Abstract
Members of the ETS family of transcription factors are among the first genes expressed in the developing vasculature, but loss-of-function experiments for individual ETS factors in mice have not uncovered important early functional roles for these genes. However, multiple ETS factors are expressed in spatially and temporally overlapping patterns in the developing vasculature, suggesting possible functional overlap. We have taken a comprehensive approach to exploring the function of these factors during vascular development by employing the genetic and experimental tools available in the zebrafish to analyze four ETS family members expressed together in the zebrafish vasculature; fli1, fli1b, ets1, and etsrp. We isolated and characterized an ENU-induced mutant with defects in trunk angiogenesis and positionally cloned the defective gene from this mutant, etsrp. Using the etsrp morpholinos targeting each of the four genes, we show that the four ETS factors function combinatorially during vascular and hematopoietic development. Reduction of etsrp or any of the other genes alone results in either partial or no defects in endothelial differentiation, while combined reduction in the function of all four genes causes dramatic loss of endothelial cells. Our results demonstrate that combinatorial ETS factor function is essential for early endothelial specification and differentiation.
Keywords: zebrafish, ETS transcription factors, intersegmental vessels, vascular development, angiogenesis
INTRODUCTION
The ETS factors are a large family of transcriptional regulatory proteins involved in a wide variety of developmental and postnatal processes, in an equally diverse array of tissues. The first known ETS family member was the proto-oncogene Ets-1, the cellular progenitor of v-ets, a viral oncogene found in the genome of the E26 acute leukemia retrovirus (Leprince et al., 1983; Nunn et al., 1983). More than 50 additional members of this family have now been identified, all of which share an 85 amino acid conserved DNA binding domain, the “ETS domain,” a winged helix-turn-helix motif generally located in the c-terminal half of the protein. The ETS domain binds to DNA sequence consisting of a core GGAA/T motif. Most ETS family members are transcriptional activators except for a few (Erf, Net, Tel) that have been shown to have repressor activity. The activity of ETS transcription factors is regulated through their interaction with a large number of different structurally unrelated transcription factors such as AP1, MafB, and CBP (for comprehensive list see (Lelievre et al., 2001; Verger and Duterque-Coquillaud, 2002).
Recent studies have suggested that a number of different ETS factors play important roles in hematopoietic and vascular development during early embryogenesis, although most in vivo loss-of-function data have not provided compelling evidence for essential roles for these genes in the specification or differentiation of these tissues (reviewed in (Lelievre et al., 2001; Sato, 2001)). During murine development, the Ets1 gene is initially expressed broadly in ventral mesoderm, becoming progressively restricted to hemangiogenic mesoderm and then endothelium (Pardanaud and Dieterlen-Lievre, 1993; Queva et al., 1993). Antisense oligonucleotides against Ets-1 inhibit angiogenesis in the chick chorioallantoic membrane (CAM) assay (Wernert et al., 1999), and dominant-negative Ets1 inhibits angiogenesis in endothelial cells in vitro and in implanted matrigel plugs (Nakano et al., 2000), all supporting the idea that this factor plays an important role in angiogenesis. However, Ets1 null mice are viable and fertile and have no detectable vascular defects (Barton et al., 1998; Bories et al., 1995; Muthusamy et al., 1995). In zebrafish, however, a recent study reported that morpholino knock-down of the novel ets1 related protein etsrp does cause defects in blood vessel formation (Sumanas and Lin, 2005).
Like Ets1, The Fli-1 and closely related ERG (Ets-related) genes become progressively restricted to endothelial cells in the trunk during zebrafish and Xenopus development (Brown et al., 2000; Meyer et al., 1995). Overexpression of either Fli-1 or ERG by microinjection into Xenopus laevis embryos causes ectopic endothelial differentiation, in addition to other defects (Baltzinger et al., 1999; Remy et al., 1996). However, transgenic mice overexpressing Fli-1 in a variety of tissues under H-2Kk promoter control do not exhibit vascular abnormalities, although they do develop a lethal renal immunologic disease (Zhang et al., 1995). Furthermore, mice homozygous for a targeted disruption of Fli-1 form a functional network of blood vessels, indicating that vasculogenesis and angiogenesis proceed normally, although they develop CNS hemorrhage (Hart et al., 2000; Spyropoulos et al., 2000). Mice with targeted disruption of the TEL repressor have normal vasculogenesis, with histologically normal dorsal aorta, intersomitic vessels and head veins at E9.5. However, they exhibit defective yolk sac angiogenic remodeling as well as intra-embryonic apoptosis of mesenchymal and neural cells (Wang et al., 1997). It has been suggested that Tel functions in the maintenance of the developing vascular network rather in the specification, differentiation, or proliferation of endothelial cells.
The early and specific expression of ETS factors in vascular tissues and their mesodermal progenitors has led to speculation that these factors might be important in the establishment of this lineage and the differentiation of angioblasts and endothelial cells, but as noted above in vivo evidence for this has been difficult to obtain from loss-of-function experiments. It is likely that a significant difficulty in probing the functional roles of ETS factors in the vasculature is the extensive overlap between these factors in this tissue, making it difficult to perform effective loss-of-ETS-function experiments. We have taken a comprehensive approach to exploring ETS factor function in the vasculature by using the zebrafish to simultaneously reduce the levels of multiple ETS family members. We identified four different vascular ETS factors in the zebrafish, and characterized a genetic mutant in one of these genes, etsrp. We show that the four genes function cooperatively in the differentiation and maintenance of endothelium.
MATERIALS AND METHODS
Zebrafish Methods
Zebrafish (Danio rerio) embryos were obtained and raised and fish maintained as described (Kimmel et al., 1995; Westerfield, 1995). The Tg(fli1:EGFP)y1 transgenic zebrafish line was described previously (Lawson and Weinstein, 2002). An ENU F3 genetic screen was performed using this line to isolate the etsrpy11 mutant (Lawson et al., unpublished results). Embryos imaged post-1.5 dpf were treated with 1-phenyl-2-thiourea (PTU) to inhibit pigment formation (Westerfield, 1995).
Meiotic and physical mapping of the y11 mutation
Meiotic and physical mapping was performed essentially as described previously (Roman et al., 2002) using an EK tg(fli1:egfp)y1; y11 /TL polymorphic mapping cross. Bulked segregant analysis was performed using a 192-marker panel of CA repeat markers (the list of markers in this panel is available upon request). Oligonucleotide sequences for the markers noted in Fig. 2 are available at http://zebrafish.mgh.harvard.edu/zebrafish/ssrQuery.aspx. BAC clones were identified by screening the CHORI 211 Zebrafish BAC library filters (Children’s Hospital Oakland Research Institute) using 32P labeled oligonucleotide probes. PAC clones were identified from DNA pools by PCR as described by the supplier (Chris Amemiya Lab). BAC and PAC DNAs were prepared using Nucleobond columns (Clontech). BAC end sequences were obtained from http://trace.ensembl.org/perl/ssahaview?server=danio_rerio. PAC end sequences were determined by sequencing. Additional genomic sequences were obtained through SSAHA2 searches of trace data from the Sanger Institute (http://www.ensembl.org/Danio_rerio/blastview). PCR primers designed against non-repetitive regions of PAC and BAC ends and genomic sequences were used to establish physical contigs and look for polymorphisms for use in meiotic mapping. To identify polymorphisms, PCR products amplified from genomic DNA from in individual wild type, heterozygous, and mutant embryos (genotyped based on flanking markers) were sequenced. SNPs were assayed as RFLPs, when possible, or were converted to RFLPs using derived cleaved amplified polymorphic sequence (dCAPS) analysis using a dCAPS Finder program (Neff et al., 2002), http://helix.wustl.edu/dcaps/dcaps.html.
Figure 2.
