Abstract
Memory B cells and the antibodies they encode are important for protective immunity against infectious pathogens. Characterization of naïve and memory B cell antibody repertoires will elucidate the molecular basis for the generation of antibody diversity in human B cells and the optimization of antibody structures that bind microbial antigens. In this study we aimed to investigate the influence of antigenic selection on the antibody genes of the two CD27+ memory B cell subsets, comparing them with the naïve repertoire in CD27− cells. We analyzed and compared the Ig heavy chain gene transcripts in three recently defined circulating naïve and memory B cell subsets (CD19+IgD+CD27− [naive], CD19+IgD+CD27+ [un-class-switched memory] or CD19+IgD− CD27+ [class-switched memory]) at the single cell level. We found similar biased patterns of variable, diversity and joining heavy chain gene usages in all three groups of cells. CD19+IgD+CD27+ memory B cells harbored as diverse an antibody gene repertoire as CD19+IgD−CD27+ memory B cells. Interestingly, CD19+IgD+CD27+ memory B cells possessed a lower frequency of somatic mutations, a higher incidence of exonuclease activity at the 3′ end of D regions, and a lower frequency of N and P nucleotide additions at both VH-D and D-JH junctions of CDR3 regions compared to CD19+IgD−CD27+ memory B cells. These data suggest distinct functional mechanisms underlying selection of this unique subset of un-class switched memory B cells.
Keywords: Human, B cells, Antibodies, Memory, Repertoire development
Introduction
The generation of highly diverse antibodies with high affinity is a key element of acquired immunity (Bernasconi et al., 2002; Crotty and Ahmed, 2004). However, the molecular and cellular basis for development of diverse and effective antibody repertoires remains only partially elucidated. Classically, memory B cells were considered B cells that had class-switched from the initial expression of surface IgD and IgM to that of other Ig classes, resulting in surface expression of IgG, IgA, or IgE, with a lack of IgD. Recent progress in B cell phenotyping has clarified that an isotype-switched phenotype identifies only a subset population of memory B cells. On the basis of recent research, adult circulating B cells can be separated on the basis of memory markers and isotypes into at least three subpopulations: 1) IgD+CD27− naïve B cells, 2) IgD+CD27+ un-class-switched memory B cells, and 3) IgD−CD27+ class-switched memory B cells (Agematsu, 2000; Agematsu et al., 2000; Klein et al., 1998; Shi et al., 2003).
The molecule CD27, a type I glycoprotein expressed on memory B cells and the majority of T cells, is a member of the tumor necrosis factor receptor family (Camerini et al., 1991; Prasad et al., 1997). Accumulating data about CD27 indicate that this molecule is present on the surface of memory B cells but not naïve B cells, and CD27 signaling promotes the differentiation of memory B cells into plasma cells (Agematsu, 2000; Agematsu et al., 2000; Klein et al., 1998; Nagumo et al., 1998; Raman et al., 2003; Shi et al., 2003). IgD+CD27+ un-class-switched memory B cells in peripheral blood have been shown to be a distinct subpopulation of memory B cells that may play a crucial role in secondary immune responses because of their prompt synthesis of high-affinity IgM molecules (Shi et al., 2003). In vivo, the origin and function of IgD+CD27+ B cells are still uncertain. It has been demonstrated that IgD+ B cells residing in human tonsils can acquire somatic mutations in variable genes without switching isotypes (Liu et al., 1996). Hyper-IgM (HIGM) patients who carry an invalidating mutation of the CD40L gene do not possess normally developed germinal centers or switched memory B cells but still have a subpopulation of circulating IgD+CD27+ B cells, suggesting that the IgD+CD27+ B cells might form a B cell subset distinct from classical germinal center-derived memory B cells (Weller et al., 2001). A recent study suggested that IgD+CD27+ B cells correspond to circulating splenic marginal zone B cells, based on phenotypic analysis, complementarity determining region 3 (CDR3) spectra-typing and gene-expression profiling of blood and splenic B cell subsets (Weller et al., 2004). Analysis of this peripheral subset of B cells in healthy children younger than 2 years further indicated that these B cells could develop and mutate their Ig receptor during ontogeny even before a functional splenic marginal zone matures.
To date, detailed molecular characterization of antibody genes expressed in these naïve and memory B cell subsets is limited. Klein et al. reported mutational analysis of 67 rearranged VH genes isolated from IgD+CD27+ memory B cells and 32 rearranged VH genes from IgD+CD27− naïve B cells by using genomic PCR specific for only 3 of the 7 VH gene families, VH 1, 3, and 4 (Klein et al., 1998). In that study, the mutation frequency of IgD+CD27+ memory B cells was 3.7%, 5.0%, and 5.9% respectively for 3 healthy adult donors. In a more recent report, Weller et al. studied the mutation frequency of one VH gene, VH3-23, in the two memory B cell groups and showed a lower mutation frequency of B cells (3.8% versus 5.7% in IgD−CD27+ memory the VH3-23 gene in IgD+CD27+ memory B cells) (Weller et al., 2004). Detailed characterization of naïve and memory B cell antibody gene repertoires will facilitate better understanding of molecular mechanisms underlying the regulation of memory B cells responses, and the generation and expansion of antibody diversity.
In this study, we simultaneously isolated single cells from the three subsets of human circulating naïve and memory B cells from healthy adult volunteers based on the surface expression of IgD and CD27. Using single B cell culture and multiplex reverse transcription PCR designed to amplify all Ig variable region gene segments in the VBASE complete database of genomic variable gene sequences, individual Ig variable region genes of Ig heavy chains from single cells were cloned and analyzed. We found a similar pattern of biased Ig gene usages among the three subsets of naïve and memory B cells, suggesting a highly conserved biased antibody repertoire in human memory B cells despite previous antigen exposure. Our data confirmed the differences in mutation frequencies in the two memory B cell groups. Furthermore, characteristics in the CDR3 regions observed in the IgD+CD27+ memory B cells compared with IgD−CD27+ memory B cells suggested differential levels of terminal deoxynucleotidyl transferase (TdT) and exonuclease activities during the generation of the two subsets of memory B cells.
Materials and Methods
Subjects
Peripheral blood samples (n=10) from healthy adult volunteers, aged 20 – 40 years, were used for study. All samples were obtained following informed consent under approval from the Vanderbilt University Medical Center Institutional Review Board.
Isolation of naïve and memory B cells from blood
Peripheral blood mononuclear cells (PBMCs) were isolated from blood samples by Ficoll-Hypaque density gradient centrifugation, then stained for 30 minutes at 4 °C in the dark using fluorescent conjugated mouse anti-human antibodies, including anti-CD19-PE-Cy7, anti-IgD-PE, anti-CD27-APC, anti-CD3/CD14-APC-Cy7 (Beckton Dickinson, San Jose, CA). Cells were processed immediately for flow cytometric analysis and cell sorting using a FACSAria cytometer (Beckton Dickinson). Cells expressing CD3 or CD14 (T cell or monocyte markers) were excluded from sorting. After each experiment, a portion of the sorted sample was analyzed to determine the post-sort purity. All sorted naïve (CD19+IgD+CD27−) and memory (CD19+IgD+CD27+ or CD19+IgD−CD27+) B cell samples exhibited a >95% purity. Data analysis was performed using FlowJo software, version 6.1 or above (Tree Star, Inc., Ashland, OR). Representative sorting data are shown in Figure 1A.
