Abstract
Eukaryotic nuclei contain three different types of RNA polymerases (RNAPs), each consisting of 12–18 different subunits. The evolutionarily highly conserved RNAP subunit RPB5 is shared by all three enzymes and therefore represents a key structural/functional component of all eukaryotic RNAPs. Here we present the crystal structure of the RPB5 subunit from Saccharomyces cerevisiae. The bipartite structure includes a eukaryote-specific N-terminal domain and a C-terminal domain resembling the archaeal RNAP subunit H. RPB5 has been implicated in direct protein-protein contacts with transcription factor IIB, one of the components of the RNAPII basal transcriptional machinery, and gene-specific activator proteins, such as the hepatitis B virus transactivator protein X. The experimentally mapped regions of RPB5 involved in these interactions correspond to distinct and surface-exposed α-helical structures.
RNA polymerases (RNAPs) are key enzymes responsible for the regulated expression of all genes within the eukaryotic nucleus. Despite their central role in all transcription processes, many functional aspects of multisubunit RNAPs are still only poorly understood. By obtaining more detailed insights into the structure of individual RNAP subunits and their quaternary arrangement within intact enzymes it should, however, be possible to understand in greater detail how RNAPs are recruited to promoters and interact with other components of the various transcriptional machineries to control the steps involved in specific transcript initiation, elongation, and termination.
Although prokaryotic cells contain only a single RNAP that is responsible for the transcription of all genes, eukaryotes have evolved three different enzymes (RNAPI, RNAPII, RNAPIII) that carry out type-specific transcription programs in conjunction with a number of biochemically distinct accessory factors (1). RNAPI transcribes the 18S-5.8S-28S ribosomal RNA precursor, RNAPII transcribes all pre-mRNAs, and RNAPIII is involved in the production of tRNAs and a variety of other small RNAs. The subunit composition of the three nuclear RNAPs has been most extensively investigated in Saccharomyces cerevisiae. These studies have revealed the presence of two large subunits (RPA1/RPB1/RPC1 and RPA2/RPB2/RPC2, respectively) that are homologous to the β′ and β in the bacterial enzymes and contain the main catalytic center (2). In addition, eukaryotic RNAPs contain 10–16 additional smaller polypeptides whose contributions to overall RNAP function are currently not well understood (1, 3). None of these subunits displays any substantial degree of primary sequence homology to other proteins of known function. The presence of closely related homologues of genes encoding these yeast polypeptides in all higher eukaryotes (including man) shows that many of the fundamental aspects of RNA polymerase architecture are essentially invariant and have been highly conserved throughout evolution of the eukaryotic domain. This view has been directly confirmed by experimental evidence showing that many human RNAP subunits can fully complement the functions of their yeast counterparts in vivo (4, 5).
During the last decade, it has also become obvious that archaea, a prokaryotic life form distinct from the eukaryotic as well as the bacterial domains (6), contain an extensive array of eukaryote-like RNAP subunits. Many of these subunits display a clear sequence homology to their eukaryotic counterparts (7, 8) and, in those cases analyzed, interact with each other in a comparable manner (9). These results indicate that the main structural features of the archaeal/eukaryotic RNAPs became firmly established more than 1.8 billion years ago (10) and have remained remarkably unchanged ever since.
Although the RNAP present in the common ancestor of the archaeal/eukaryotic evolutionary lineage would presumably have been capable of transcribing every gene in the genome, subsequent evolution of eukaryotic cells led to the emergence of specialized RNAPs containing type-specific subunits. Such subunits are likely to play important roles in ensuring the type-specific assembly of the different RNAPs in vivo and in facilitating their communication with a range of specialized transcription factors. Nevertheless, five subunits have remained universal and are present in all three enzymes (11). They are not thought to make any direct contribution to the catalytic RNAP activity but are likely to play a pivotal role by providing a structural scaffold for assembling and maintaining a common quaternary structure.
To gain further insights into the function of RNA polymerases, a number of structural studies of individual recombinant subunits have been carried out, including NMR structures of human RPB6 (12), yeast RPB8 (13), archaeal RPB5 (14), and the C-terminal domain of archaeal RPB9 (15). This detailed information on individual subunits has recently been complemented by comprehensive x-ray structures of intact RNAPs, such as a bacterial core RNAP (16) and a low resolution electron density map of yeast RNAPII (17).
