant and has been found in hundreds of wells across the United States (5).
Phytoremediation, the use of plants for remediation of soil and groundwater pollution, is a less expensive, low maintenance, and more aesthetically appealing method of remediating contaminated sites than are the conventional methods of active cleanup. Plants have been shown to take up and, in some cases, to metabolize organic pollutants (6), excess nutrients (6, 7), 2,4,6-trinitrotoluene (TNT) (8, 9), and metals (10, 11). Work in our laboratory has demonstrated that poplar trees are able to take up and metabolize TCE both under laboratory conditions and in a field test (12–14).
Although plants have been found that will naturally take up and metabolize pollutants, transgenic plants have enhanced capabilities. Transgenic yellow poplars containing a bacterial mercuric reductase were able to grow in normally toxic levels of ionic mercury and convert it to less toxic elemental mercury at a rate 10 times higher than that in nontransgenic plants (15). French and coworkers (16) developed transgenic plants able to degrade explosive nitrate esters and nitroaromatics by introducing the bacterial enzyme pentaerythritol tetranitrate reductase.
P450 2E1 (CYP2E1) is an intensively studied mammalian cytochrome that oxidizes a wide range of compounds, including TCE, EDB, carbon tetrachloride, benzene, styrene, chloroform, 1,2-dichloropropane, vinyl chloride, and others (17). Because it has activity on several major soil contaminants, P450 2E1 was a good candidate for enhancing phytoremediation. Other mammalian P450s have been introduced successfully into plants to confer herbicide resistance (18). In this study, we demonstrate that introduction of human P450 2E1 into tobacco results in a significant increase in metabolism of both TCE and EDB. This research shows that plants have been engineered to address the most widespread of groundwater contaminants, the halogenated hydrocarbons.
Experimental Procedures
Plasmid Constructions.
A 1.7-kb EcoRI fragment containing the P450 2E1 cDNA (19) kindly provided by Frank Gonzales (National Institutes of Health, Bethesda, MD) was subcloned between the Mac promoter (20) and the mas terminator (21) in the vector pLAY112, which was kindly provided by Luca Comai (University of Washington). The 3.8-kb BglII fragment containing Mac-CYP2E1-mas 3′ was subcloned into the BamHI site of the binary vector pCGN1578 (21), which was provided by Luca Comai. The resulting plasmid, pSLD3, was introduced into Agrobacterium by electroporation (22, 23).
Plant Transformation and Regeneration.
Tobacco leaf disks were transformed with the nopaline-type Agrobacterium strain C58C1 (pGV3850) (24), containing pTVK291 (25) for enhanced induction of the virulence genes, and either pCGN1578 (21) or pSLD3. Transformation and regeneration were done as described (26). Kanamycin resistant plants were verified by PCR to be transgenic. Expression of the gene was verified by reverse transcription–PCR analysis. Control plants containing the null vector, pCGN1578 (binary vector without P450 2E1), also were generated.
Western Blotting.
Total root protein was extracted by mixing powdered roots with 1.5× SDS loading buffer (2 ml/gm of fresh weight), boiling for 5 min, and centrifuging at 12,000 × g for 5 min. SDS/PAGE gels were run with a Bio-Rad miniProtean II, and the proteins were transferred to poly(vinylidene difluoride) membranes with the Amersham Pharmacia Mighty-Small Transfer Unit at 400 mA for 1 h. After blocking for 1 h with 5% nonfat milk in Tris-buffered saline (TBS), blots were incubated with diluted rabbit anti-human P450 2E1 that was kindly provided by K. E. Thummel (University of Washington) or goat anti-rat 2E1 (Gentest, Woburn, MA) for 1 h. Blots were washed three times with TBS/Tween 20 (0.1%) before incubation with the appropriate peroxidase-conjugated secondary antibody. Blots were washed four times with wash buffer (20 mM Tris, pH7.4/0.5 M NaCl/0.5% Tween 20). Blots were developed by using Amersham Pharmacia's Enhanced Chemiluminescence or Enhanced Chemiluminescence Plus according to the manufacturer's instructions. For quantitation, the blots were dried and analyzed with the Storm PhosphorImager from Molecular Dynamics. A standard curve was made with 2–20 ng of purified human P450 2E1 protein (27), generously provided by S. D. Nelson (University of Washington).