FIGURE 1. A. Representative data from flow cytometric analyses demonstrating human naïve and memory B cells were isolated from adult blood samples with high purity.
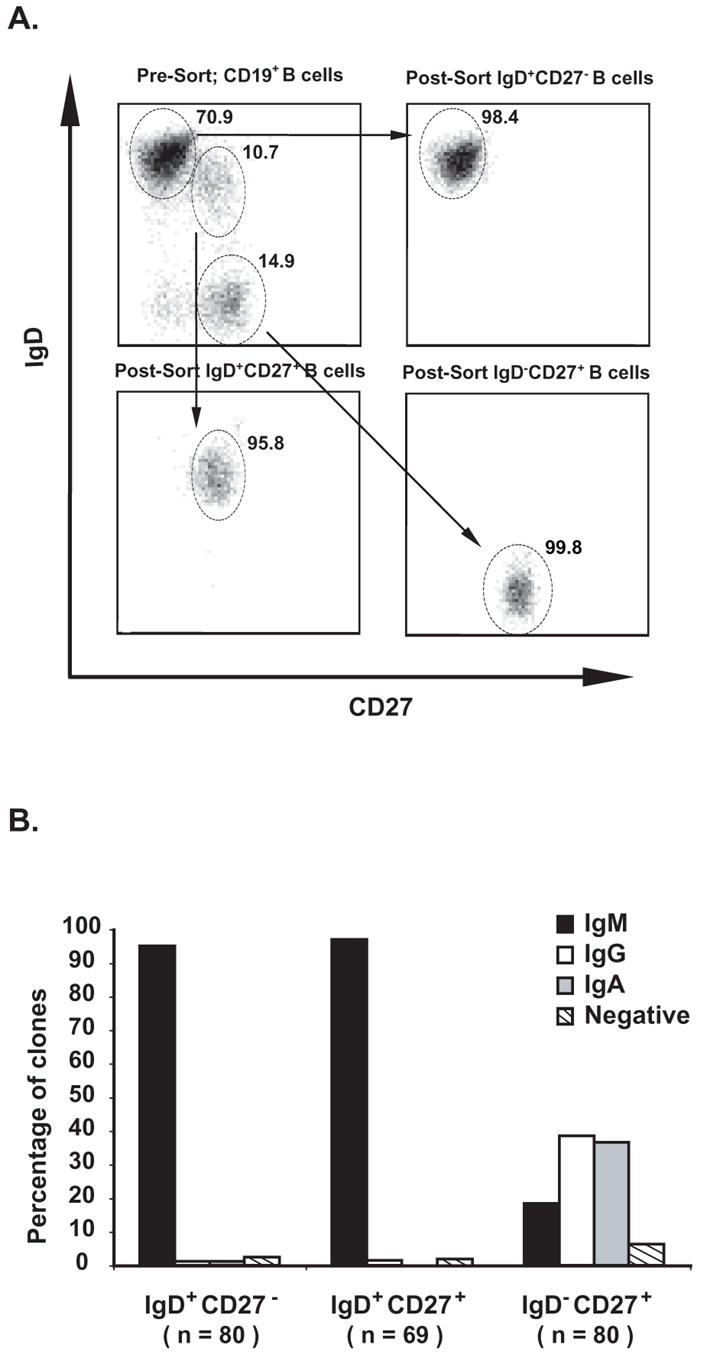
The numbers indicate the percentage of cells that were of naïve or memory phenotypes (IgD+CD27−, IgD+CD27+ or IgD−CD27+) as indicated by the oval gates. All cells plotted were gated to be CD19+. All post-sort samples contained >99% CD19+ B cells. B. Isotypes of antibody produced by single B cell clones were determined by Ig capture ELISA. Culture supernatants from 80 IgD+CD27− naïve B cells, 69 IgD+CD27+ memory B cells and 80 IgD−CD27+ memory B cell clones were used for determining isotypes of antibody produced at 21 days post culture. Approximately 95% of IgD+CD27− naïve B cells and 97% of IgD+CD27+ memory B cell clones produced IgM. The IgD−CD27+ memory B cell clones produced IgM, IgG or IgA (19%, 39%, or 36%, respectively).
Expansion of single B cells into clones
We used a culture system as previously described for the expansion of single B cells into clones (Weitkamp et al., 2003b). Briefly, 50,000 irradiated (50 Gy) EL-4-B5 mouse thymoma cells (kindly provided by Dr. R.H. Zubler) per well of 96-well culture plates were used as feeder cells immediately following single-cell isolation of B cells. A combination of 100 U recombinant human IL-2, 5 ng/ml PMA, and 5% (v/v) of supernatant from pokeweed mitogen-activated human T cells (T cell replacing factor) was added. The culture plates were incubated for 7 days at 37 °C in an atmosphere of 8% CO2. After 7 days, we removed 100 μl of supernatant and added 10,000 irradiated (50 Gy) fibroblastic L cells stably transfected with human CD154 (CD40L) to each well. This cell line was kindly provided by DNAX via the ATCC (CRL 12095). We also added 5 ng/ml recombinant human IL-4 in addition to the B cell culture media described above. The cultures were kept for another 2 weeks with a second addition of CD154 fibroblasts, cytokines, PMA and T cell replacing factor on day 14. Prior to RNA isolation on day 21, human Ig capture ELISA was performed to identify the B cell clones that were secreting antibodies on day 14.
Determination of isotypes of Ab produced by single B cell-derived clones
We used a human Ig capture ELISA for determining the isotype of the Ab produced by single B cell-derived clones. Un-conjugated goat anti-human Ig (H&L) antibody (Southern Biotechnology, Birmingham, AL), diluted 1:1000 in sodium carbonate buffer was used as capture antibody. Alkaline phophatase-labeled goat anti-human IgM, IgG or IgA antibody (Southern Biotechnology) diluted 1:1,000 in sodium carbonate buffer was used separately as secondary Ab in the replicate assays. Purified human IgM, IgG, or IgA (Biodesign International, Saco, Maine) was used as positive control, and culture supernatant from wells lacking B cells but containing all other medium additives was used as negative control.
RNA extraction and RT-PCR amplification
We used an oligo-dT based mRNA capture kit to isolate mRNA from the suspension generated by lysis of cells in the single B-cell-derived clones secreting human Ig. We used a single-tube strategy for reverse transcription (RT) PCR amplification of the VH region (Titan One Tube RT-PCR System, Roche Diagnostics, Mannheim, Germany). The pooled PCR primer mixture that was designed to amplify antibody genes from all antibody gene families was described previously (Weitkamp et al., 2003b).