Here we describe the 1.9 Å structure of S. cerevisiae RPB5 (yRPB5), which is the largest of the universal RNAP subunits. This subunit is particularly intriguing because of the relative wealth of information available about its possible functional role. Previous studies have shown that RPB5 is in close contact to promoter DNA when RNAPII is recruited into the preinitiation complex (18). Furthermore, RPB5 has been shown to interact with the basal RNAPII transcription factor IIB (TFIIB) and has been implicated as a potential interaction target for transcriptional activators (19–21). There is conflicting evidence on the stoichiometry of RPB5 in the RNAP core. Various experimental observations suggested that RPB5 might be present as a homodimer in endogenous RNAPII (21, 22). The structure of yRPB5 presented here sheds light on these findings by providing a structural basis for the interpretation of the experimental data.
Methods
Expression, Purification, and Crystallization of yRPB5.
Recombinant full-length yRPB5 was expressed as a fusion with glutathione S-transferase in Escherichia coli BL21 (DE3) cells and was purified as described (23). Crystals were grown at 4°C by using the vapor diffusion hanging-drop method with a protein concentration of 15–20 mg/ml. Initially, monoclinic crystals were obtained from a solution containing 20–24% polyethylene glycol 8000, 15% glycerol, 0.2 M ammonium sulfate, and 0.1 M sodium cacodylate (pH 6.5). These crystals were very temperature-sensitive but could be stabilized in a solution containing 30% polyethylene glycol 8000, 15% glycerol, and 0.1 M sodium cacodylate (pH 6.5) and used for data collection at cryogenic temperatures. On harvesting, a slight rearrangement of the lattice caused a change in space group, and the crystals showed a diffraction pattern consistent with the orthorhombic space group C2221 with cell dimensions of approximately a = 44 Å, b = 82 Å, and c = 135 Å. Attempts to find heavy atoms derivatives were unsuccessful because of lack of isomorphism between native crystals, with Rderiv up to 20% between different data sets. The structure was therefore solved by the multiple anomalous dispersion method using selenomethionyl-substituted protein, which was expressed in the E. coli Met auxotroph B834 (DE3) strain and was purified by using the same protocol.
Data Collection.
All data were collected at 100 K in a stream of cold nitrogen produced by a Cryostream Cooler (Oxford Cryosystems, Oxford, U.K.). A native diffraction data set to 1.9 Å resolution was obtained on station 9.6 at the Synchrotron Radiation Source (Daresbury, U.K.), on an ADSC Quantum-4 CCD detector. Multiple wavelength anomalous dispersion data to 2.3 Å were recorded from a single SeMet crystal at beam line 5.2R at ELETTRA (Trieste, Italy) on a Mar345 imaging plate system. Three wavelengths were collected: λ1 at the maximum of the selenium absorption edge, λ2 at the inflection point, and λ3 on the high energy side remote from the absorption edge. The images were processed and integrated by using the program mosflm (A. G. W. Leslie, personal communication), and all data handling leading to electron density maps was done by using the CCP4 program package (24). Data collection statistics are summarized in Table 1.
Table 1.
Data collection, phasing, and refinement statistics
| Native | White line, λ1 | Inflection point, λ2 | Remote, λ3 | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength, Å | 0.87 | 0.9790 | 0.9792 | 0.935 |
| Resolution, Å | 15–1.9 | 20–2.3 | 20–2.3 | 20–2.3 |
| Unique reflections | 19,245 | 10,977 | 10,990 | 11,006 |
| Multiplicity, % | 4.7 | 4.2 | 3.9 | 4.0 |
| Completeness, %* | 96.9 (98.2) | 99.2 (98.5) | 99.3 (99.3) | 99.6 (99.6) |
| Rmerge, %*† | 5.4 (22.6) | 4.1 (17.8) | 3.9 (20.0) | 4.2 (25.6) |
| Intensity/σ* | 8.1 (2.9) | 12.1 (3.9) | 13 (3.5) | 10.3 (2.7) |
| Phasing, 10–2.5 Å | ||||
| RCullis, centric/acentric‡ | 0.80/0.79 | 0.62/0.67 | ||
| RCullis, anomalous§ | 0.49 | 0.61 | 0.68 | |
| Phasing power, centric/acentric¶ | 0.78/1.12 | 1.18/1.71 | ||
| Mean figure of merit | 0.69 | |||
| Refinement, 15–1.9 Å | ||||
| R factor, %‖ | 21.7 | |||
| Rfree, %‖ | 27.1 | |||
| rmsd bond lengths, Å | 0.009 | |||
| rmsd bond angles, ° | 1.363 | |||
| rmsd B factors, Å2 | 1.901 |
Values in parentheses refer to the highest resolution bin.