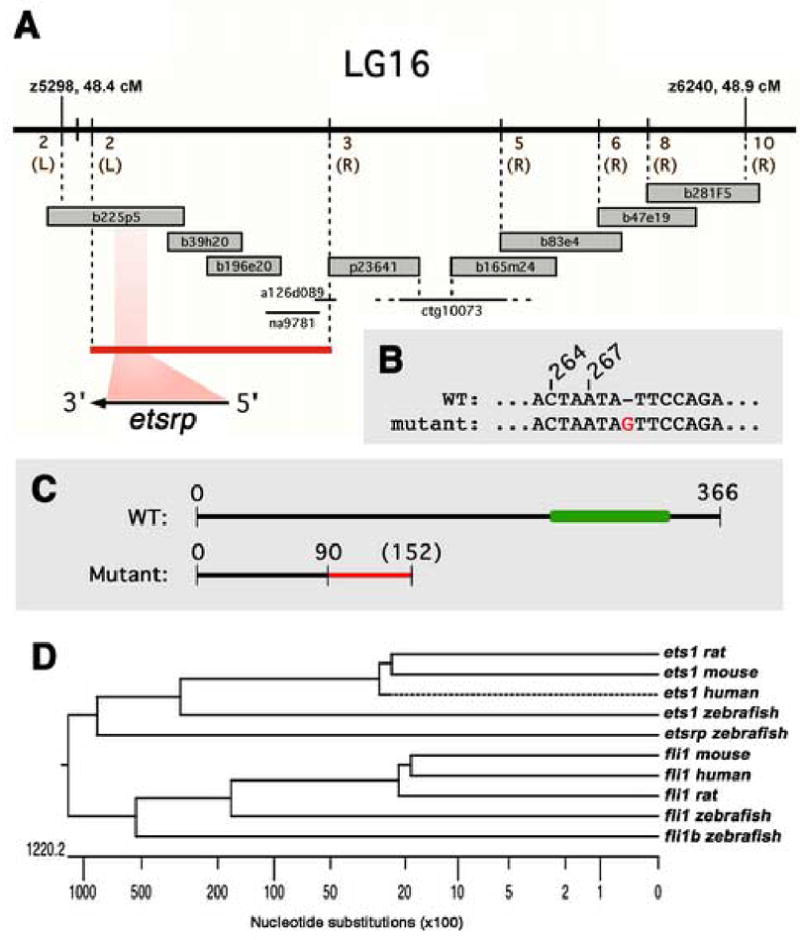
Positional cloning of etsrp, the defective gene in y11 mutants. (A) Genetic and physical map of the y11 interval. Genetic map is shown at top, with the number of recombinants in approximately 2000 meioses noted in brown (and whether recombinants are on the right or left sides, in parentheses). BAC and PAC clones are shown as large grey bars, genomic sequence contigs and traces are shown as black bars. The red bar shows the y11 critical interval containing the entire etsrp transcript. (B) In y11 mutants a G is inserted after nucleotide 269 in the etsrp coding sequence. (C) The y11 insertion mutation results in an altered reading frame after amino acid 90 of etsrp coding for an additional novel 62 amino acids (shown in red) before terminating at a stop codon. The y11 mutant protein lacks the highly conserved ETS DNA binding domain (shown in green in the wild type protein). See results and methods for additional details. (D) ETS family members expressed in the zebrafish vasculature and related mammalian orthologs. Dendogram of nucleotide simiilarity across the entire coding region of zebrafish ETS family members expressed in the zebrafish vasculature and the most closely related mammalian genes. The zebrafish ets1 and fli1 genes are more closely related to their mammalian counterparts than to other zebrafish ETS family members. The etsrp and fli1b genes are most closely related to ets1 and fli1 genes, respectively.
Cloning of full length zebrafish etsrp, fli1b, and ets1 cDNAs
etsrp
Sequences encompassing the 5′ UTR to 3′ UTR of etsrp were obtained using available zebrafish EST (GenBank #s AI877585, AL915831, AI 793509, AI793542) and genomic trace sequences. 5′ RACE was performed from 24 hpf cDNA to obtain the complete 5′ UTR sequence. The full-length cDNA of etsrp was obtained by high fidelity PCR on 24 hpf cDNA using Pfu Ultra High-Fidelity DNA polymerase (Stratagene) and the primers 5′-CTTTAAGATATGGAAATGTACCAATCGG - 3′’ and 5′-CCAATCCTTCGATTCCTCCTCTA-3′. A single product was obtained, cloned into pCRII-TOPO (Invitrogen) and verified by sequencing.
fli1b
The 3′end of fli1b were obtained using available zebrafish EST sequence (GenBank # CA472045). 5′ RACE was performed from 24 hpf cDNA to obtain the complete 5′ UTR and coding sequence. The full-length coding sequence of zebrafish fli1b was obtained by high fidelity PCR as above with primers 5′-CAGAAATCTGCAATGGACT-3′ and 5′-GTGACTGTTTTAATAATAAGTGTTC-3′. A single product was obtained, cloned into pCRII-TOPO (Invitrogen) and verified by sequencing.
ets1
The full-length coding sequence of zebrafish ets1 (GenBank # BC092935) was amplified by high fidelity PCR as above using primers 5′-ACAGACTCTGTACGTTTGAATGCGT-3′ and 5′-GTCCAGACTTTACTCGTCCGTGTC-3′. A single product was obtained, cloned into pCRII-TOPO (Invitrogen) and verified by sequencing.
Expression constructs, in vitro transcription/translation, and RNA injection
pCS2/etsrp
The etsrp pCRII-TOPO clones described above were used as templates in a PCR reaction with Pfu Ultra High-Fidelity DNA polymerase (Strategene) and primers extended with a BamHI site (5′-AGGGATCCCAGAGCTGAGGCACTGTCAAAACC 3′) and an XbaI site (5′-GCTCTAGACCAATCCTTCGATTCCTCCTCTA-3′). The resulting PCR product was digested with BamHI and XbaI and cloned into Bam/XbaI-digested pCS2+. The construct was verified by sequencing.
pCS2+/fli1
A full-length fli1 cDNA clone (Genbank NM131348)) was ordered (Open Biosystems, Inc.) and used as template in a PCR reaction with PfuUltra High-Fidelity DNA polymerase (Strategene) and primers extended with an EcoRI site (5′-CGGAATTCTAATTCAGACGCGTGTCATGT-3′), and an XhoI site (5′-CCGCTCGAGCCATCTTCGAGTGCAGTTCAAG-3′). The resulting PCR product was digested with EcoRI and XhoI and cloned into EcoRI/XhoI digested pCS2+. The construct was verified by sequencing.
pCS2+/ets1 and pCS2+/fli1b
The ets1 pCRII-TOPO and fli1b pCRII-TOPO clones described above were digested with BamHI and XbaI and cloned into BamHI/XbaI digested pCS2+.
pFliL/etsrp
The etsrp pCRII-TOPO clones described above were used as templates in a PCR reacdtion with PfuUltra High-Fidelity DNA polymerase (Strategene) and primers 5′-GCCACCATGGAAATGTACCAATCTGG-3′ and 5′-ATGTGTCCAGGACTCTGTGGTTTCCT-3′. A single product was obtained, cloned into pcDNA3.1 (Invitrogen), and verified by sequencing. The etsrp pcDNA3.1 construct was used as template in a PCR reaction with PfuUltra High-Fidelity DNA polymerase (Strategene) and primers extended with an NheI site (5′-CTAGCTAGCTAGGCCACCATGGAAATGTACCAA-3′), and an AscI site (5′-TTGGCGCGCCAAACTCAGACAATGCGATGC-3′). The resulting PCR product was digested with NheI and AscI and cloned into NheI/AscI-digested pFliL. The construct was verified by sequencing.
Capped mRNA for injection was synthesized from NotI-digested PCS2 expression plasmids using the mMessage mMachine SP6 kit according to the provider’s protocol (Ambion). Capped mRNA was injected into 1–4-cell embryos either into a single blastomere or into the streaming yolk cytoplasm, just beneath the blastomeres, using a pneumatic picopump and micromanipulator (World Precision Instruments), as described previously (Westerfield, 1995). In vitro transcription/translation assays were performed using the TNT SP6 Quick Coupled Transcription Translation System (Promega), following the manufacturer’s instructions.