TCE Exposure.
The stems of cuttings were surface-sterilized with 10% bleach for 12 min and then rinsed three times with sterile water. Cuttings were grown hydroponically for 4–6 weeks. The plants then were moved to 500-ml flasks with a sidearm sealed with a Teflon-lined septa cap. Two sets of glass plates with small center holes were placed around the plant stem and sealed with Fluorolube (Fisher) and plumber's putty. Exposure to TCE was done in a fume hood. The hydroponic solution was dosed with 50 ml of TCE-saturated half-strength Hoagland's solution to a level of 118 μg/ml TCE. After 5 days, roots, stems, and leaves were frozen in liquid nitrogen and stored at −80°C.
EDB Exposure.
Plants were treated the same as for TCE exposure but were dosed to a level of only 2.18 μg/ml EDB because of the high toxicity of this compound (15 μg/ml EDB caused wilting, but at the levels used, the plants were healthy). The sidearms of the flasks were fitted with a mini-nert valve. Samples of the solution were taken through the mini-nert valve ≈15 min after dosing (time 0) and again after 5 days.
Extractions of TCE-Exposed Plant Tissues.
Tissues were ground with mortar and pestle in liquid nitrogen. Samples (1 g) were weighed out and transferred to chilled glass centrifuge tubes. To standardize the extractions, 500 μl of a diluted EDB solution (0.00109 μg) was added to the powdered tissues on ice. Samples (2 ml) of 1 M H2SO4/10% NaCl were added to each tube and the tubes were shaken vigorously for 1 min. Samples (10 ml) of tert-butyl methyl ether were added, and the tubes were again shaken for 1 min. Centrifugation at 4°C was done for 10 min at 8,000 × g, and 7 ml of the supernatant was transferred to vials containing 2 g of Na2SO4. After 1 h, 1-ml samples of the extract were placed in GC autosampler vials.
GC Analysis.
The GC system used consisted of a Perkin–Elmer Autosystem gas chromatograph equipped with an electron capture detector (ECD). An XTI-5 (Restek, Bellefonte, PA) 30-m, 0.25-mm internal diameter column with a stationary phase thickness of 1.0 μm was used. The oven temperature was ramped from 40°C to 200°C. Analysis was done with the turbochrom navigator (Perkin–Elmer) Version 6.1.0.2. Calibration curves were made for trichloroethanol by using commercially obtained trichloroethanol (Sigma).
The trichloroethanol peak from the 3–1 plant was verified to be trichloroethanol by using mass spectroscopy.
Duplicate extractions of the same tissue powder varied by an average of 2.7%. Some undosed samples were spiked with trichloroethanol, TCE, and chloral to verify the retention times of these compounds.
EDB Detection in the Solution.
Samples (40 μl) were diluted in 40 ml of HPLC-grade water (Aldrich) in triplicate and analyzed by using a Tekmar (Cincinnati) Purge and Trap concentrator in line with a Perkin–Elmer Autosystem XL GC with ECD.
Bromide Ion Detection.
Samples of hydroponic solution were passed through a 0.22-μM filter, and three 5-ml samples of each were run on a Dionex DX-120 ion chromatograph equipped with a Dionex AS40 autosampler. Data were analyzed with Dionex peaknet 5.01 software. Dilutions of sodium bromide were run to make a standard curve for quantification.
Results
Transformation of Tobacco.
The cDNA encoding P450 2E1 was subcloned under the control of the MAC promoter (20), which is highly expressed constitutively throughout the plant but especially in the roots. The construct was cloned into the binary vector, pCGN1578 (21), and the resulting plasmid, pSLD3, was introduced into tobacco through Agrobacterium tumefaciens-mediated transformation of tobacco leaf disks. Kanamycin-resistant plants were regenerated and the presence of the transgene was confirmed by PCR analysis. Expression of the P450 2E1 gene was verified by reverse transcription–PCR analysis. Control plants containing the null vector, pCGN1578 (binary vector without P450 2E1), also were generated.