Rmerge = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where Ii(h) is the ith measurement of reflection h and 〈I(h)〉 is the weighted mean of all measurements of I(h).
RCullis is defined as the isomorphous lack-of-closure over the isomorphous difference.
RCullis (anomalous) is calculated as the anomalous lack-of-closure over the anomalous difference.
The phasing power is defined as (rms Fh/rms lack-of-closure) summed over all reflections used in the heavy atom refinement.
R factor = Σh|Fobs − Fcalc|/ΣhFobs, where Fobs and Fcalc are the observed and calculated structure factors.
Structure Determination and Refinement.
The structure was solved by using the multiple anomalous dispersion method with a MIR-like approach using the data set collected at the inflection point wavelength (λ2) as pseudonative. Five of the nine selenium atoms were initially located by direct methods using shelx (25) and were refined with the program mlphare (26). An additional selenium site was located by using difference Fourier techniques. The calculated phases at 2.5 Å had a mean figure of merit of 0.69. The initial density map, further improved by density modification, was of excellent quality and allowed the trace of an almost complete initial model with the interactive graphics program o (27). The model was subsequently refined against the high resolution native data set (between 15 and 1.9 Å) by using the program xplor (28). Low resolution data to 15 Å were included, and a bulk solvent correction was applied throughout the refinement procedure. A random sample containing 5% of the data was excluded from the refinement, and the agreement between calculated and observed structure factors for those reflections (Rfree) was used to monitor the course of the refinement (29). The final model, comprising residues 5–215 (1,728 protein atoms) and 169 water molecules, has a crystallographic R factor of 21.7% (Rfree = 27.1%). The loop between strands C3 and C4 in the C-terminal domain (residues 203–208) is not well ordered. Analysis of the model with procheck (30) shows good geometry, with 92.6% of the residues in the most favored region of the Ramachandran plot and 7.4% in the allowed region. Phasing and refinement statistics are shown in Table 1.
Results and Discussion
Overall Structure.
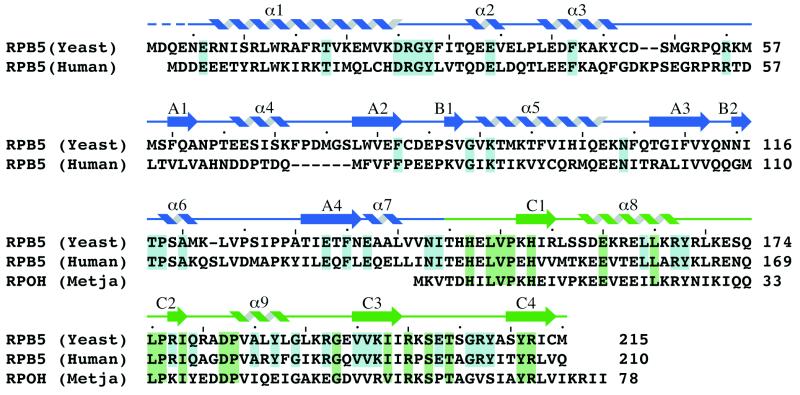
Inspection of the available primary sequences of RPB5 homologues from archaeal and eukaryotic organisms suggests that the origins of RPB5, similar to those of many other RNAP subunits, can be traced back to a time preceding the divergence of the archaeal and eukaryotic evolutionary domains. All archaeal RPB5 homologues are compact proteins with a size of approximately 7 kDa. Sequences highly similar to the archaeal RPB5 subunits are also present in all eukaryotic RPB5 subunits identified so far, but all eukaryotic RPB5 proteins feature an additional large N-terminal domain (Fig. 1). The aligned primary sequence data supports the idea of a bipartite organization of all members of the eukaryotic family of RPB5 proteins, containing both an evolutionary ancient C-terminal domain and a more recently evolved N-terminal domain displaying a lower degree of primary sequence conservation.