Morpholino injections
Morpholino-modified antisense oligonucleotides (Genetools) used in thius study included: a standard control morpholino (5′-CCTCTTACCTCAGTTACAATTTATA-3′); etsrp (5′-GGTTTTGACAGTGCCTCAGCTCTGC-3′) targeting −10 to −34 of the 5′ UTR of etsrp; etsrpE/I-2 (5′ AAATAAGATATTACCATATGAACTG 3′) targeting the exon 2 / intron 2 boundary; etsrpE/I-3 (5′-TGAGATGCTCACCTTTGTGCAACAG-3′) targeting the exon 3 / intron 3 boundary; ets1 (5′-GTCATGGTCACGCATTCAAACGTAC-3′) targeting −10 to +15 of the 5′ UTR and coding region of ets1; fli1b (5′-GGTTAAACTTGAGCTATGTAAACCC 3′) targeting −57 to −81 of the 5′ UTR of fli1b; fli1 (5′-GTTCCGTCCATTTTCCGCAATTTTC-3′) targeting −14 to +11 of the 5′ UTR and coding region of fli1. Morpholinos were used at the following “high” doses (see text): etsrp 2ng; etsrpE/I-2, 15ng; etsrpE/I-3, 12ng; combined etsrpE/I-2 + etsrpE/I-3, 10ng each (the combination gives a stronger phenotype similar to the “etsrp” morpholino, the individual “etsrpE/I” morpholinos also give similar but less severe phenotypes); ets1, 2ng, fli1b, 15ng; fli1, 15ng. “Medium” and “low” doses of morpholinos used in this study were one-half and one-quarter of the “high” dose, respectively. All morpholinos used in this study were shown to specifically and strongly reduce the levels of the targeted gene product in an in vitro transcription-translation assay without affecting the levels of the other three ETS factors (Supplemental Fig. 1). Morpholino injections were performed on 1–4-cell embryos as described previously (Lawson et al., 2002).
Whole mount in situ hybridization and TUNEL staining
Antisense mRNA probes for fli1, flt4, vecdn, flk1, plxnd1, efnb2, gata1 and gata2 were prepared as described (Brown et al., 2000; Detrich et al., 1995; Larson et al., 2004; Lawson et al., 2001; Thompson et al., 1998; Torres-Vazquez et al., 2004). To derive etsrp, fli1b, and ets1 riboprobes, the corresponding pCRII-TOPO constructs were linearized with XhoI, BamHI, or SpeI, respectively, and transcribed using T7 RNA polymerase. Whole-mount in situ hybridization was performed essentially as described elsewhere (Hauptmann and Gerster, 1994). TUNEL staining of 24 hpf zebrafish embryos was performed as described previously (Parng et al., 2004).
Microscopic imaging methods
Transmitted light images were obtained with a Leica MZ12 microscope equipped with a ProgRes mF digital camera (Jenoptik, Eching, Germany). Two photon microscopy of Tg(fli1:EGFP)y1 zebrafish embryos and larvae was performed using a Radiance 2000 imaging system (BioRad) with 960 nm pulsed mode-locked laser emission from a tunable Ti-Sapphire laser (Tsunami laser, Spectra Physics Inc.). Stacks of frame-averaged (5 frames) multiphoton optical slices were collected digitally, and 2-D or 3-D reconstructions of image data were prepared using the LaserSharp (BioRad) or Metamorph (Universal Imaging) software packages. The fluorescence images shown in this paper are single-view 2-D reconstructions of collected image Z-series stacks, reconstructed at an angle of zero degrees.
Electron microscopy
For transmission electron microscopy zebrafish embryos were fixed at room temperature in buffered 2.5% glutaraldehyde (pH7.3), post-fixed in 2% osmium tetroxide, and processed into Spurr’s epoxy via increasing concentrations of ethanol followed by propylene oxide. Semithin 1 μm plastic sections were cut from at least six plastic blocks and stained with Toluidine Blue O stain. Thin sections of at least two blocks were prepared and stained with uranyl acetate and lead citrate and examined in a JEOL 1010 electron microscope operating at 80kV. Anesthetized adult zebrafish were cut transversely into 6 parts and fixed at 4°C in 2.5 glutaraldehyde-2% paraformaldehyde– 0.1M phosphate buffer (pH7.4) for 2 hours, post-fixed at 4 °C in 1% osmium tetroxide– 0.1M phosphate buffer (pH7.4) for 2 hours, and processed into Agar 100 resin (Agar Scientific Limited, UK) via increasing concentrations of ethanol followed by QY-2. Semithin 0.5 μm plastic sections were cut and stained with Toluidine Blue O stain. Thin sections were stained with uranyl acetate and lead citrate and examined in a Hitachi H-7100 electron microscope operating at 100kV.
RESULTS
Identification of y11, a mutant defective in vascular morphogenesis and angiogenesis
We identified the y11 mutant in an F2 haploid screen for mutants with trunk vascular defects carried out in the Tg(fli1:EGFP)y1 transgenic background (Lawson and Weinstein, 2002) and unpublished results). Homozygous y11 mutants have defects in both vascular morphogenesis and angiogenesis. Mutants have a beating heart, but they lack trunk circulation at 24 hpf and 48 hpf. No reduction in initial numbers of EGFP-positive cells is apparent in the trunk at 24 hpf, suggesting there is no defect in specification of angioblasts during initial vasculogenesis (data not shown). However, increased TUNEL staining is detected in the axial vessels of y11 mutants at 24 hpf (Fig. 1A,B), and the primary vasculogenic vessels (including the trunk dorsal aorta and posterior cardinal vein) fail to undergo proper morphogenesis to form defined tubular vessels (Fig. 1C,D,F,G). Transmission electron microscopy reveals that mutant angioblasts fail to form an ordered epithelium and appear disorganized (Fig. 1J,K). Angiogenic intersegmental vessel sprouts are absent at 24 hpf in mutant trunks (Fig. 1C,D), but aberrant stunted sprouts appear by 48 hpf (Fig. 1H,I). Mutant intersegmental vessel sprouts fail to lumenize properly and form inappropriate branches (Fig. 1H,I), although the overall patterning of the trunk vasculature is not grossly perturbed as it is in vascular patterning mutants such as out of bounds (Torres-Vazquez et al., 2004). Inspection of y11 mutants under transmitted light reveals no developmental delay or detectable deficits in the patterning, size, or shape of non-vascular tissues including the somites, notochord, brain, neural tube, and gut (data not shown). These results indicate that the function of y11 is restricted to the circulatory system.
Figure 1.