TCE Metabolism in P450 2E1 Tobacco.
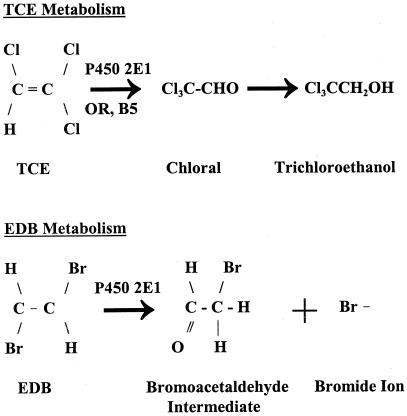
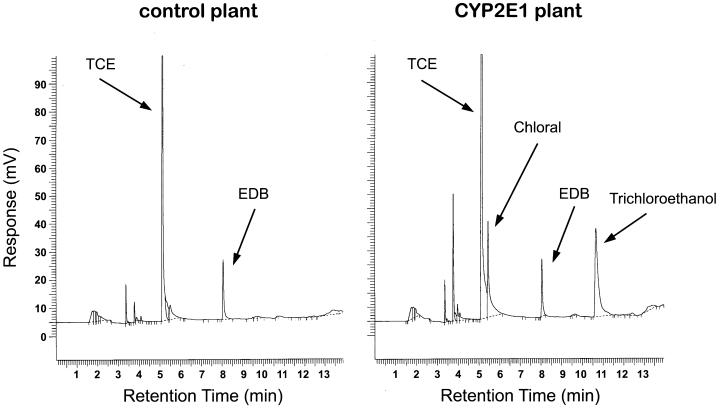
To determine the effect of P450 2E1 on TCE metabolism, plants containing P450 2E1 and those containing the vector control were dosed with TCE, and the amounts of TCE metabolites were compared. The initial detectable product of TCE oxidation by P450 2E1 is chloral (2,2,2-trichloroacetaldehyde), which is further metabolized to trichloroethanol or trichloroacetic acid (28) (Fig. 1). We chose to use hydroponically grown plants so as to study the metabolism of the plants apart from that of soil microbes. Surface-sterilized transgenic plants grown hydroponically for about 1 month were sealed into flasks and dosed to a level of 118 μg/ml TCE. Both vector control plants and P450 2E1 plants were healthy at this exposure level. After a 5-day exposure, the plant tissues were collected, extracted, and analyzed by a GC-ECD. The P450 2E1 plant roots clearly contained the early TCE metabolites, chloral and trichloroethanol, whereas in the null-vector plant root, these compounds were barely detectable (Fig. 2).
Figure 1.
P450 2E1-mediated pathways for the metabolism of TCE and EDB.
Figure 2.
Gas chromatograph of root extracts from a vector control plant (Left) and from a P450 2E1 plant (Right). The hydroponically grown plants were exposed to 118 μg/ml TCE for 5 days before the tissues were collected, frozen in liquid nitrogen, and processed. Powdered root samples were spiked with equal amounts of EDB to serve as an internal standard. The samples were then extracted with tert-butyl methyl ether and run on a GC-ECD. Peaks representing the internal standard EDB and TCE and its metabolites, chloral and trichloroethanol, are indicated by arrows.