Figure 1.
Amino acid sequence alignment of the eukaryotic S. cerevisiae and human RPB5 subunits with the archaeal M. jannaschii RpoH subunit. Highlighted in blue are the amino acid residues that are strictly conserved in the six available eukaryotic sequences whereas the residues that are conserved in 15 of 17 homologues of the C-terminal domain, including both eukaryotic and archaeal sequences, are shown in green. The position of secondary structure elements determined by using the algorithm of Kabsch and Sander (40) is indicated above the sequence. The secondary structure elements are colored in blue when belonging to the N-terminal domain and in green when belonging to the C-terminal domain. A dashed line indicates residues omitted from the final model.
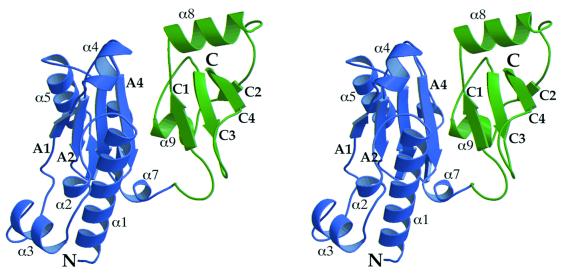
The 1.9 Å resolution structure of yRPB5 confirms this interpretation by revealing two structurally distinct domains (Fig. 2). The larger N-terminal domain includes residues 1–144 and folds into a four-stranded mixed β-sheet (A1–A4), surrounded by a number of helices (α1–α7). Two short β strands (B1 and B2) form an additional small parallel β-sheet that lies perpendicular to the central strands of the A sheet. The C-terminal domain (residues 145–215) is formed by a twisted four-stranded mixed β-sheet (C1–C4) flanked by two helices (α8 and α9).
Figure 2.
Stereo diagram showing a ribbon representation of the overall structure of RPB5. The eukaryote-specific N-terminal domain is shown in blue whereas the C-terminal domain, also present in archaea, is shown in green. Selected secondary structure elements are labeled as in Fig. 1.
An automated search of the structures deposited in the Protein Data Bank was performed for the two domains separately by using the program dali (31). The closest match to the yRPB5 N-terminal domain is the catalytic domain of the restriction endonuclease FokI (32), with 99 residues of 137 giving a rms separation of 4 Å. The common fold includes β-sheet A and the surrounding helices but lacks the three initial helices present in yRPB5. The C-terminal domain of yRPB5 lacks any significant structural similarity with any other protein in the database.
The Conserved C-Terminal Domain.
The previously determined solution structure of RNAP subunit H from Methanococcus jannaschii showed similar secondary structure elements to the C-terminal domain of yRPB5, as expected from the high degree of sequence conservation (47% identity), but suggested that both α-helices were positioned on the same side of the β-sheet (14). It is very unlikely that the archaeal subunit has a different arrangement of secondary structure elements to that seen in yRPB5. The discrepancy is probably attributable to the shortage of long-range nuclear Overhauser distance measurements in the NMR data. The placement of helix α8 on the opposite side of the β-sheet is compatible with an alternative interpretation of the NMR distance data (S. Matthews, personal communication).
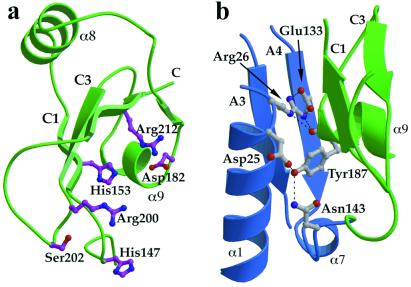
The majority of the polar amino acid residues that are invariant in the archaeal H subunits and the C-terminal domain of eukaryotic RPB5 polypeptides (Fig. 1) are clustered on one side of the domain (Fig. 3a). The cluster consists of hydrophilic and mostly positively charged amino acid residues (His 147, His 153, Asp 182, Arg 200, Ser 202, and Arg 212). This invariant surface is likely to be involved in interactions with other RNAP subunits and/or nucleic acid substrates. A number of experiments, including Far-Western blot analysis of Schizosaccharomyces pombe RNAPII (33) and two-hybrid studies in S. cerevisiae systems (34), indicate that RPB5 interacts with the conserved region H of RPB1. Moreover, protein-DNA photo-crosslinking experiments have identified RPB5 as the only small subunit to be located near the template DNA in the RNAPII preinitiation complex, making contact between position +5 and +15 on a single face of the DNA helix (18). This is the same side to which RPB1 crosslinks.