DNA sequence analysis
We ligated gel-extracted PCR products into a TA cloning vector (Promega, Madison, WI), generated bacterial clones, and purified plasmid DNA from overnight bacterial cultures. Plasmid DNA was digested with restriction enzymes to identify clones with proper ligation. The nucleotide sequence of plasmid DNAs that contained a VH insert was determined using vector-specific primers and an automated DNA sequencer (Applied Biosystems, Foster City, CA).
Criteria for the analysis of antibody sequences
We analyzed VH region sequences by comparison with the international ImMunoGeneTics (IMGT) information system database (http://imgt.cines.fr:8104) (Ruiz et al., 2000). The characteristics of individual sequences for VH regions are shown in the online supplemental material (Supplemental Tables I–VI). All sequences were submitted to GenBank (225 sequences, accession numbers DQ535261 through DQ535485). We characterized somatic mutations as replacement (R) or silent (S) mutations by comparing variable region sequences with the corresponding IMGT germ-line sequences. The first 24 nucleotides of FR1 were encoded by PCR primers and therefore were not analyzed for mutations. Hotspot mutations were identified by comparing the nucleotide sequences surrounding mutations with known hotspot motifs (DGYW and its complement WRCH; or WA and its complement TW).
Statistical analysis
Chi-square tests, with p-values computed using Monte Carlo simulation with 500,000 replications, were used to compare proportions. Kruskal-Wallis tests were used to compare continuous variables (e.g., frequency of mutations, CDR3 lengths, numbers of insertions). The unique donor exclusion method was used for examining the influence of inter-individual heterogeneity on antibody repertoire analysis of VH, D, and JH family and gene segments usage.
Results
Frequency of circulating naïve and memory B cells in healthy blood donors
We examined the frequency of circulating naïve and memory B cells in peripheral blood from 10 healthy blood donors. The mean frequency of B cells with naïve phenotype in these donors was 68.6% (range 27.4 –89.4%). A mean of 15.1% (range 3.68 –49.2%) of circulating B cells in our analysis were IgD+CD27+ memory B cells, and a mean of 13.9% (range 5.04 –21.5%) of circulating B cells were IgD−CD27+ memory B cells.
Isotypes of Ab produced by single B cell clones were uniformly IgM in the naïve and IgD+CD27+ memory B cells, and were IgM, IgG or IgA in the IgD−CD27+ memory B cells
Isotypes of Ab produced by single B cell clones were determined for 80 IgD+CD27− naïve B cells, 69 IgD+CD27+ memory B cells and 80 IgD−CD27+ memory B cells clones. As shown in Figure 1B, more than 95% of IgD+CD27− naïve B cells and IgD+CD27+ memory B cells clones produced IgM. In contrast, the IgD−CD27+ memory B cell clones produced IgM, IgG or IgA (19%, 39%, or 36%, respectively)(Figure 1B).
The VH antibody repertoires in circulating naïve and memory B cells were all VH3 dominant
All statistical analyses performed here assumed independence between cells (i.e., ignored any correlation that might exist between measurements taken on different cells from the same individual). It was reasonable to assume the independence of individual cells, as analysis of the V-D-J combinations revealed that none of the recombined variable regions were clonally related. We determined the nucleotide sequences of VH gene segments from 77 naïve B cells, 63 IgD+CD27+ memory B cells and 85 IgD−CD27+ memory B cell clones isolated from seven individuals (Figure 2A) (See supplemental tables for number of Ig heavy chain sequences and corresponding gene segments analyzed from each donor). 51% of VH gene segments in IgD+CD27− naïve B cells, 54% of VH gene segments in IgD+CD27+ memory B cells and 45% of VH gene segments in IgD−CD27+ memory B cells clones belonged to the VH3 family. The second and third most common VH families in all B subsets cells were VH4 and VH1. VH4 segments were found in 22%, 18% and 17% of IgD+CD27− naïve B cells, IgD+CD27+ memory B cells and IgD−CD27− memory B cells respectively. VH1 segments were found in 21% of IgD+CD27− naïve B cells, 18% of IgD+CD27+ memory B cells and 16% of IgD−CD27+ memory B cells. A minor trend of increased VH5 family usage in the IgD+CD27+ and IgD−CD27+ memory B cells was noted, with approximately 3% of IgD+CD27− naïve B cells, 8% of IgD+CD27+ memory B cells and 10% of IgD−CD27− memory B cell clones harboring VH5 family genes. However, we found no statistically significant difference in the distribution of VH families among the three types of naïve and memory B cells. A unique donor exclusion method was used to evaluate the influences of inter-individual heterogeneity in VH family gene usage analysis. Similar results were obtained in all unique donor exclusion analyses compared with the analysis result using the whole data set. Therefore, there was no apparent biased VH family gene usage in any particular donor that influenced the overall data analysis.
FIGURE 2. Similar VH, D and JH gene family utilization in circulating human naïve or memory B cells.

Single circulating naïve or memory B cells from healthy blood donors were sorted, amplified in culture, and tested for antibody production by ELISA followed by sequence analysis of the expressed VH gene segment. Frequencies of VH, D and JH gene family utilization in all three circulating naïve and memory B cells are presented in 2A, 2B and 2C, respectively. No difference was found in the distribution of VH, D or JH gene family utilization between the three types of B cells (p>0.5 for all comparisons, Chi-square tests).
The D and JH antibody repertoires in circulating naïve and memory B cells were all D-3 and JH-4-dominant
D and JH family gene usages also were analyzed in the three groups of naïve and memory B cells. As shown in Figure 2B and 2C, striking similarities were found in the distributions of D and JH family genes. D3 was identified as the most common D family in all three groups, and was found in 31% of IgD+CD27− naïve B cells, 32% of IgD+CD27+ memory B cells and 34% of IgD−CD27− memory B cell clones respectively. JH4 was found to be the most commonly used JH family in all groups. 42% of IgD+CD27− naïve B cells, 52% of IgD+CD27+ memory B cells and 58% of IgD−CD27− memory B cell clones used JH4 family genes. A minor trend of increase of D6 usage was noted for IgD+CD27+ memory B cells. In addition, increased frequency of non-assignable D and JH genes were detected in the memory B cell groups, reflecting greater frequency of somatic mutations in these cells. The unique donor exclusion method also was used to evaluate the influences of inter-individual heterogeneity in D and JH family gene usage analysis. There was no apparent biased D or JH family gene usage in any particular donor that influenced the overall data analysis.