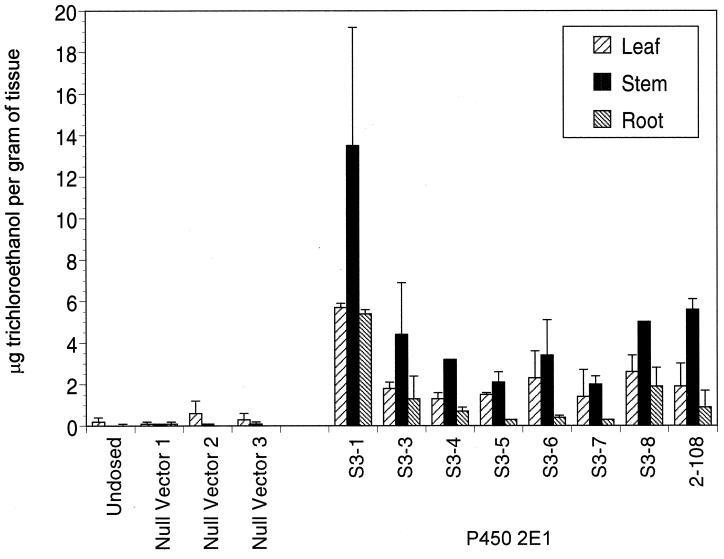
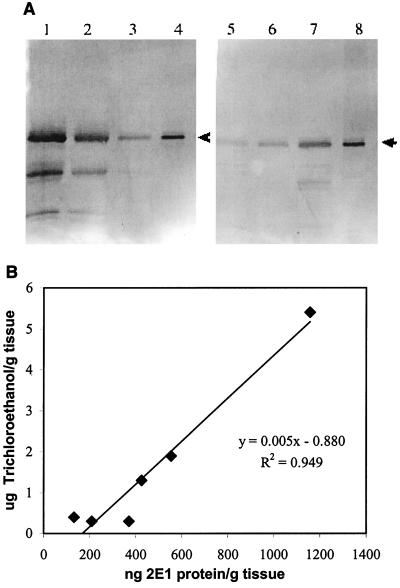
Eight plants with P450 2E1 and three null-vector plants were exposed to TCE for 5 days and the tissues were analyzed for metabolites. The amount of the TCE metabolite trichloroethanol was greatly increased in all of the P450 2E1 plants as compared with that in the vector control plants (Fig. 3). The amount of trichloroethanol in the highest-expressing plant, 3–1, was elevated by 642-fold in roots, 171-fold in stems, and 140-fold in leaves as compared with the null-vector plants. The average increase in the other P450 2E1 plants was 149-fold in roots, 44-fold in stem, and 72-fold in leaves. Quantitative Western blotting analysis was performed on the same root tissue as in the above experiment. There was a direct correlation between P450 2E1 protein levels and trichloroethanol production (Fig. 4).
Figure 3.
Trichloroethanol in tissues of transgenic tobacco plants after exposure to TCE for 5 days. Three transgenic plants containing the null vector and eight transgenic plants containing P450 2E1 were tested for the ability to metabolize TCE. One cutting of each plant was grown hydroponically under sterile conditions before being exposed to 118 μg/ml TCE for 5 days. Tissues were extracted and, the samples were run on a GC-ECD. Values were normalized to an internal standard. Averages and SDs from two independent experiments are presented as recovery of trichloroethanol per g of tissue.
Figure 4.
(A) Western blots of root protein extracts from transgenic tobacco plants. Rabbit polyclonal anti-P450 2E1 was used as the primary antibody. Lanes 1 to 8 are extracts from plants 3–1, 3–3, 3–5, human P450 2E1 (2 ng), 3–6, 3–7, 3–8, respectively. A 15-μl aliquot of extract (equivalent to 7.5 mg of fresh weight) was loaded in each lane except for the P450 2E1 standards. The arrowheads indicate the position of P450 2E1. (B) Correlation between trichloroethanol and P450 2E1 concentrations in transgenic tobacco roots. Amounts of P450 2E1 protein in root extracts were calculated based on a quantative Western blot standard curve (R2 = 0.988), and the nanogram values were converted to concentrations based on the fresh weight of the tissue from which the extracts were prepared.
To determine whether TCE metabolism was blocked at the trichloroethanol step or whether the trichloroethanol was further metabolized, six 3–1 plants were dosed with TCE for 5 days. Three of the plants then were analyzed for trichloroethanol content. The roots of the remaining three plants were rinsed in sterile water, and the plants were placed in fresh solution without TCE for another 5 days. All of the plant tissues were extracted and analyzed by GC-ECD. Most of the trichloroethanol from root and stem was gone (91% and 94%, respectively) after 5 days but only 37% was removed from leaves. The experiment was repeated with plant 2–108 with a longer recovery period. After 10 days, 89% of the trichloroethanol was gone from roots, 93% from stem, and 81% from leaves.