Figure 3.
(a) The C-terminal domain contains a number of residues that are highly conserved from archaea to humans. A cluster of invariant polar amino acids (mostly positively charged) are located on one surface of the domain. (b) The subunit interface contains few hydrophobic residues and is instead stabilized by polar interactions between a number of invariant residues in the N-terminal domain and Tyr 187 in the C-terminal domain. The figure shows the conserved residues at the interface and the hydrogen-bonding interactions. The secondary structure elements are colored as in Fig. 2.
The Domain Interface.
There are relatively few direct contacts between residues located in the two domains, and many of the interactions are mediated by water molecules. The interface region close to the connecting peptide is stabilized by hydrogen bonds between residues that are invariant in the six available eukaryotic sequences (Figs. 1 and 3b). Tyr 187 on helix α9 in the C-terminal domain plays a key role in these polar interactions. The side-chain hydroxyl group is hydrogen-bonded to both the carboxylate of Asp 25 and the side-chain amide of Asn 143 on the connecting peptide whereas the main-chain carbonyl oxygen forms a hydrogen bond with the side chain of Arg 26. Asp 25 and Arg 26 form part of the conserved DRGY motif and lie on a loop at the end of helix α1. Although Tyr 187 is invariant in eukaryotes, it is not conserved in archaea (Fig. 1) in which the N-terminal domain is absent. The fact that the interface interactions are mostly polar and often mediated by water molecules suggests that some interdomain movements may occur. In fact, optimal fitting of our model to a 3.2 Å resolution map of yeast RNAPII requires a small relative movement of the two domains to match the conformation of the yRPB5 subunit present in the endogenous enzyme (R. Kornberg, personal communication).
The Eukaryote-Specific N-Terminal Domain.
The N-terminal domain in eukaryotes may have specifically evolved to adopt novel and additional functions. Intriguingly, evidence for such functions has been found during the last few years in several independent studies.
A mutant of yRPB5 has been described (19) in which the substitution of Val 111 with a glycine led to defects in transcriptional activation with no changes in the level of basal transcription, similarly to what has been seen in mutants of the C-terminal domain of RPB1. Our structure reveals that this residue is not solvent exposed, being located on the face of strand A3 that packs against helix α1 (Fig. 2), and therefore is unlikely to be directly involved in any specific interaction with activators. The observed effect on transcriptional activation is likely to be caused by destabilization of the hydrophobic core of this region.
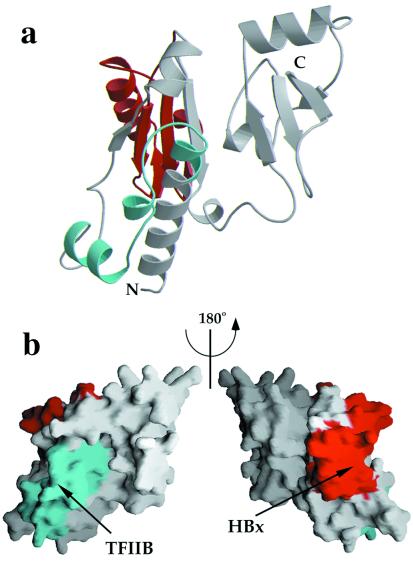
More direct evidence for the role of RPB5 in activated transcription has been presented in hepatitis B virus infected cells. This virus transactivates a variety of promoters through the action of the virally encoded transcriptional activator protein X (HBx). The human homologue of RPB5 (hRPB5) specifically associates in vitro and in vivo with HBx, and this interaction appears to be stabilized through additional contacts with the basal RNAPII-specific transcription factor TFIIB (20, 21). Deletion mutant studies have narrowed down the regions of RPB5 interaction with both HBx and TFIIB to specific fragments of the N-terminal domain. The high sequence homology between the human and yeast RPB5 (41% identity) suggests that the structure of the two proteins is directly comparable, and thus the yRPB5 structure can be used as a framework to interpret the available biochemical data. The HBx-binding site maps to the region including amino acid residues 73–120, which folds into β-strands A2 and A3 and helix α5. The TFIIB binding site has been located in the region between residues 21 and 47, which includes two solvent-exposed helices (α2 and α3) in addition to the strictly conserved loop DRGY (Fig. 4). Both regions remain solvent-exposed when RPB5 is placed in the RNAP complex (R. Kornberg, personal communication).