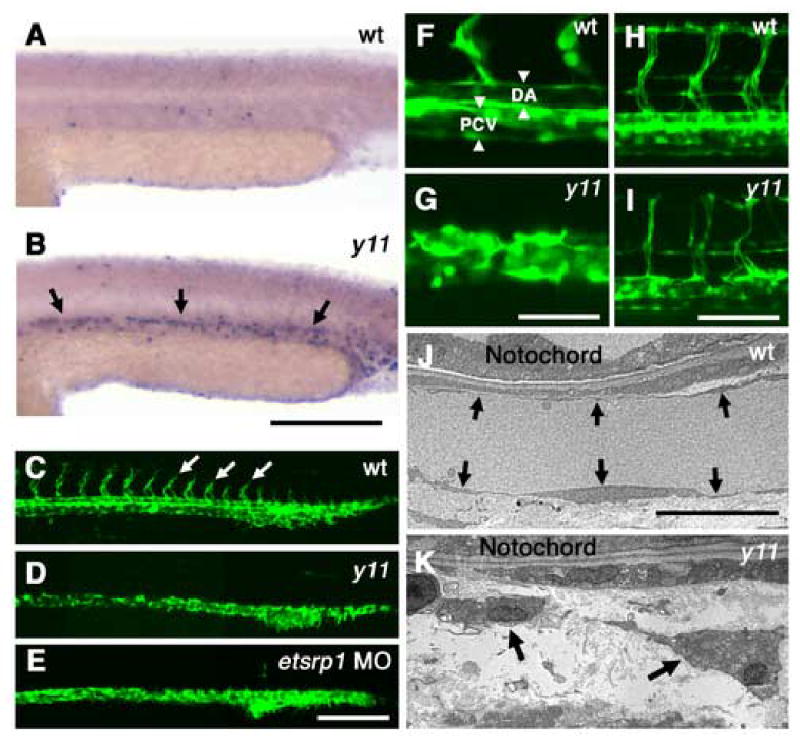
y11 mutants have defects in angiogenesis and vascular morphogenesis. (A,B) TUNEL staining of the trunks of 24 hpf wild type (A) and y11 mutant (B) animals, low background in wild type animals and substantially increased staining in the developing axial vessels of y11 mutants (arrows). (C–I) Two-photon images of Tg(fli1:EGFP)y1 transgenic wild type or y11 mutant animals at 24 hpf (C–G) and 48 hpf (H,I). Wild type siblings (C,F,H) have intersegmental vessel sprouts (arrows in C) and properly lumenized axial vessels (arrowheads in F) at 24 hpf. Intersegmental vessels have formed a lumenized, functional vascular network by 48 hpf (H). In y11 mutants (D,G,I) intersegmental vessel sprouts are absent at 24 hpf (D) and axial vessels fail to undergo proper tubular morphogenesis (G). Intersegmental vessels are similarly absent in etsrp morphants at 24 hpf (E). By 48 hpf y11 mutants have aberrant intersegmental vessel sprouts that are not fully extended and have some branching and pathfinding errors (I). (J,K) Electron microscopy of 24 hpf zebrafish shows defects in vascular morphogenesis in y11 mutants. Wild type siblings have normal single-cell thick endothelial epithelium (arrows) around the dorsal aorta (J), but endothelial cells in y11 mutants do not form a proper epithelium (K). Anterior is to the left, dorsal up in all panels. Rostral is to the left and dorsal is up in panels A–I. Scale bars = 200 μm (A,B), 200 μm (C–E), 50 μm (F,G), 100 μm (H,I), 10 μm (J,K)
Molecular cloning of etsrp, the defective gene in y11 mutants
To identify the defective gene in y11 mutants, we performed meiotic mapping to localize y11 in the zebrafish genome. Initial bulked segregant mapping placed y11 on LG16, and fine genetic mapping using a panel of 1445 mutant embryos from a polymorphic mapping cross localized y11 to a 0.5 cM interval between markers z5298 and z6240 (Fig. 2A). Chromosome walks initiated from both markers refined this to a 270 Kb, 0.35 cM critical interval defined by 2 recombinants on the z5298 end and 3 recombinants on the z6240 end. This interval included the acsI, pycard, etsrp, fli1b, and fbl genes, as well as genes with homology to mammalian DYRK, ZPdomain, and GIRK2. The complete coding sequence of each of these genes was obtained from cDNA from y11 mutants and their wild type siblings. Comparison of wild type and mutant sequences for each of the genes showed that only one gene has a sequence alteration resulting in a change in the protein coding sequence in y11 mutants, the ETS-related gene etsrp. In y11 mutants a G is inserted after nucleotide 269 of the etsrp coding sequence, resulting in a +1 frame shift (Fig. 2B). The y11 mutant polypeptide goes out of frame after amino acid 90, continuing out-of-frame for an additional 62 amino acids before terminating. The predicted polypeptide lacks 276 amino acids or approximately three-fourths of the wild type etsrp protein, including the conserved ETS DNA binding domain shown to be essential for the function of other ETS genes (Fig. 2C). The etsrp gene is most closely related to the ets1 gene implicated in vascular and hematopoietic development in other species (Fig. 2D). We performed in situ hybridization to determine the expression pattern of etsrp and found that its expression is restricted to blood vessels and cells within them (see below), further supporting a function for this gene in vascular and hematopoietic development. To confirm that the defect in the etsrp gene is indeed responsible for the y11 mutant phenotype, we injected morpholino antisense oligonucleotides either targeting the translation initiation site or a combination of two morpholinos targeting the first plus second splice donor sites of the etsrp gene into wild type Tg(fli1:EGFP)y1 embryos. The translation intiation site (“ATG”) morpholino fully phenocopied the trunk circulation and intersegmental vessel sprouting defects of y11 mutants (Fig. 1D,E). The combined splice site morpholino-injected animals gave similar but less dramatic vascular defects than y11 mutants or the ATG morpholino (data not shown), so the ATG morpholino was used for all further analysis.
We examined whether wild type etsrp gene product could support endothelial differentiation. Injection of wild type etsrp mRNA into either y11 mutants or wild type animals at 100 pg doses resulted in early lethality during gastrulation. However, when Tg(fli1:EGFP)y1 or Tg(flk1:GFP) animals were injected at the 4–8 cell stage with lower doses (20 pg) of etsrp mRNA directed to only 1 or 2 cells, most embryos developed to 24 hpf and older with few abnormalities. Broad ectopic expression of EGFP-positive cells could be seen in most Tg(fli1:EGFP)y1 animals during mid-somitogenesis (Fig. 3B), with more limited ectopic expression of GFP noted in a smaller number of Tg(flk1:GFP) animals (Fig. 3D). Expression of the transgenes was greatly reduced or extinguished by 48 hpf, however, and no ectopic blood vessels were noted (data not shown). These results indicate that the etsrp gene product can initiate endothelial differentiation, as noted for overexpression of ETS factors in Xenopus laevis (Baltzinger et al., 1999; Remy et al., 1996).
Figure 3.
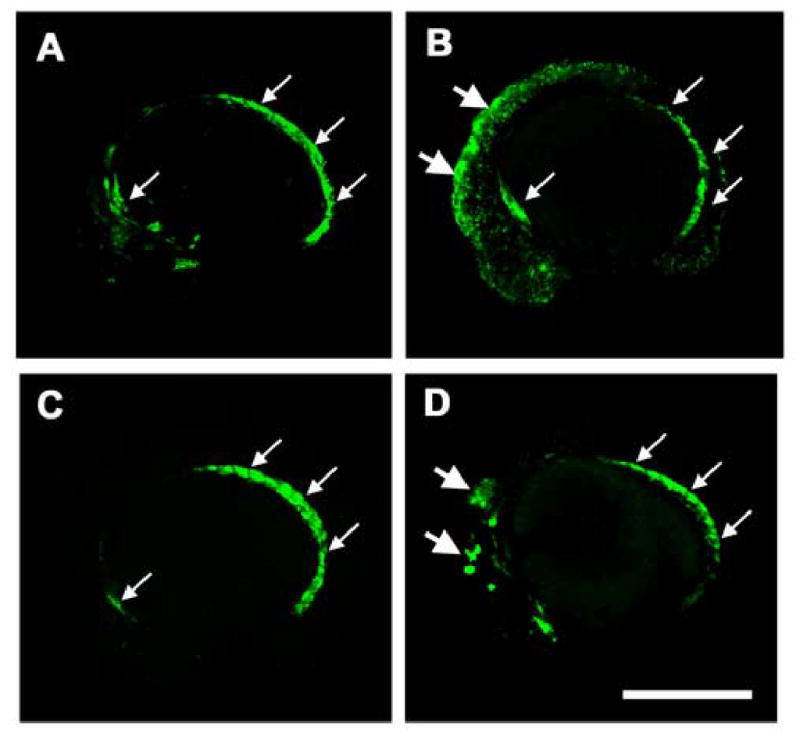
Induction of ectopic fli1-EGFP and flk1-GFP expression by etsrp mRNA injection. (A–D) Confocal images of Tg(fli1:EGFP)y1 (A,B) and Tg(flk1:GFP) (C,D) transgenic animals at approximately 14 somite stage/16 hours post-fertilization. (A,C) Uninjected control animals. (B,D) Animals injected at the 4–8 cell stage with 20 pg etsrp mRNA, targeting a limited number of blastomeres. Ectopic expression of the fli1-EGFP and flk1-GFP transgenes is noted by large arrows; small arrows show normal expression domains. Rostral is to the left and dorsal is up in all panels. Scale bar = 400 μm.
To test whether endothelial-specific expression of etsrp could rescue the y11 mutant phenotype we constructed a vector in which the fli1 promoter drives etsrp expression. We injected either fli1-etsrp DNA or control fli1-mRFP1 DNA into embryos from an incross of y11/+, Tg(fli1:EGFP)y1 animals, scoring for the presence or absence of intersegmental vessel sprouts at 24 hpf. As expected, 25% (8/33 in 2 separate experiments) of control fli1-mRFP1 injected animals lacked all intersegmental vessel sprouts at 24 hpf. In contrast, 100% (137/137 in 2 separate experiments) of fli1-etsrp injected animals had intersegmental vessel sprouts, although some spouts were missing or less well developed in some animals, as would be expected from the mosaic expression from injected DNA. This shows that wild-type etsrp expressed in endothelium can at least partially rescue the phenotype of y11 mutants. Together, all of these results indicate that the defective gene responsible for the y11 mutant phenotype is the ETS family member etsrp, and the mutation is thus designated etsrpy11.