Broad and similar usage of VH, D and JH family gene segments in circulating naive and memory B cells
As shown in Figure 3, broad usages of VH, D and JH gene segments were noted in all three naïve and memory B cell groups. Of the 39 to 46 known functional VH genes (depending on the polymorphisms present in humans), 15 different VH3 family gene segments, 5 different VH4 family gene segments, 5 different VH1 family gene segments, and 1 of each the VH5, VH6, VH2, and VH7 family gene segments were identified within 77 IgD+CD27− naïve B cell clones. Within 63 IgD+CD27+ memory B cell clones, 12 different VH3 family gene segments, 5 different VH4 family gene segments, 5 different VH1 family gene segments, and 1 of each of the VH5, and VH2 family gene segments were identified. Within 85 IgD−CD27+ memory B cell clones, 16 different VH3 family gene segments, 5 different VH4 family gene segments, 7 different VH1 family gene segments, 2 VH5 family gene segments, and 1 of each of the VH6, VH2, and VH7 family gene segments were identified. Of 23 known functional D genes, 20, 18 and 21 were identified in IgD+CD27−naïve B cells, IgD+CD27+ memory B cells and IgD−CD27− memory B cells respectively. All 6 known functional JH genes were found in all groups of naïve or memory B cells. VH, D and JH gene segments identified in naïve or memory B cell clones were selected from across the entire gene locus and appeared to distribute in a similar and biased fashion, i.e., some gene segments were overrepresented, while others were not used at all (Figure 3A, 3B and 3C). Numbers of VH, D and JH gene segments identified in all three naïve and memory B cell clones were similar (Figure 3D, 3E and 3F). Statistically significant differences were not found in the distributions of VH, D or JH family gene segments among the three types of B cells. The donor exclusion method also was used to evaluate the influences of inter-individual heterogeneity in the VH, D or JH family gene segments usage analysis. There was no apparent biased VH, D or JH family gene segments usage in any particular donor that influenced the overall data analysis.
FIGURE 3. Similar VH, D and JH gene segment utilization in circulating human naïve or memory B cells.

Frequencies of VH, D and JH gene segments utilized in three circulating naïve and memory B cells are shown in 3A, 3B and 3C. No significant difference between cell types was found for any of the VH, D or JH gene segment utilizations (p>0.2). Numbers of VH, D or JH gene segments identified (used at least once within a group of cells) in all three circulating naïve and memory B cells are presented in 3D, 3E and 3F.
CD19+IgD+CD27+ memory B cells harbor shorter CDR3s in the VH gene segments
Analysis of heavy chain CDR3 regions suggested shorter CDR3 lengths in IgD+CD27+ memory B cells. As shown in Figure 4A, the CDR3 lengths for IgD+CD27+ memory B cells were shorter than those of IgD+CD27− naïve B cells (p=0.022). The median CDR3 length for naïve B cells was 45 bp (range from 21 to 96 bp), whereas the median CDR3 length for IgD+CD27+ memory B cells was 42 bp (range from 21 to 78 bp), and the median CDR3 length for IgD−CD27+ memory B cells was 45 bp (range from 15 to 84 bp).
FIGURE 4. Shorter variable heavy chain CDR3 regions in IgD+CD27+ memory B cells.

A. The CDR3 lengths for three circulating naïve and memory B cells are shown with medians and 25th and 75th percentiles (interquartile range) indicated as bars. The CDR3 lengths for IgD+CD27+ memory B cells were shorter than those of IgD+CD27− naïve B cells, p=0.022. B. Total N and P nucleotides in both the VH-D and D-JH junctions of naïve and memory B cells are presented with medians and 25th and 75th percentiles indicated as bars. Total N and P nucleotides in IgD+CD27+ memory B cells were significantly less than those in IgD−CD27+ memory B cells, p = 0.0425.
Evidence for differing levels of TdT activities in the VH-D and D-JH junctions of naïve and memory B cells
We analyzed the VH-D and D-JH junctional sequences of naïve and memory B cells for evidence of the activity of insertion mechanisms. As indicated in Table I, the percentages of sequences that lacked N additions at either VH-D or D-JH junctions were lower in the IgD−CD27+ memory B cells (13.8% and 13.8%) than those in IgD+CD27− naïve B cells (24.3% and 21.4%) or those in IgD+CD27+ memory B cells (25.0% and 28.3%). Less N nucleotide additions also were noted in D-JH junctions for IgD+CD27+ memory B cells. Further analyses indicated that the shorter CDR3 lengths in IgD+CD27+ memory B cells were a consequence of reduced numbers of N and P nucleotide insertions in both the VH-D and D-JH junctions. As indicated in Figure 4B, the median number of total N and P nucleotides in both the VH-D and D-JH junctions for IgD+CD27+ memory B cells was 6.5 bp (range from 0 to 34 bp), whereas the median number of total N and P nucleotides in both the VH-D and D-JH junctions for IgD+CD27− naïve B cells was 8 bp (range from 0 to 36 bp), and the median of total N and P nucleotides in both the VH-D and D-JH junctions for IgD−CD27+ memory B cells was 10 bp (range from 0 to 34 bp). The total number of N and P nucleotides in IgD+CD27+ memory B cells was significantly less than that in IgD−CD27+ memory B cells (p = 0.04).
Table I.
N nucleotide additions in the VH-D or D-JH junctions of the heavy chain CDR3 sequences of naïve or memory B cells.
| Type of B cells | VH-D junctions lacking additions (% of sequences) | D-JH junctions lacking additions (% of sequences) | VH-D N-nuleotide additions; Median (IQR*) | D-JH N-nuleotide additions; Median (IQR) |
|---|---|---|---|---|
| IgD+CD27− naïve | 24.3 | 21.4 | 3.0 (1.0, 6.0) | 4.0 (1.0, 7.0) |
| IgD+CD27+ memory | 25.0 | 28.3 | 3.0 (0.7, 6.0) | 3.0 (0.0, 6.0) |
| IgD−CD27+ memory | 13.8 | 13.8 | 3.0 (1.0, 7.3) | 4.0 (1.0, 6.3) |
IQR=Interquartile range.
Caption: Lower percentages of sequences that lacked N additions at either VH-D or D-JH junctions were found in the IgD−CD27+ memory B cells than those in IgD+CD27− naïve B cells or those in IgD+CD27+ memory B cells. Less N nucleotide additions were noted in the D-JH junctions for IgD+CD27+ memory B cells.
Exonuclease activities in the VH-D and D-JH junctions of naïve and memory B cells
Examination of VH-D and D-JH junctions of the variable heavy chain genes revealed significant differences in the extent of exonuclease removal events between the VH, D and JH gene ends in B cell groups. As indicated in Table II, the number of exonuclease removals in the 3′ ends of the D region was greater in IgD+CD27+ memory B cells than in IgD+CD27− naive B cells (p< 0.05), while the number of exonuclease removals in the 3′ ends of the D region in IgD−CD27+ memory B cells was similar to the one in IgD+CD27− naïve B cells. In IgD+CD27+ memory B cells and IgD−CD27+ memory B cells, a similar increased level of exonuclease removals in VH region ends was found compared with that in IgD+CD27− naïve B cells (p< 0.05). In addition, exonuclease removals from VH region ends in all three types of B cells were lower than removals from D or JH region ends (p<0.0001).