Figure 4.
Location of regions that have been proposed to interact with TFIIB and hepatitis B virus protein X (HBx). (a) Ribbon diagram representation of yRPB5 (in the same orientation as in Fig. 2). The fragment corresponding to the homologous sequence in hRPB5 that has been shown to interact with hTFIIB is shown in cyan whereas the region that is responsible for HBx interaction is shown in red. (b) Representation of the molecular surface of yRPB5 in the same orientation as Fig. 2 (Left) and rotated by 180° around the vertical axis (Right), showing the exposed surfaces of the above regions.
An additional transcriptional modulator facilitating the recruitment of RNAPs to the preinitiation complex, TIP120, has been shown to specifically interact with RPB5 and the TATA-box binding protein TPB (35). Taken together, all of these results emphasize the role of RPB5 in facilitating communication between the RNAP core and a variety of basal and gene-specific transcription factors.
No Evidence for RPB5 Homodimerization.
In vivo labeling studies of endogenous yeast RNAPII with [35S]methionine suggested the presence of two copies of yRPB5 in each RNAPII molecule (22). Subsequent far Western blotting studies identified a putative “homodimerization” domain within the N-terminal part of hRPB5 (21), leading to the overall conclusion that hRPB5 could be present as a homodimer within eukaryotic RNAPs. Archaeal RPB5 homologues, on the other hand, lack this particular domain, and no evidence for homodimerization of subunit H from M. jannaschii has been found (14). Size exclusion column chromatography of the recombinant yRPB5 used for our crystallization trials similarly failed to provide support for any detectable tendency for yRPB5 to homodimerize under a number of different experimental conditions (R.O.J.W., unpublished observations). The intrinsically monomeric nature of yRPB5 is clearly confirmed by the arrangement of the molecules in the crystal lattice. Within the orthorhombic lattice, a crystallographic dyad axis relates two RPB5 molecules, but the only dimer interface, involving helix α3 and strand A1, is rather small (600 Å2) and does not include highly conserved residues. It is therefore likely that the evidence for RPB5 homodimerization is artefactual, and we favor a model in which RPB5 occurs as a monomer in both archaeal and eukaryotic RNAPs.
Acknowledgments
This work was supported by a Wellcome Trust Grant to S.O., P.B. and R.O.J.W. Figs. 2–4 were prepared by using raster3d (36), molscript (37, 38), and grasp (39).
Abbreviations
- RNAP
RNA polymerase
- TFIIB
transcription factor IIB
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1dzf).