EDB Metabolism.
To determine whether the halogenated hydrocarbons were being dehalogenated by the plants, the amount of the halide ion released was determined. Analysis of free chloride ion released on metabolism of TCE was not practical, because the chloride was rapidly taken up by the plants from the hydroponic solution (data not shown). Bromide ion, however, was not transported and there was no background level in the hydroponic solution of undosed plants. Thus, the transformation of EDB, a known substrate of P450 2E1, was studied.
Cuttings of the P450 2E1 plant 3–1 and null-vector control plants were grown in hydroponics and then dosed to a level of 2.18 μg/ml. After 5 days, the hydroponic solution was analyzed by ion chromatography. The level of bromide ion released in the solution by plant 3–1 was markedly increased as compared with the null-vector plants (Table 1). There was no detectable bromide ion (<0.1 mg/liter) in the hydroponic solutions of the undosed plant and of the control without a plant. There was also an increased uptake of EDB from the hydroponic solution of the P450 2E1 plant. There was an average loss of 98% of the EDB from the hydroponic solutions of the 3–1 plants as compared with 63% from the null-vector control plant solutions. These data demonstrate that the EDB was being taken up and debrominated at an increased rate in the P450 2E1 transgenic plants.
Table 1.
Enhanced uptake and degradation of EDB by transgenic tobacco containing P450 2E1
| Plant | Percentage EDB loss* | Br− released, μg/ml† |
|---|---|---|
| Null vector controls | 63.2 ± 13.3 | 0.054 ± 0.058 |
| 3-1 | 98.1 ± 1.1 | 0.243 ± 0.142 |
Four 3-1 plants and three vector control plants were exposed to EDB for 5 days, and the hydroponic solutions were analyzed. Data shown are from one experiment. Similar results were obtained in three independent experiments.
Percentage EDB loss from the hydroponic solution after 5 days.
Bromide ion release into the hydroponic solution after 5 days.
Discussion
Transgenic tobacco plants expressing P450 2E1 metabolized the environmental pollutants TCE and EDB at an enhanced rate. The largest increase in TCE metabolism when compared with the control plants was found in the roots. This finding is understandable because the MAC promoter is expressed most strongly in the roots (20). It is anticipated that the increased metabolism of TCE in transgenic plants will result in an increased uptake in the field with less time and planted area required for remediation.
By using a rat P450 1A1 fused to a yeast NADPH-P450 oxidoreductase, Shiota and coworkers (29) made transgenic tobacco plants that metabolize the herbicide chlortoluron. Like other cytochrome P450 enzymes, P450 2E1 requires the NADPH oxidoreductase and also cytochrome B5 for efficient transfer of electrons from NADPH (27, 29). It is interesting in our case that the human P450 presumably was able to interact with the tobacco NADPH-cytochrome P450 oxidoreductase and cytochrome B5. In animals and microorganisms, a single, highly conserved oxidoreductase is believed to transfer electrons to all of the different P450s. The genes encoding oxidoreductases from plants have been cloned from several plant species and have been sequenced. They show a high degree of sequence similarity with the animal oxidoreductase (30). Apparently, the mammalian and plant oxidoreductases are similar enough that a human P450 can interact with a tobacco oxidoreductase.
Although trichloroethanol was elevated in the plant tissues, it was not an end product that remained in plant tissues, which may have posed a problem for the ultimate disposal or environmental dispersion of plant materials. When tobacco plants were allowed to recover for 5 days from exposure to TCE, the amount of trichloroethanol was rapidly reduced to low levels in the roots and stems, but significant levels of the metabolite remained in leaves for several more days before these levels also dropped. The sharp decline in the amount of trichloroethanol in roots and stems suggests that trichloroethanol was transported to the leaves, where it was further metabolized. Another possibility is that the trichloroethanol was being transported out of the roots and into the solution. We have detected trace amounts (about 0.01 μg/ml) of trichloroethanol in the hydroponic solution after the plants have been dosed with 20 μg/ml TCE. However, this amount of trichloroethanol does not account for the near complete loss of trichloroethanol in the tissues. A third possibility as to why the trichloroethanol disappearance is slightly delayed in leaves is that metabolism of trichloroethanol simply may be faster in the stem and root. More experiments will need to be performed to determine the downstream metabolic products of TCE in plants.