Table II.
Nucleotide removals in the VH-D or D-JH junctions of the heavy chain CDR3 sequences of naïve or memory B cells.
| Type of B cells | VH deletion; Median (IQR*) | 5′ end D deletion; Median (IQR*) | D segment length; Median (IQR) | 3′ end D deletion; Median (IQR) | JH deletion; Median (IQR) |
|---|---|---|---|---|---|
| IgD+CD27− naïve | 0.0 (0,1.0)† | 3.5 (1.0, 7.0) | 16.0 (13.0, 20.0) | 3.0 (1.0, 6.0) | 4.0 (1.3, 6.0) |
| IgD+CD27+ memory | 1.0 (0,2.0)# † | 4.0 (1.8, 7.0) | 14.5 (11.0, 18.0) | 4.0 (2.0, 9.0)# | 4.0 (2.8, 7.0) |
| IgD−CD27+ memory | 1.0 (0,2.0)# † | 3.0 (1.0, 7.0) | 15.0 (13.0, 19.0) | 4.0 (1.0, 7.0) | 4.0 (1.0, 6.3) |
p<0.05 compared to naïve cells.
, p<0.0001 compared to removals from D or JH region ends.
IQR=Interquartile range.
Caption: The number of exonuclease removals in the 3′ ends of the D region was greater in IgD+CD27+ memory B cells. In IgD+CD27+ memory B cells and IgD−CD27+ memory B cells, a similar increased level of exonuclease removals in VH region ends was found. Exonuclease removals from VH region ends in all three types of B cells were lower than removals from D or JH region ends.
CD19+IgD+CD27+ memory B cells contained significantly fewer somatic mutations in VH genes than CD19+IgD−CD27+ memory B cells
Over 94% of VH genes isolated from IgD+CD27− naïve B cells were either unmutated (72.7%) or contained a single mutation (22%). In contrast, only 6% of VH genes isolated from IgD+CD27+ memory B cells and none of VH genes isolated from IgD−CD27+ memory B cells were un-mutated. Consistent with a previous report about the mutation frequency of cells using a single VH gene segment (VH3-23), our data confirmed that the un-class-switched IgD+CD27+ memory B cells possessed significantly fewer mutations in their Ig heavy chain variable regions than IgD−CD27+ memory B cells in all segments identified in the repertoire (p<0.0001). Further analysis demonstrated that mutations in both groups of memory B cells were distributed across the variable region of heavy chain genes with an increased frequency of replacement mutations, especially in the CDR1, CDR2 and FR3 regions. As expected, higher replacement/silent ratios (>3) were observed in the CDR regions of the memory B cell groups than in the FR regions (Figure 5B).
FIGURE 5. A. Lower frequency of somatic mutations in IgD+CD27+ memory B cells.
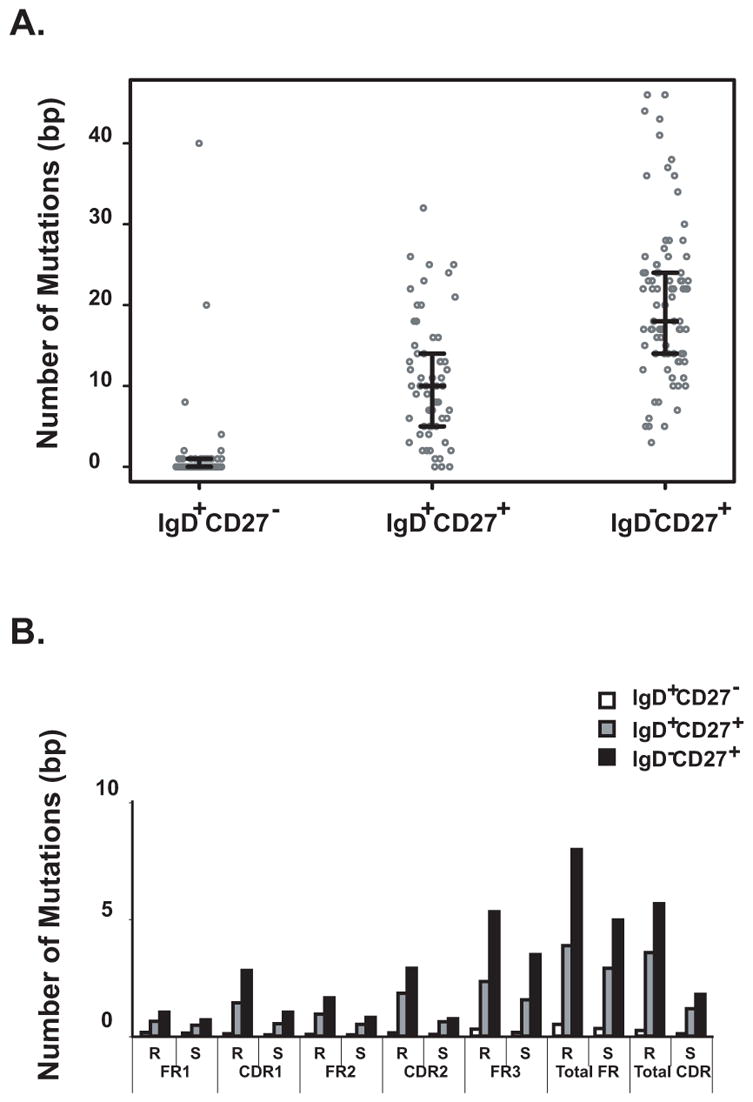
The number of mutations identified in variable heavy chain genes for circulating naïve or memory B cells is shown. Medians and 25th and 75th percentiles interquartile values are indicated. Naïve B cells possessed fewer mutations (median 0 bp) than did IgD+CD27+ (median 10 bp) or IgD−CD27+ memory B cells (median 18 bp) (p<0.0001). IgD+CD27+ memory B cells possessed fewer mutations than IgD−CD27+ memory B cells (p<0.0001). B. Distributions of total somatic mutations and replacement mutations were similar in IgD+CD27+ and IgD−CD27+ memory B cells. Mutations in both types of memory B cells were distributed across the variable region of heavy chain genes with an increased frequency of replacement mutations (R), especially in the CDR1, CDR2 and FR3 regions. Higher replacement mutation (R) /silent mutation (S) ratios (>3) are detected in the CDR regions of the two memory B cell groups than in the FR regions.
Hotspot motif analysis
Higher numbers of mutations in the context of hotspot motifs were found in the CDR regions compared with the FR regions of both sets of memory B cells. As indicated in Figure 6, the frequency of mutations within hotspot motifs and the distribution of hotspot mutations along the variable regions did not differ between IgD+CD27+ and IgD−CD27+ memory B cells. These data suggest that although the number of somatic mutations is reduced in IgD+CD27+ memory B cells compared with IgD−CD27+ memory B cells, the hotspot targeting function of the somatic mutation related machinery is fully developed in IgD+CD27+ memory B cells.