References
- 1.Thuriaux P, Sentenac A. The Molecular Biology of the Yeast Saccharomyces: Gene Expression. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 1–48. [Google Scholar]
- 2.Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Goldfarb A, Mustaev A. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 3.Young R A. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 4.McKune K, Moore P A, Hull M W, Woychik N A. Mol Cell Biol. 1995;15:6895–6900. doi: 10.1128/mcb.15.12.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shpakovski G V, Acker J, Wintzerith M, Lacroix J F, Thuriaux P, Vigneron M. Mol Cell Biol. 1995;15:4702–4710. doi: 10.1128/mcb.15.9.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woese C R, Kandler O, Wheelis M L. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zillig W, Palm P, Klenk H-P, Langer D, Hudepohl U, Hain J, Lanzendorfer M, Holz I. In: The Biochemistry of Archaea (Archaebacteria) Kates M, Kushner D J, Matheson A T, editors. Amsterdam: Elsevier; 1993. pp. 367–391. [Google Scholar]
- 8.Darcy T J, Hausner W, Awery D E, Edwards A M, Thomm M, Reeve J N. J Bacteriol. 1999;181:4424–4429. doi: 10.1128/jb.181.14.4424-4429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eloranta J J, Kato A, Teng M S, Weinzierl R O J. Nucleic Acids Res. 1998;26:5562–5567. doi: 10.1093/nar/26.24.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doolittle R F, Feng D F, Tsang S, Cho G, Little E. Science. 1996;271:470–477. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- 11.Woychik N A, Liao S M, Kolodziej P A, Young R A. Genes Dev. 1990;4:313–323. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]
- 12.del Rio-Portilla F, Gaskell A, Gilbert D, Ladias J A A, Wagner G. Nat Struct Biol. 1999;6:1039–1042. doi: 10.1038/14923. [DOI] [PubMed] [Google Scholar]
- 13.Krapp S, Kelly G, Reischl J, Weinzierl R O, Matthews S. Nat Struct Biol. 1998;5:110–114. doi: 10.1038/nsb0298-110. [DOI] [PubMed] [Google Scholar]
- 14.Thiru A, Hodach M, Eloranta J J, Kostourou V, Weinzierl R O, Matthews S. J Mol Biol. 1999;287:753–760. doi: 10.1006/jmbi.1999.2638. [DOI] [PubMed] [Google Scholar]
- 15.Wang B, Jones D N M, Kaine B P, Weiss M A. Structure (London) 1998;6:555–569. doi: 10.1016/s0969-2126(98)00058-6. [DOI] [PubMed] [Google Scholar]
- 16.Zhang G, Campbell E A, Minakhin L, Richter C, Severinov K, Darst S A. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 17.Fu J, Gnatt A L, Bushnell D A, Jensen G J, Thompson N E, Burgess R R, David P R, Kornberg R D. Cell. 1999;98:799–810. doi: 10.1016/s0092-8674(00)81514-7. [DOI] [PubMed] [Google Scholar]
- 18.Kim T K, Lagrange T, Wang Y H, Griffith J D, Reinberg D, Erbright R H. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyao T, Woychik N A. Proc Natl Acad Sci USA. 1998;95:15281–15286. doi: 10.1073/pnas.95.26.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheong J H, Yi M, Lin Y, Murakami S. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y, Nomura T, Cheong J, Dorjsuren D, Iida K, Murakami S. J Biol Chem. 1997;272:7132–7139. doi: 10.1074/jbc.272.11.7132. [DOI] [PubMed] [Google Scholar]
- 22.Kolodziej P A, Woychik N, Liao S M, Young R A. Mol Cell Biol. 1990;10:1915–1920. doi: 10.1128/mcb.10.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodach M, Todone F, Eloranta J J, Onesti S, Weinzierl R O J. Acta Crystallogr D. 1999;55:1373–1374. doi: 10.1107/s0907444999007271. [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 25.Sheldrick G M. Acta Crystallogr A. 1990;46:467–473. [Google Scholar]
- 26.Otwinowski Z. Isomorphous Replacement and Anomalous Scattering: Proceedings of the CCP4 Study Weekend. Daresbury, U.K.: SERC Daresbury Laboratory; 1991. pp. 80–86. [Google Scholar]
- 27.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Brünger A T, Kuriyan J, Karplus M. Science. 1987;235:458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 29.Brünger A T. Nature (London) 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski R A, MacArthur M V, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 31.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 32.Wah D A, Hirsch J A, Dorner L F, Schildkraut I, Aggarwal A K. Nature (London) 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 33.Miyao T, Yasui K, Sakurai H, Yamagishi M, Ishihama A. Genes Cells. 1996;1:843–854. doi: 10.1046/j.1365-2443.1996.730274.x. [DOI] [PubMed] [Google Scholar]
- 34.Miyao T, Honda A, Qu Z, Ishihama A. Mol Gen Genet. 1998;259:123–129. doi: 10.1007/s004380050796. [DOI] [PubMed] [Google Scholar]
- 35.Makino Y, Yogosawa S, Kayukawa K, Coin F, Egly J M, Wang Z, Roeder R G, Yamamoto K, Muramatsu M, Tamura T. Mol Cell Biol. 1999;19:7951–7960. doi: 10.1128/mcb.19.12.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merritt E A, Bacon D J. Methods Enzymol. 1997;227:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 37.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 38.Esnouf R M. J Mol Graphics. 1997;15:133–138. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 39.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 40.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]