Multiple ETS family members are expressed in the zebrafish vasculature
The functional role of ETS transcription factors in blood vessel formation has not been fully explored in mice and other species due to the expression of multiple ETS factors within the vasculature. We identified and obtained full-length cDNA sequences for four zebrafish ETS family members with vascular-restricted expression during early development- two Fli1 related genes, fli1 and fli1b, and two Ets1 related genes, ets1 and etsrp. Extensive database searches and analysis by in situ hybridization of a number of other ETS-related candidate genes failed to reveal additional genes with vascular expression (data not shown). The sequence similarity between these genes and related mammalian ETS family members is shown in Fig. 2D. All four genes begin to be expressed during early somitogenesis in the trunk lateral mesoderm (Fig. 4A–D). Expression of ETS genes in vascular progenitors precedes the expression of vascular markers such as vecdn1 (vecadherin) and flk1 (Fouquet et al., 1997; Larson et al., 2004; Liao et al., 1997), which are not detected at the 5 somite stage (data not shown), but are apparent by the 10 somite stage (Fig. 4I,J). The etsrp and fli1 genes both show robust expression at the 5 somite stage (Fig. 4A,C), although rostral expression of fli1 is not apparent until the 10 somite stage (Fig. 4G). At 24 hpf, all four ETS family genes are expressed in the vasculature (Fig. 4Q–T,W–Z), as is evident from comparison to the expression patterns of vascular markers flk1 and vecdn1 (Fig. 4U,V,A’,B’). All four ETS genes are expressed in both the vasculogenic axial vessels (dorsal aorta and posterior cardinal vein) and in the angiogenic intersegmental vessel sprouts (Fig. 4W–Z). The overlapping expression of four different related ETS genes in developing zebrafish blood vessels suggests that these genes might have overlapping roles in the vasculature.
Figure 4.
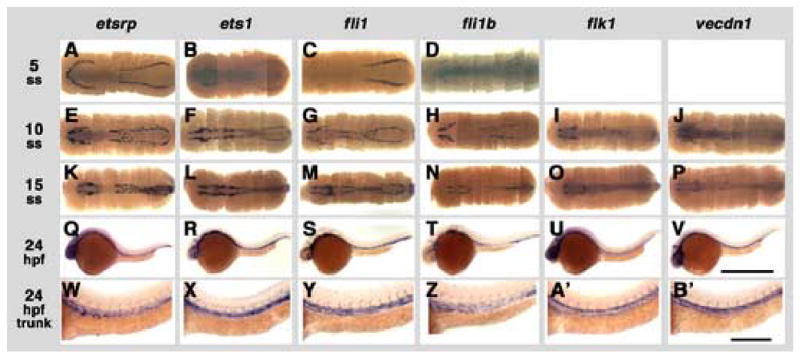
Expression of zebrafish ETS transcription factors during somitogenesis, with flk1 and vecdn1 shown for comparison. Whole mount in situ hybridization using probes for zebrafish etsrp (A, E, K, Q, W), ets1 (B, F, L, R, X), fli1 (C, G, M, S, Y), fli1b (D, H, N, T, Z), flk1 (I, O, U, A’), and ve-cdn (J, P, V, B’). Embryos are probed at the 5 somite stage (ss)/11.5 hours post-fertilization (hpf) (A–D), 10 ss/14 hpf (E–J), 15 ss/16.5 hpf (K–P), and 24 hours post-fertilization (Q-B’). Images in A–P are complete rostral to caudal collages of multiple dorsal-view images, for a “virtual flat mount.” Images in Q–V are lateral views of the entire 24 hpf animal, while W-B’ are higher magnification lateral views of 24 hpf trunks. Rostral is to the left in all panels, dorsal is up in Q-B’. Scale bars = 1000 μm (A–V), and 250 μm (W–B’).
etsrpy11 mutants have defects in vascular and hematopoietic gene expression
To further probe the nature of the vascular defects in etsrpy11 mutants, we compared the expression of a variety of vascular and hematopoietic genes in wild type and mutant embryos, including all four ETS family members. Whole mount in situ hybridization with fli1, fli1b, ets1, and etsrp showed no apparent difference in expression levels or pattern at the 10 somite stage (data not shown). However, expression of a number of endothelial-specific markers was already affected at this stage of development in etsrpy11. flk1, flt4 (Thompson et al., 1998), vecdn1, and plxnD1 (Torres-Vazquez et al., 2004) expression levels were all reduced at the 10 somite stage, the earliest time point examined (Fig. 5). By 24 hpf, trunk axial vessel expression of fli1b and etsrp was strongly reduced (Fig. 6B,D), fli1 appeared slightly reduced (Fig. 6A), while ets1 appeared unchanged (Fig. 6C). These results suggest that etsrp is not required for the initial expression of ETS genes within angioblast progenitors, but is required to maintain expression of both fli1b and its own transcript. Expression of the four endothelial marker genes was still substantially reduced, although clearly detectable, within the vasculature of 24 hpf animals (Fig. 6E–H). The arterial vascular marker efnb2 (Lawson et al., 2001) was also reduced in the dorsal aorta but not in non-vascular tissues at 24 hpf (Fig. 6I). Thus, etsrp appears to be required for proper expression of markers of differentiated endothelial cell fate.
Figure 5.
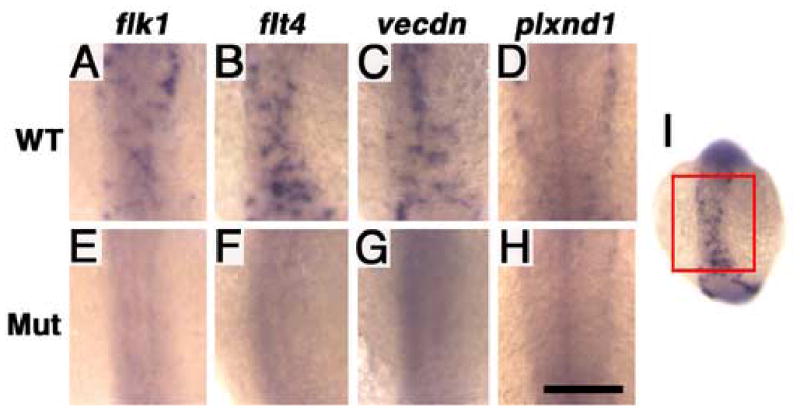
Expression of vascular marker genes in y11 mutants at 14 hpf (approximately 10 somite stage). Panels show whole mount in situ hybridization of 14 hpf wild type siblings (A–D) and y11 mutants (E–H) probed for flk1 (A,E), flt4 (B,F,I), vecdn (C,G), and plxnd1 (D,H). The developing trunk region imaged in panels A–H is noted by the box on the whole flt4-stained wild type embryo in panel I. Rostral is up in all panels. Scale bar = 200 μm (A–H).
Figure 6.