FIGURE 6. Frequency of mutations within hotspot motifs and the distribution of hotspot mutations along the variable regions were similar between IgD+CD27+ and IgD−CD27+ memory B cells.

A. Numbers of total mutations and hotspot-targeted replacement or silent mutations within CDRs and FRs are shown. B. Frequency of hotspot-targeted mutations in the CDRs or FRs for the three subsets of circulating naïve and memory B cells are similar.
Discussion
The success of vaccination for disease prevention depends on the immunological memory that is carried by memory B and T cells. One useful approach to characterize immune memory is to study the generation, diversity and function of memory B cells that are responsible for human immunity. In this study, we analyzed human antibody variable heavy chain genes isolated simultaneously from three naïve and memory B cell groups in healthy human adult blood samples. The central findings of this study are that: memory B cells in peripheral blood share the preferential Ig gene usages with that of naïve B cells; and distinct characteristics in CDR3 regions of Ig gene rearrangements in these naïve and memory B cells indicate differential levels of activity of enzymes leading to somatic mutations and junctional diversity during the ontogeny or selection of these groups of B cells.
Antibody gene analysis at the single-cell level revealed that the VH antibody repertoire in the three naïve and memory B cells groups were all VH3-dominated. We also observed a similar bias towards the D3 and JH4 gene segment families in these three groups. When we analyzed individual VH gene segment usages, we found further evidence for similarity of combinatorial diversity in the three groups of naïve and memory B cells. These data suggested that the antibody repertoire in human B cells is regulated tightly during the ontogeny of naive B cells, and is not altered significantly following antigenic exposure. The broad diversity that is present in human naïve B cells and maintained in memory cells suggests a well-developed human antibody repertoire that is established and regulated prior to, and independently of, antigen exposure.
Another important finding of this study is the delineation of fine molecular features of the Ig heavy chain genes of the recently defined un-class switched memory B cell group, IgD+CD27+ B cells. Studies from several groups have suggested unique biological functions and possibly a unique developmental pathway of this group of cells (Agematsu et al., 1998; Di Sabatino et al., 2004; Kruetzmann et al., 2003; Ma et al., 2006; Weller et al., 2004; Xiao et al., 2004; Zandvoort et al., 2001). Our data demonstrated that VH, D and JH gene segment usage in this type of memory B cells are broad and very similar to those of both naïve B cells and IgD−CD27+ memory B cells. Consistent with previous reports, our data indicates a significantly lower frequency of somatic mutations in the VH genes of these cells than in IgD−CD27+ memory B cells. This finding is in line with the hypothesis that IgD+CD27+ B cells are formed in the absence of a robust T cell help environment, and may be derived from an extra-follicular pathway (Weller et al., 2001). Since the up-regulation of transcription of genes involved in somatic hypermutation, such as activation induced cytidine deaminase (AID) occurs exclusively in germinal centers in response to T cell interaction (Muramatsu et al., 1999), the lower frequency of somatic mutations in the VH genes of IgD+CD27+ cells suggests that these B cells may mutate their antigen receptors in the absence of robust classical CD40-CD40L mediated T-B interaction characteristic of germinal center formation.
Despite the lower frequency of mutations observed in the IgD+CD27+ memory B cells, the ratio and distribution of replacement, silent or hotspot-targeted mutations across the variable regions were very similar to those in IgD−CD27+ memory B cells. Hotspot-directed mutations reflect the activity of particular error-prone DNA polymerases. As reported, somatic mutation hotspots WA/TW correspond to the error spectrum of DNA polymerase η and the consensus motifs DGYW (D =A/G/T) and its complement WRCH (H= T/C/A) are predictors of mutability in AID-induced immunoglobulin hypermutation (Rogozin and Diaz, 2004; Rogozin et al., 2001). Additional error-prone polymerases, such as κ, ι, μ and ζ are likely responsible for some of the observed somatic mutations that occur outside the described hotspot motifs (Faili et al., 2002; Papavasiliou and Schatz, 2002; Storb, 2001). Our findings of similar targeting features suggested that the somatic mutations that accumulated in IgD+CD27+ memory B cells were induced using similar molecular mutational machinery to that used in IgD−CD27+ memory B cells.
Our finding of shorter heavy chain CDR3 lengths in the IgD+CD27+ memory B cells provides supportive evidence of unique features in the ontogeny of these cells. Rosner et al. reported shorter CDR3 regions in mutated VH6 genes rearranged to IgM constant genes when compare to un-mutated ones (Rosner et al., 2001). That study found the CDR3 region of mutated genes contained shorter V, D, J, P and N components than non-mutated genes and hypothesized that B cells bearing Ig molecules with shorter CDR3s have been selected for binding to antigen. In addition to data that is consistent with that in Rosner et al.’s report, our data indicated distinct characteristics of CDR3 regions in the two memory B cell groups. In our study, examination of the VH-D and D-JH junctions of Ig genes indicated that shorter heavy chain CDR3 lengths in the IgD+CD27+ memory B cells were due to reduced N and P nucleotide additions at both junctions. Reduced TdT-mediated extension activity and an increase of exonuclease activity at the 3′ end of D genes were suggested by the nucleotide sequences of junctions in the IgD+CD27+ memory B cells compared with those of IgD−CD27+ memory B cells. The CDR3 length is determined largely in the pro-B cell stage when recombinase activating gene (RAG) 1 and 2 and TdT genes accomplish recombination and modification of the ends of gene segments (Fugmann et al., 2000; Komori et al., 1993; Lee et al., 2004). Therefore, these IgD+CD27+ memory B cells could be selectively derived from a more restricted naïve B cell population that harbored shorter CDR3 regions than did the circulating naïve B cells or IgD−CD27+ memory B cells. Interestingly, studies of germinal center human tonsilar B cells, follicular lymphoma cells or synovial B cells in rheumatoid arthritis patients have demonstrated that the progress of somatic hypermutation could introduce insertions and deletions into CDR regions of immunoglobulin V genes in addition to base-pair substitutions, although at a very low frequency (Kobrin et al., 2001; Wilson et al., 1998). It is possible that the unique features we observed in the profile of junctional nucleotide insertions and deletions of VH genes for IgD+CD27+ memory B cells were due partially to a lower frequency of somatic hypermutation events that occurred during the affinity maturation of these memory B cells.