The increased metabolism of chlorinated hydrocarbons by transgenic tobacco suggests that introduction of P450 2E1 into deep-rooted, fast-growing trees could result in a significantly increased uptake and destruction of these important pollutants. Thorough study of transgenic trees will be required to verify that toxic intermediates are not released into the environment. Because nontransgenic poplars have the same TCE metabolites as do the transgenic plants, it is likely that the plants would use the existing pathways to complete the degradation of TCE to nontoxic substances. Consumption of leaves from nontransgenic poplar exposed to TCE had no harmful effects on herbivorous insects (31).
Previous work with untransformed poplar trees has shown that chlorinated hydrocarbons such as TCE are taken up along with the water transpired (14). There is little selective uptake of TCE from groundwater; therefore, reductions in the concentration of TCE are insignificant. Our present work shows that increases in the metabolism of halogenated hydrocarbons in plants could result in significant reductions in the concentration of pollutants in the water surrounding the roots. Use of the greatly increased degradative ability of transgenic plants is likely to significantly reduce both the land area and the number of plants required for phytoremediation, increasing the number of contaminated sites on which this very economical technology can be used.
Acknowledgments
We thank Dr. Luca Comai and Dr. Sidney Nelson for their critical review of this manuscript. We also want to thank Raimund Peter for helpful discussions, Bill Howland for mass spectroscopy analysis, and Katrina Gery for plant maintenance. This work was funded by grants from the Department of Energy and the National Institute of Environmental Health Sciences.
Abbreviations
- TCE
trichloroethylene
- EDB
ethylene dibromide
- ECD
electron capture detector
References
- 1.Costa A K, Katz I D, Ivanetich K M. Biochem Pharmacol. 1980;29:433–439. doi: 10.1016/0006-2952(80)90524-9. [DOI] [PubMed] [Google Scholar]
- 2.Wackett L P, Brusseau G A, Householder S R, Hanson R S. Appl Environ Microbiol. 1989;55:2960–2964. doi: 10.1128/aem.55.11.2960-2964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ensley B D. Annu Rev Microbiol. 1991;45:283–299. doi: 10.1146/annurev.mi.45.100191.001435. [DOI] [PubMed] [Google Scholar]
- 4.Ploemen J H, Wormhoudt L W, Haenen G R, Oudshoorn M J, Commandeur J N, Vermeulen N P, de Waziers I, Beaune P H, Watabe T, Van Bladeren P J. Toxicol Appl Pharmacol. 1997;143:56–69. doi: 10.1006/taap.1996.8004. [DOI] [PubMed] [Google Scholar]
- 5.Ritter W F. J Environ Sci Health B. 1990;25:1–29. doi: 10.1080/03601239009372674. [DOI] [PubMed] [Google Scholar]
- 6.Schnoor J L, Licht L A, McCutcheon S C, Wolfe N L, Carreira L H. Environ Sci Technol. 1995;29:318–323. doi: 10.1021/es00007a747. [DOI] [PubMed] [Google Scholar]
- 7.Song B J, Gelboin H V, Park S S, Yang C S, Gonzalez F J. J Biol Chem. 1986;261:16689–16697. [PubMed] [Google Scholar]
- 8.Hughes J B, Shanks J, Vanderford M, Lauritzen J, Bhadra R. Environ Sci Technol. 1997;31:266–271. [Google Scholar]
- 9.Thompson P L, Ramer L A, Schnoor J L. Environ Sci Technol. 1998;32:975–980. [Google Scholar]