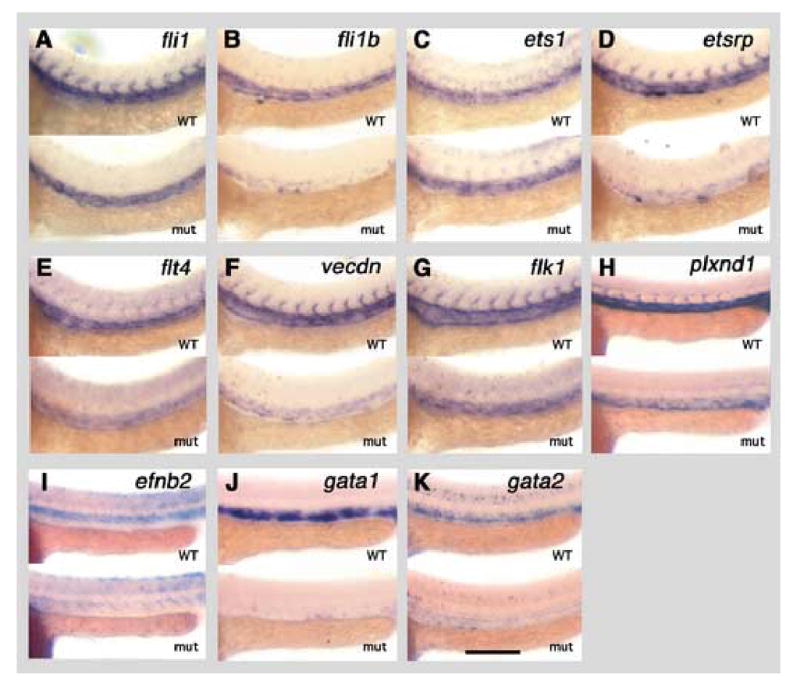
Expression of ETS transcription factors and vascular and hematopoietic marker genes in y11 mutants at 24 hpf (approximately 30 somite stage). Panels show whole mount in situ hybridization of the trunks of 24 hpf wild type siblings (top of each panel) and y11 mutants (bottom of each panel) probed for fli1 (A), fli1b (B), ets1 (C), etsrp (D), flt4 (E), vecdn (F), flk1 (G), plxnd1 (H), efnb2 (I), gata1 (J), and gata2 (K). Rostral is to the left and dorsal is up in all panels. Scale bar = 200 μm.
Since ETS genes play a role in hematopoietic development in other species (Bartel et al., 2000; Barton et al., 1998; Maroulakou and Bowe, 2000; Remy and Baltzinger, 2000; Spyropoulos et al., 2000), we also examined the expression of two markers of the hematopoietic lineage in the zebrafish, gata1 and gata2 (Detrich et al., 1995). The gata1 gene has been shown to be critical for erythroid development in zebrafish (Lyons et al., 2002) and other species. The gata2 gene is important for hematopoietic stem cell production and expansion in mice (Ling et al., 2004), and loss of zebrafish gata2 results in modest decreases in erythroid and myeloid marker expression (Galloway et al., 2005). Whole mount in situ hybridization at the 10 somite stage with gata1 and gata2 probes revealed no apparent difference in the pattern or level of expression of either gene between mutant and wild type animals (data not shown). By 24 hpf, however, gata1 and gata2 expression were strongly reduced (Fig. 6J,K). These results suggest that similarly to ets1 and etsrp expression, maintenance but not initial induction of hematopoietic gene expression requires etsrp function.
Functional overlap between ETS family members in the zebrafish vasculature
Since we identified four different ETS-related family members expressed within the vasculature, it seemed likely that these genes might show functional redundancy, and that the phenotype of loss of etsrp function in etsrpy11 mutants might reflect a partial loss of ETS transcription factor function within the vasculature. In order to probe the vascular role of ETS transcription factors within the vasculature more fully, we performed morpholino knockdown of different combinations of the four genes (Fig. 7). Translation initiation-or splice site-targeting morpholinos were designed for each of the four genes. The efficacy and specificity of each of these morpholinos for their target ETS factor was verified by in vitro translation assays (Supp. Fig. 1). These assays showed that each morpholino quantitatively reduced the levels of its targeted translation product without affecting the other three genes. Different combinations of the four oligos were injected into single cell embryos at three different dose levels (“low,” “medium,” and “high” - see Materials and Methods). At 24 and 48 hpf, animals were scored for the presence of trunk circulation and the number of primary intersegmental vessel sprouts that had emerged. These data are shown in Fig. 7. The high morpholino doses used were titrated to just below the dose level at which gross morphological defects and lethality began to occur. High-dose injection of morpholinos against different individual ETS factors led to phenotypes ranging from relatively mild for fli1 and fli1b (partial loss of trunk circulation at 24 hpf with full recovery of circulation in all animals by 48 hpf) to most severe for etsrp (complete loss of trunk circulation through 48 hpf and absence of nearly all intersegmental vessel sprouts at 24 hpf). However, with any of the single MO injections 100% of intersegmental vessel sprouts emerged by 48 hpf. The different morpholinos were also injected in different combinations at medium (1/2 of the high dose) and low (1/4 of the high dose) dose levels. Strong synergy was seen between the morpholinos in the circulation and sprouting defects they caused. When all four morpholinos were injected together at the low dose, the resulting intersegmental sprouting phenotype was more severe than that found for any single morpholino at the (four times higher) high dose. Experiments were also carried out in which morpholinos targeting three out of the four ETS genes were injected at the intermediate or low dose levels. In addition to confirming synergy between ETS factors, these injections revealed a similar ranking of the relative importance of these genes compared to what was found in single-MO injection experiments, that is, etsrp > ets1 > fli1 ≥ fli1b.
Figure 7.

Trunk circulation and intersegmental vessel sprouting defects in zebrafish injected with morpholinos targeting vascular ETS factors. Morpholinos targeting fli1, fli1b, ets1, or etsrp were injected into Tg(fli1:EGFP)y1 zebrafish either alone, in combinations of three morpholinos, or all together, as noted by “x” marks at the top and bottom of the figure. The number of animals for which phenotypes were assessed for each injection is shown at the extreme bottom of the Fig. (N=). The upper graph shows the % of animals with active blood circulation in the trunk at 24 hpf (purple bars) and 48 hpf (red bars). The middle graph shows the % of animals at 24 hpf with 1–15 (orange bars) or >15 (blue bars) intersegmental vessel sprouts in the trunk proper, as visualized by microscopic examination of fli1:EGFP transgene expression in developing blood vessels. The lower graph shows the % of animals at 48 hpf with 1–15 (orange bars) or >15 (blue bars) intersegmental vessel sprouts in the trunk proper. The graphs also include tabulation of circulation and intersegmental vessel sprout phenotypes measured in control morpholino injected animals, all of which had trunk circulation and a full complement of trunk intersegmental vessel sprouts.
The synergy between ETS factors can also be seen in their effects on vascular marker expression as visualized by whole mount in situ hybridization (Fig. 8). Co-injection of MOs targeting all four vascular ETS factors at “medium” dose levels resulted in very strong reduction in transcription of either the ETS factors themselves (Fig. 8A–D) or endothelial markers (Fig. 8E–I). The near-complete loss of many of these markers in animals injected with the four MOs (Fig. 8A–I) was more pronounced than the reduced levels of expression seen in etsrpy11 null mutants (Fig. 6A–I). Furthermore, unlike etsrpy11 mutants, a strong reduction in the number of endothelial cells was seen in 36 hpf four-morpholino injected Tg(fli1:nEGFP)y7 animals, suggesting that a significant part of the decrease in marker expression was due to fewer expressing cells (Fig. 8L,M;). These results, together with those in Fig. 7, suggest that the four ETS factors function combinatorially in endothelial specification and differentiation. Interestingly, a completely different result was observed for expression of hematopoietic markers gata1 and gata2. Both of these genes showed little or no reduction in their expression in animals injected with “medium” doses of MO against all four ETS factors (Fig. 8J,K), as compared to the reduced expression observed in etsrpy11 null mutants (Fig. 6J,K), suggesting that the hematopoietic lineage has a more specific requirement for etsrp function.
Figure 8.