Restriction of Ig gene usages in antigen-specific B cells have been reported in both animal and human studies of antibody responses towards a variety of antigens, ranging from hapten, carbohydrate, viral protein, Rhesus D antigens, to other self proteins (Berek and Milstein, 1987; Casadevall and Scharff, 1991; Minnerath et al., 1995; Perera et al., 2000a; Perera et al., 2000b; Zhang et al., 2006), while others reported diverse antibody responses to confined regions of foreign proteins or homologs of self proteins (Andria et al., 1990; Caton et al., 1991; Kalinke et al., 1996). We previously showed that antibody gene usages of rotavirus (RV)-specific B cells differed from that of randomly selected B cells, with a distinct bias in the usage of gene segments in the VH1 and VH4 families. The biased RV-specific B cell heavy chain gene repertoire was shared by both human infants and adults (Weitkamp et al., 2003a). A similar antibody gene bias was found in RV-specific IgD− memory B cells expressing the intestinal homing marker α4β7 (Weitkamp et al., 2003a; Weitkamp et al., 2005). Investigating antibody variable gene usages and molecular characteristics in naïve and memory antibody responses to a particular antigen would help to better understand the influence of affinity maturation in the maintenance, expansion, or narrowing of antigen-specific repertoires in memory B cells.
As suggested by Rosner et al., shorter CDR3 regions in IgM expressing memory B cells may play a critical role in the proper interaction of antibody with antigen. Since the CDR3 loop often is situated in the center of the antigen-binding site, antibodies with shorter CDR3s may have more space in the antigen-binding pocket for the antigen to enter and make contact with the CDR1 and 2. Our observation that IgD−CD27+ isotype-switched memory B cells do not share the same molecular features of CDR3 regions with IgD+CD27+ memory B cells is interesting. The findings suggest a distinct structural determinant for Ig heavy chain CDR3 lengths that may be related to the isotypes of antibodies that are expressed in memory B cells.
In conclusion, our data revealed a highly regulated VH gene segment repertoire that develops early in the ontogeny of human memory B cells. Our findings of distinct mutational and junctional features of antibody variable genes in IgD+CD27+ un-class switched memory B cells support the idea that these memory B cells develop or become selected in a manner distinct from that of class-switched memory B cells. Differential TdT mediated nucleotide addition and exonuclease activities in VH-D and D-JH junctions appear to contribute largely to the varied CDR3 lengths among these groups of naïve and memory B cells.
Supplementary Material
Acknowledgments
This work was supported by a grant from the NIAID (R01 AI-57933). C.T. was a 2003–2004 fellow of the NIAID Molecular Basis in Infectious Diseases Training Program (T32 AI-07474). The Vanderbilt Flow Cytometry Core Laboratory was supported by the Vanderbilt Ingram Cancer Center (P30 CA68485). Clinical support was provided by the Vanderbilt General Clinical Research Center (grant M01 RR-00095, National Center for Research Resources, National Institute of Health).
We thank all the volunteers who participated in this study for their generosity. We thank DNAX for use of the CD154-expressing cell line and Dr. Rudolf H. Zubler for the EL4-B5 cell line.
Abbreviations
- AID
activation induced cytidine deaminase
- CDR
complementarity determining region
- FR
framework region
- Ig
immunoglobulin
- PBMC
peripheral blood mononuclear cell
- R
replacement mutation
- RAG
recombinase activating genes
- S
silent mutation
- TdT
terminal deoxynucleotidyl transferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agematsu K. Memory B cells and CD27. Histol Histopathol. 2000;15:573–6. doi: 10.14670/HH-15.573. [DOI] [PubMed] [Google Scholar]
- Agematsu K, Hokibara S, Nagumo H, Komiyama A. CD27: a memory B-cell marker. Immunol Today. 2000;21:204–6. doi: 10.1016/s0167-5699(00)01605-4. [DOI] [PubMed] [Google Scholar]
- Agematsu K, Nagumo H, Shinozaki K, Hokibara S, Yasui K, Terada K, Kawamura N, Toba T, Nonoyama S, Ochs HD, Komiyama A. Absence of IgD-CD27(+) memory B cell population in X-linked hyper-IgM syndrome. J Clin Invest. 1998;102:853–60. doi: 10.1172/JCI3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andria ML, Levy S, Benjamini E. Diverse VH and VL genes are used to produce antibodies against a defined protein epitope. J Immunol. 1990;144:2614–9. [PubMed] [Google Scholar]
- Berek C, Milstein C. Mutation drift and repertoire shift in the maturation of the immune response. Immunol Rev. 1987;96:23–41. doi: 10.1111/j.1600-065x.1987.tb00507.x. [DOI] [PubMed] [Google Scholar]
- Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298:2199–202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- Camerini D, Walz G, Loenen WA, Borst J, Seed B. The T cell activation antigen CD27 is a member of the nerve growth factor/tumor necrosis factor receptor gene family. J Immunol. 1991;147:3165–9. [PubMed] [Google Scholar]
- Casadevall A, Scharff MD. The mouse antibody response to infection with Cryptococcus neoformans: VH and VL usage in polysaccharide binding antibodies. J Exp Med. 1991;174:151–60. doi: 10.1084/jem.174.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton AJ, Stark SE, Kavaler J, Staudt LM, Schwartz D, Gerhard W. Many variable region genes are utilized in the antibody response of BALB/c mice to the influenza virus A/PR/8/34 hemagglutinin. J Immunol. 1991;147:1675–86. [PubMed] [Google Scholar]
- Crotty S, Ahmed R. Immunological memory in humans. Semin Immunol. 2004;16:197–203. doi: 10.1016/j.smim.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Di Sabatino A, Carsetti R, Rosado MM, Ciccocioppo R, Cazzola P, Morera R, Tinozzi FP, Tinozzi S, Corazza GR. Immunoglobulin M memory B cell decrease in inflammatory bowel disease. Eur Rev Med Pharmacol Sci. 2004;8:199–203. [PubMed] [Google Scholar]
- Faili A, Aoufouchi S, Flatter E, Gueranger Q, Reynaud CA, Weill JC. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature. 2002;419:944–7. doi: 10.1038/nature01117. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Kalinke U, Bucher EM, Ernst B, Oxenius A, Roost HP, Geley S, Kofler R, Zinkernagel RM, Hengartner H. The role of somatic mutation in the generation of the protective humoral immune response against vesicular stomatitis virus. Immunity. 1996;5:639–52. doi: 10.1016/s1074-7613(00)80277-0. [DOI] [PubMed] [Google Scholar]
- Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188:1679–89. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobrin C, Bendandi M, Kwak L. Novel secondary Ig VH gene rearrangement and in-frame Ig heavy chain complementarity-determining region III insertion/deletion variants in de novo follicular lymphoma. J Immunol. 2001;166:2235–43. doi: 10.4049/jimmunol.166.4.2235. [DOI] [PubMed] [Google Scholar]
- Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–5. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- Kruetzmann S, Rosado MM, Weber H, Germing U, Tournilhac O, Peter HH, Berner R, Peters A, Boehm T, Plebani A, Quinti I, Carsetti R. Human immunoglobulin M memory B cells controlling Streptococcus pneumoniae infections are generated in the spleen. J Exp Med. 2003;197:939–45. doi: 10.1084/jem.20022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–84. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- Liu YJ, de Bouteiller O, Arpin C, Briere F, Galibert L, Ho S, Martinez-Valdez H, Banchereau J, Lebecque S. Normal human IgD+IgM- germinal center B cells can express up to 80 mutations in the variable region of their IgD transcripts. Immunity. 1996;4:603–13. doi: 10.1016/s1074-7613(00)80486-0. [DOI] [PubMed] [Google Scholar]
- Ma CS, Pittaluga S, Avery DT, Hare NJ, Maric I, Klion AD, Nichols KE, Tangye SG. Selective generation of functional somatically mutated IgM+CD27+, but not Ig isotype-switched, memory B cells in X-linked lymphoproliferative disease. J Clin Invest. 2006;116:322–33. doi: 10.1172/JCI25720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnerath JM, Mueller CM, Buron S, Jemmerson R. B lymphocyte recognition of cytochrome c: higher frequency of cells specific for self versus foreign antigen early in the immune response and V gene usage in the response to self antigen. Eur J Immunol. 1995;25:784–91. doi: 10.1002/eji.1830250324. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–6. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Nagumo H, Agematsu K, Shinozaki K, Hokibara S, Ito S, Takamoto M, Nikaido T, Yasui K, Uehara Y, Yachie A, Komiyama A. CD27/CD70 interaction augments IgE secretion by promoting the differentiation of memory B cells into plasma cells. J Immunol. 1998;161:6496–502. [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. Somatic hypermutation of immunoglobulin genes: merging mechanisms for genetic diversity. Cell. 2002;109(Suppl):S35–44. doi: 10.1016/s0092-8674(02)00706-7. [DOI] [PubMed] [Google Scholar]
- Perera WS, Moss MT, Urbaniak SJ. Comparison between hybridoma and Fab/phage anti-RhD: their V gene usage and pairings. Dis Markers. 2000a;16:15–9. doi: 10.1155/2000/451713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera WS, Moss MT, Urbaniak SJ. V(D)J germline gene repertoire analysis of monoclonal D antibodies and the implications for D epitope specificity. Transfusion. 2000b;40:846–55. doi: 10.1046/j.1537-2995.2000.40070846.x. [DOI] [PubMed] [Google Scholar]
- Prasad KV, Ao Z, Yoon Y, Wu MX, Rizk M, Jacquot S, Schlossman SF. CD27, a member of the tumor necrosis factor receptor family, induces apoptosis and binds to Siva, a proapoptotic protein. Proc Natl Acad Sci U S A. 1997;94:6346–51. doi: 10.1073/pnas.94.12.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman VS, Akondy RS, Rath S, Bal V, George A. Ligation of CD27 on B cells in vivo during primary immunization enhances commitment to memory B cell responses. J Immunol. 2003;171:5876–81. doi: 10.4049/jimmunol.171.11.5876. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Diaz M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J Immunol. 2004;172:3382–4. doi: 10.4049/jimmunol.172.6.3382. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat Immunol. 2001;2:530–6. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- Rosner K, Winter DB, Tarone RE, Skovgaard GL, Bohr VA, Gearhart PJ. Third complementarity-determining region of mutated VH immunoglobulin genes contains shorter V, D, J, P, and N components than non-mutated genes. Immunology. 2001;103:179–87. doi: 10.1046/j.1365-2567.2001.01220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M, Giudicelli V, Ginestoux C, Stoehr P, Robinson J, Bodmer J, Marsh SG, Bontrop R, Lemaitre M, Lefranc G, Chaume D, Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2000;28:219–21. doi: 10.1093/nar/28.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Agematsu K, Ochs HD, Sugane K. Functional analysis of human memory B-cell subpopulations: IgD+CD27+ B cells are crucial in secondary immune response by producing high affinity IgM. Clin Immunol. 2003;108:128–37. doi: 10.1016/s1521-6616(03)00092-5. [DOI] [PubMed] [Google Scholar]
- Storb U. DNA polymerases in immunity: profiting from errors. Nat Immunol. 2001;2:484–5. doi: 10.1038/88673. [DOI] [PubMed] [Google Scholar]
- Weitkamp JH, Kallewaard N, Kusuhara K, Bures E, Williams JV, LaFleur B, Greenberg HB, Crowe JE., Jr Infant and adult human B cell responses to rotavirus share common immunodominant variable gene repertoires. J Immunol. 2003a;171:4680–8. doi: 10.4049/jimmunol.171.9.4680. [DOI] [PubMed] [Google Scholar]
- Weitkamp JH, Kallewaard N, Kusuhara K, Feigelstock D, Feng N, Greenberg HB, Crowe JE., Jr Generation of recombinant human monoclonal antibodies to rotavirus from single antigen-specific B cells selected with fluorescent virus-like particles. J Immunol Methods. 2003b;275:223–37. doi: 10.1016/s0022-1759(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Weitkamp JH, Kallewaard NL, Bowen AL, Lafleur BJ, Greenberg HB, Crowe JE., Jr VH1–46 is the dominant immunoglobulin heavy chain gene segment in rotavirus-specific memory B cells expressing the intestinal homing receptor alpha4beta7. J Immunol. 2005;174:3454–60. doi: 10.4049/jimmunol.174.6.3454. [DOI] [PubMed] [Google Scholar]
- Weller S, Braun MC, Tan BK, Rosenwald A, Cordier C, Conley ME, Plebani A, Kumararatne DS, Bonnet D, Tournilhac O, Tchernia G, Steiniger B, Staudt LM, Casanova JL, Reynaud CA, Weill JC. Human blood IgM "memory" B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood. 2004;104:3647–54. doi: 10.1182/blood-2004-01-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S, Faili A, Garcia C, Braun MC, Le Deist FF, de Saint Basile GG, Hermine O, Fischer A, Reynaud CA, Weill JC. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc Natl Acad Sci U S A. 2001;98:1166–70. doi: 10.1073/pnas.98.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PC, de Bouteiller O, Liu YJ, Potter K, Banchereau J, Capra JD, Pascual V. Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. J Exp Med. 1998;187:59–70. doi: 10.1084/jem.187.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Hendriks J, Langerak P, Jacobs H, Borst J. CD27 is acquired by primed B cells at the centroblast stage and promotes germinal center formation. J Immunol. 2004;172:7432–41. doi: 10.4049/jimmunol.172.12.7432. [DOI] [PubMed] [Google Scholar]
- Zandvoort A, Lodewijk ME, de Boer NK, Dammers PM, Kroese FG, Timens W. CD27 expression in the human splenic marginal zone: the infant marginal zone is populated by naive B cells. Tissue Antigens. 2001;58:234–42. doi: 10.1034/j.1399-0039.2001.580403.x. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zharikova D, Mozdzanowska K, Otvos L, Gerhard W. Fine specificity and sequence of antibodies directed against the ectodomain of matrix protein 2 of influenza A virus. Mol Immunol. 2006;43:2195–206. doi: 10.1016/j.molimm.2005.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