- 10.Chaney R L. Curr Opin Biotechnol. 1997;8:279–284. doi: 10.1016/s0958-1669(97)80004-3. [DOI] [PubMed] [Google Scholar]
- 11.Salt D E, Blaylock M, Kumar N P B A, Dushenkov V, Ensley B D, Chet I, Raskin I. Bio/Technology. 1995;13:468–474. doi: 10.1038/nbt0595-468. [DOI] [PubMed] [Google Scholar]
- 12.Gordon M P, Choe N, Duffy J, Ekuan G, Heilman P, Muiznieks I, Newman L, Ruszaj M, Shurtleff B B, Strand S, et al. In: Phytoremediation of Soil and Water Contaminants. Kruger E L, Anderson T A, Coats J R, editors. Washington DC: Am. Chem. Soc.; 1997. pp. 177–185. [Google Scholar]
- 13.Newman L, Strand S, Choe N, Duffy J, Ekuan G, Ruszaj M, Shurtleff B B, Wilmoth J, Heilman P, Gordon M P. Environ Sci Technol. 1997;31:1062–1067. doi: 10.1289/ehp.98106s41001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman L, Wang X, Muiznieks I, Ekuan G, Ruszaj M, Cortellucci R, Domroes D, Karscig G, Newman T, Crampton R S, et al. Environ Sci Technol. 1999;33:2257–2265. [Google Scholar]
- 15.Rugh C L, Senecoff J F, Meagher R B, Merkle S A. Nat Biotechnol. 1998;16:925–928. doi: 10.1038/nbt1098-925. [DOI] [PubMed] [Google Scholar]
- 16.French C E, Rosser S J, Davies G J, Nicklin S, Bruce N C. Nat Biotechnol. 1999;17:491–494. doi: 10.1038/8673. [DOI] [PubMed] [Google Scholar]
- 17.Guengerich F P, Kim D H, Iwasaki M. Chem Res Toxicol. 1991;4:168–179. doi: 10.1021/tx00020a008. [DOI] [PubMed] [Google Scholar]
- 18.Ohkawa H, Imaishi H, Shiota N. In: Pesticide Chemistry and Bioscience: The Food-Environment Challenge. Brooks J T, Roberts T R, editors. Cambridge, U.K.: R. Soc. Chem.; 1999. [Google Scholar]
- 19.Umeno M, McBride O W, Yang C S, Gelboin H V, Gonzalez F J. Biochemistry. 1988;27:9006–9013. doi: 10.1021/bi00425a019. [DOI] [PubMed] [Google Scholar]
- 20.Comai L, Moran P, Maslyar D. Plant Mol Biol. 1990;15:373–381. doi: 10.1007/BF00019155. [DOI] [PubMed] [Google Scholar]
- 21.McBride K E, Summerfelt K R. Plant Mol Biol. 1990;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- 22.Cangelosi G A, Best E A, Martinetti G, Nester E W. Methods Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- 23.Rogers S O, Bendich A J. In: Plant Molecular Biology Manual. Gelvin S, Schilperoort R A, editors. Dordrecht, The Netherlands: Kluwer; 1994. , Sect. D1, pp. 1–8. [Google Scholar]
- 24.Zambryski P, Joos H, Genetello C, Leemans J, Montagu M V, Schell J. EMBO J. 1983;2:2143–2150. doi: 10.1002/j.1460-2075.1983.tb01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knauf V C, Nester E W. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 26.Mathis N L, Hinchee M A W. In: Plant Molecular Biology Manual. Gelvin S, Schilperoort R A, editors. Dordrecht, The Netherlands: Kluwer; 1994. , Sect. B6, pp. 1–9. [Google Scholar]
- 27.Chen W, Peter R M, McArdle S, Thummel K E, Sigle R O, Nelson S D. Arch Biochem Biophys. 1996;335:123–130. doi: 10.1006/abbi.1996.0489. [DOI] [PubMed] [Google Scholar]
- 28.Dekant W. Dev Toxicol Environ Sci. 1986;12:211–221. [PubMed] [Google Scholar]
- 29.Shiota N, Nagasawa A, Sakaki T, Yabusaki Y, Ohkawa H. Plant Physiol. 1994;106:17–23. doi: 10.1104/pp.106.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lesot A, Hasenfratz M P, Batard Y, Durst F, Benveniste I. Plant Physiol Biochem. 1995;33:751–757. [Google Scholar]
- 31.Sorbet M. M.S. thesis. Clemson, SC: Clemson University; 1998. [Google Scholar]