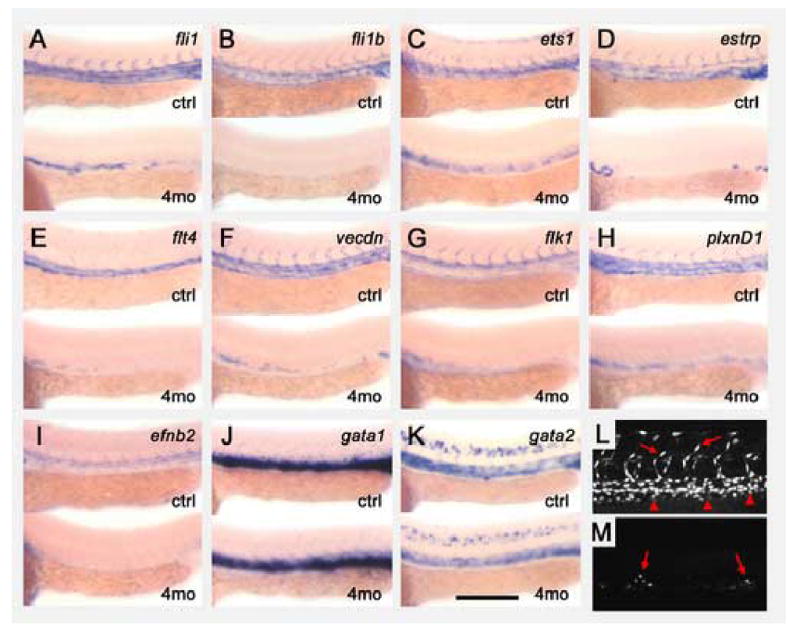
Reduction of all four vascular ETS transcription factors by morpholino knockdown. (A–K) Expression of ETS transcription factors and vascular and hematopoietic marker genes in animals injected with either a control morpholino (top of each panel) or a cocktail of four morpholinos targeting the fli1, fli1b, ets1, and etsrp genes (bottom of each panel). The four morpholino cocktail was injected at the “medium” dose, and an equivalent dosage of morpholino (ng) was injected in control animals. Panels show whole mount in situ hybridization of the trunks of 24 hpf morpholino-injected animals probed for zebrafish fli1 (A), fli1b (B), ets1 (C), etsrp (D), flt4 (E), vecdn (F), flk1 (G), plxnd1 (H), efnb2 (I), gata1 (J), and gata2 (K). (L,M) Confocal images of the mid-trunk of 36 hpf control- (L) and four morpholino cocktail- (M) injected Tg(fli1:nEGFP)y7 animals, collected using identical microscope settings. (L) Control animals display numerous brightly EGFP-positive endothelial nuclei in both axial (arrowheads) and intersegmental (arrows) vessels. (M) Only a small number of residual moderately EGFP-positive cells are seen in the position of the axial vessel (arrows) in four morpholino-injected animals. Anterior is to the left in all images. Scale bar = 250 μm.
DISCUSSION
We have taken advantage of the ease with which one can effect combined loss-of-function on multiple members of gene families in the zebrafish to examine the role of the ETS transcription factor family during vascular development. Gain-of-function studies in mice have suggested an important role for these factors in endothelial specification and/or differentiation, but loss-of-function studies of individual ETS factors have generally yielded less dramatic defects in vascular development, possibly because of extensively overlapping expression and functional overlap between a number of different ETS factors within the vasculature, including Ets1, Fli1, ERG, and TEL (reviewed in (Sato, 2001)). Reduction in levels of the ETS-domain protein etsrp has been reported to cause vascular defects in zebrafish (Sumanas and Lin, 2005).
As we show in this study, there are at least three additional ETS factors expressed within the zebrafish vasculature. We sought to determine whether these other factors also contribute to formation of the vasculature and what the consequences are of combined reduction in levels of multiple vascular ETS transcription factors. We show that the four vascular ETS factors function in a combinatorial fashion in the endothelium, by using morpholinos targeting etsrp and three other vascular ETS factors (ets1, fli1, and fli1b. Simultaneous reduction in levels of all four ETS factors by combined morpholino knock-down leads to a near-total loss of endothelial differentiation.
Morpholino injections targeting the four vascular ETS factors show that they are not equivalent, (Fig. 7), with a relative importance etsrp > ets1 > fli1 ≥ fli1b. Reduction of etsrp by morpholino injection causes the most severe defects in trunk circulation and intersegmental vessel sprouting, and etsrp is able to support some vascular differentiation on its own, in animals injected with morpholinos against the other three factors. In contrast, knock-down of fli1 or fli1b causes only mild vascular defects and they have little or no capacity to support vascular development on their own (i. e. when the other ETS factors are knocked down). These differences could reflect differential functional capabilities of the different ETS factors. Despite a high degree of sequence homology, some ETS factors do bind slightly different sites. In addition, ETS factors have been shown to be differentially regulated by post-translational modification and interaction with a large number of accessory factors. Some of the differences in vascular ETS factor requirement that we have observed could also reflect the relative timing or amount of expression of each of the genes. The etsrp gene is expressed strongly in both anterior and posterior expression domains at the 5 somite stage, unlike the other vascular ETS genes at this stage (Fig. 4). Finally, cross-regulation of the ETS factors could also account for differences in the relative requirement for each gene. Loss of etsrp in etsrpy11 mutants results in strong reduction in not only etsrp but also fli1b message levels at 24 hpf (Fig. 6). Despite the substantial quantitative differences in relative importance of the four ETS factors, there do not appear to be obvious qualitative differences in their morpholino vascular phenotypes, and for the strongest vascular phenotype reduction in multiple factors is required. In contrast, our data suggest that etsrp plays a more critical role in maintenance of hematopoietic development. Although gata1 and gata2 expression are normal in etsrpy11 mutants during early somitogenesis (14hpf/10 somites, data not shown), nearly complete loss of the expression of gata1 and strong reduction in gata2 is observed at 24 hpf (Fig. 6). In contrast, injection of morpholinos against all four vascular ETS factors at “medium” dose levels results in little or no loss of expression of these genes despite dramatic effects on vascular markers (Fig. 8). This presumably reflects residual etsrp protein present in animals injected with “medium” doses of the etsrp morpholino.
Form our molecular characterization it is clear that the etsrpy11 mutation represents a null mutant for etsrp, with an early stop codon predicted to terminate the translated peptide early in the protein, well before the essential ETS DNA binding domain. Morpholino knockdown results in a phenocopy of the mutant defect. An earlier report of etsrp morpholino knock-down described a somewhat more severe vascular phenotype, with loss of nearly all endothelial cells (as measured by flk1-GFP transgene expression) at the highest morpholino dose levels (Sumanas and Lin, 2005). The reasons for this difference are not clear, but it could partly reflect different vascular-specific transgenic lines used to visualize vessels (fli-EGFP in our study vs. flk-GFP in Sumanas et al.), differences in the developmental stages at which marker genes were assayed, or toxicity of high doses of injected morpholinos. In our hands, as noted above, a more complete defect in vascular development requires combined morpholino targeting of multiple ETS factors.
In conclusion, we have shown that ETS factors function combinatorially in the zebrafish vasculature and that the function of these factors is essential for endothelial specification and differentiation. These factors do not act equivalently, however, since the functional requirement for some of them is much greater than for others. Additional work will need to be done to further explore the differences in functional requirement for the different ETS factors, and determine whether any of these differences reflect differing functional capabilities of the proteins.
Supplementary Material
In vitro transcription-translation assays demonstrate that morpholinos efficiently knock down specific zebrafish vascular ETS factors. A combination of zebrafish ets1, etsrp, fli1a, and fli1b cDNAs were subjected to in vitro transcription-translation assays in the presence (or absence) of different morpholinos using the TNT Coupled Quick Trascription/Translation System (Promega). Lanes show: (1) Benchmark fluorescence size marker (sizes in Kd at left); (2) background control (transcription-translation mix only- no template); (3–8) in vitro transcription translation assays with all four templates plus (3) ets1 MO, (4) etsrp MO, (5) fli1a MO, (6) fli1b MO, (7) control MO, and (8) no MO added.
Acknowledgments
The authors would like to thank the members of the Weinstein lab for discussions and technical assistance. We also thank Dr. Igor B. Dawid for critical reading of this manuscript. NDL was supported by an NSF fellowship. This research was supported (in part) by the Intramural Research Program of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baltzinger M, Mager-Heckel AM, Remy P. Xl erg: expression pattern and overexpression during development plead for a role in endothelial cell differentiation. Dev Dyn. 1999;216:420–33. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<420::AID-DVDY10>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Bartel FO, Higuchi T, Spyropoulos DD. Mouse models in the study of the Ets family of transcription factors. Oncogene. 2000;19:6443–54. doi: 10.1038/sj.onc.1204038. [DOI] [PubMed] [Google Scholar]
- Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–63. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- Bories JC, Willerford DM, Grevin D, Davidson L, Camus A, Martin P, Stehelin D, Alt FW. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 1995;377:635–8. doi: 10.1038/377635a0. [DOI] [PubMed] [Google Scholar]
- Brown LA, Rodaway AR, Schilling TF, Jowett T, Ingham PW, Patient RK, Sharrocks AD. Insights into early vasculogenesis revealed by expression of the ETS-domain transcription factor Fli-1 in wild-type and mutant zebrafish embryos. Mech Dev. 2000;90:237–52. doi: 10.1016/s0925-4773(99)00256-7. [DOI] [PubMed] [Google Scholar]
- Detrich HW, 3rd, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, Pratt S, Ransom D, Zon LI. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci U S A. 1995;92:10713–7. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet B, Weinstein BM, Serluca FC, Fishman MC. Vessel patterning in the embryo of the zebrafish: guidance by notochord. Dev Biol. 1997;183:37–48. doi: 10.1006/dbio.1996.8495. [DOI] [PubMed] [Google Scholar]
- Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell. 2005;8:109–16. doi: 10.1016/j.devcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Hart A, Melet F, Grossfeld P, Chien K, Jones C, Tunnacliffe A, Favier R, Bernstein A. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity. 2000;13:167–77. doi: 10.1016/s1074-7613(00)00017-0. [DOI] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Larson JD, Wadman SA, Chen E, Kerley L, Clark KJ, Eide M, Lippert S, Nasevicius A, Ekker SC, Hackett PB, Essner JJ. Expression of VE-cadherin in zebrafish embryos: a new tool to evaluate vascular development. Dev Dyn. 2004;231:204–13. doi: 10.1002/dvdy.20102. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Scheer N, Pham VN, Kim CH, Chitnis AB, Campos-Ortega JA, Weinstein BM. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–83. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Vogel AM, Weinstein BM. sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev Cell. 2002;3:127–36. doi: 10.1016/s1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–18. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Lelievre E, Lionneton F, Soncin F, Vandenbunder B. The Ets family contains transcriptional activators and repressors involved in angiogenesis. Int J Biochem Cell Biol. 2001;33:391–407. doi: 10.1016/s1357-2725(01)00025-5. [DOI] [PubMed] [Google Scholar]
- Leprince D, Gegonne A, Coll J, de Taisne C, Schneeberger A, Lagrou C, Stehelin D. A putative second cell-derived oncogene of the avian leukaemia retrovirus E26. Nature. 1983;306:395–7. doi: 10.1038/306395a0. [DOI] [PubMed] [Google Scholar]
- Liao W, Bisgrove BW, Sawyer H, Hug B, Bell B, Peters K, Grunwald DJ, Stainier DY. The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development. 1997;124:381–9. doi: 10.1242/dev.124.2.381. [DOI] [PubMed] [Google Scholar]
- Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–82. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SE, Lawson ND, Lei L, Bennett PE, Weinstein BM, Liu PP. A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in vlad tepes. Proc Natl Acad Sci U S A. 2002;99:5454–9. doi: 10.1073/pnas.082695299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19:6432–42. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- Meyer D, Stiegler P, Hindelang C, Mager AM, Remy P. Whole-mount in situ hybridization reveals the expression of the Xl-Fli gene in several lineages of migrating cells in Xenopus embryos. Int J Dev Biol. 1995;39:909–19. [PubMed] [Google Scholar]
- Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377:639–42. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- Nakano T, Abe M, Tanaka K, Shineha R, Satomi S, Sato Y. Angiogenesis inhibition by transdominant mutant Ets-1. J Cell Physiol. 2000;184:255–62. doi: 10.1002/1097-4652(200008)184:2<255::AID-JCP14>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002;18:613–5. doi: 10.1016/s0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- Nunn MF, Seeburg PH, Moscovici C, Duesberg PH. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature. 1983;306:391–5. doi: 10.1038/306391a0. [DOI] [PubMed] [Google Scholar]
- Pardanaud L, Dieterlen-Lievre F. Expression of C-ETS1 in early chick embryo mesoderm: relationship to the hemangioblastic lineage. Cell Adhes Commun. 1993;1:151–60. doi: 10.3109/15419069309095691. [DOI] [PubMed] [Google Scholar]
- Parng C, Anderson N, Ton C, McGrath P. Zebrafish apoptosis assays for drug discovery. Methods Cell Biol. 2004;76:75–85. doi: 10.1016/s0091-679x(04)76005-7. [DOI] [PubMed] [Google Scholar]
- Queva C, Leprince D, Stehelin D, Vandenbunder B. p54c-ets-1 and p68c-ets-1, the two transcription factors encoded by the c-ets-1 locus, are differentially expressed during the development of the chick embryo. Oncogene. 1993;8:2511–20. [PubMed] [Google Scholar]
- Remy P, Baltzinger M. The Ets-transcription factor family in embryonic development: lessons from the amphibian and bird. Oncogene. 2000;19:6417–31. doi: 10.1038/sj.onc.1204044. [DOI] [PubMed] [Google Scholar]
- Remy P, Senan F, Meyer D, Mager AM, Hindelang C. Overexpression of the Xenopus Xl-fli gene during early embryogenesis leads to anomalies in head and heart development and erythroid differentiation. Int J Dev Biol. 1996;40:577–89. [PubMed] [Google Scholar]
- Roman BL, Pham VN, Lawson ND, Kulik M, Childs S, Lekven AC, Garrity DM, Moon RT, Fishman MC, Lechleider RJ, Weinstein BM. Disruption of acvrl1 increases endothelial cell number in zebrafish cranial vessels. Development. 2002;129:3009–19. doi: 10.1242/dev.129.12.3009. [DOI] [PubMed] [Google Scholar]
- Sato Y. Role of ETS family transcription factors in vascular development and angiogenesis. Cell Struct Funct. 2001;26:19–24. doi: 10.1247/csf.26.19. [DOI] [PubMed] [Google Scholar]
- Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, Watson DK. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol. 2000;20:5643–52. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanas S, Lin S. Ets1-Related Protein Is a Key Regulator of Vasculogenesis in Zebrafish. PLoS Biol. 2005;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MA, Ransom DG, Pratt SJ, MacLennan H, Kieran MW, Detrich HW, 3rd, Vail B, Huber TL, Paw B, Brownlie AJ, Oates AC, Fritz A, Gates MA, Amores A, Bahary N, Talbot WS, Her H, Beier DR, Postlethwait JH, Zon LI. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248–69. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- Torres-Vazquez J, Gitler AD, Fraser SD, Berk JD, Van NP, Fishman MC, Childs S, Epstein JA, Weinstein BM. Semaphorin-plexin signaling guides patterning of the developing vasculature. Dev Cell. 2004;7:117–23. doi: 10.1016/j.devcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Verger A, Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–70. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- Wang LC, Kuo F, Fujiwara Y, Gilliland DG, Golub TR, Orkin SH. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. Embo J. 1997;16:4374–83. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernert N, Stanjek A, Kiriakidis S, Hugel A, Jha HC, Mazitschek R, Giannis A. Inhibition of Angiogenesis In Vivo by ets-1 Antisense Oligonucleotides-Inhibition of Ets-1 Transcription Factor Expression by the Antibiotic Fumagillin. Angew Chem Int Ed Engl. 1999;38:3228–3231. doi: 10.1002/(sici)1521-3773(19991102)38:21<3228::aid-anie3228>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. University of Oregon Press; Eugene: 1995. [Google Scholar]
- Zhang L, Eddy A, Teng YT, Fritzler M, Kluppel M, Melet F, Bernstein A. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Mol Cell Biol. 1995;15:6961–70. doi: 10.1128/mcb.15.12.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vitro transcription-translation assays demonstrate that morpholinos efficiently knock down specific zebrafish vascular ETS factors. A combination of zebrafish ets1, etsrp, fli1a, and fli1b cDNAs were subjected to in vitro transcription-translation assays in the presence (or absence) of different morpholinos using the TNT Coupled Quick Trascription/Translation System (Promega). Lanes show: (1) Benchmark fluorescence size marker (sizes in Kd at left); (2) background control (transcription-translation mix only- no template); (3–8) in vitro transcription translation assays with all four templates plus (3) ets1 MO, (4) etsrp MO, (5) fli1a MO, (6) fli1b MO, (7) control MO, and (8) no MO added.


