Abstract
The prohibitin (PHB)-domain proteins are membrane proteins that regulate a variety of biological activities, including mechanosensation, osmotic homeostasis, and cell signaling, although the mechanism of this regulation is unknown. We have studied two members of this large protein family, MEC-2, which is needed for touch sensitivity in Caenorhabditis elegans, and Podocin, a protein involved in the function of the filtration barrier in the mammalian kidney, and find that both proteins bind cholesterol. This binding requires the PHB domain (including palmitoylation sites within it) and part of the N-terminally adjacent hydrophobic domain that attaches the proteins to the inner leaflet of the plasma membrane. By binding to MEC-2 and Podocin, cholesterol associates with ion-channel complexes to which these proteins bind: DEG/ENaC channels for MEC-2 and TRPC channels for Podocin. Both the MEC-2-dependent activation of mechanosensation and the Podocin-dependent activation of TRPC channels require cholesterol. Thus, MEC-2, Podocin, and probably many other PHB-domain proteins by binding to themselves, cholesterol, and target proteins regulate the formation and function of large protein–cholesterol supercomplexes in the plasma membrane.
Keywords: prohibitin-domain proteins, TRP channels, DEG/ENaC channels, slit diaphragm, mechanosensation
The prohibitin homology (PHB)-domain proteins constitute a family of ≈1,800 proteins (SMART database; http://smart.embl-heidelberg.de) (1) all of which share an ≈150-aa domain similar to that in the mitochondrial protein prohibitin (2). More than 340 of these proteins, many of which have an N-terminal adjacent hydrophobic region that places them on the inner leaflet of the lipid bilayer, have been identified in animals. These membrane-associated proteins regulate osmotic homeostasis, mechanosensation, and cell signaling (3–5). Several of the animal PHB-domain proteins including flotillin, Podocin, prohibitin, stomatin, UNC-1, UNC-24, and the UNC-24-like mammalian protein SLP-1 are found in cholesterol-rich membrane fractions derived from the plasma membrane (reviewed in ref. 2).
In this article, we investigate the function of these proteins using two members of the family, MEC-2 from Caenorhabditis elegans and Podocin from mouse. MEC-2 (6) and Podocin (7) have a single, central hydrophobic domain that embeds these proteins in the inner leaflet of the plasma membrane with their N- and C-terminal tails facing the cytoplasm (Fig. 1a). Although the two proteins contain different N and C termini, they have hydrophobic regions that are 35% identical and 75% similar and PHB-domains that are 50% identical and 80% similar (Fig. 1b). The PHB domain is critical for the action of both proteins (8, 9). The role of these conserved domains and the function of PHB-domain proteins, however, are unclear.
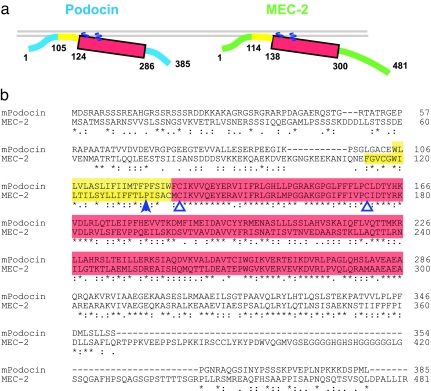
Fig. 1.
Structure and sequence of Podocin and MEC-2. (a) Membrane orientation of Podocin and MEC-2. The hydrophobic region (yellow) inserts into the inner leaflet of the plasma membrane, causing the remaining parts of the protein, including the PHB domain (red) to face the cytoplasm and the inner leaflet. Sites of palmitate attachment are indicated by blue wavy lines. (b) Alignment of mouse Podocin and C. elegans MEC-2, showing the hydrophobic region (yellow), PHB domain (red), palmitoylation sites (open blue triangles), and the site of the Pro-to-Ser mutation in the hydrophobic region that prevents cholesterol binding (filled blue triangle).
MEC-2 is part of a multiprotein-channel complex with the degenerin/epithelial Na+ channel (DEG/ENaC) proteins MEC-4 and MEC-10 that transduces gentle touch (6, 10, 11). In touch-receptor neurons, this channel complex is localized to regular puncta along the neuronal process; MEC-2 has been shown to regulate the MEC-4/MEC-10 ion channel in these puncta (9). However, the mechanism by which MEC-2 regulates ion-channel activity is elusive.
Podocin localizes specifically to the slit diaphragm of the mammalian kidney. The slit-diaphragm is a specialized intercellular junction that connects adjacent foot processes of kidney podocytes and is part of the glomerular-filtration barrier (12). Slit diaphragm proteins induce signal transduction in podocytes, the visceral epithelial cells of the kidney glomerulus, that regulates cytoskeletal rearrangement and transcriptional activity (3). Like MEC-2, Podocin is part of a multiprotein complex containing the transmembrane proteins Neph1, Neph2, and Nephrin and the cytoplasmic adaptor protein CD2AP (3). Mutations in Podocin and the other proteins of this complex result in focal segmental glomerulosclerosis and congenital nephrotic syndrome, severe genetic kidney disorders in humans characterized by disruption of the filtration barrier (7, 13, 14). Recently, mutations in the transient receptor potential C channel protein TRPC6 were also shown to cause focal segmental glomerulosclerosis (15, 16).
In this article, we show that Podocin and MEC-2 are cholesterol-binding proteins and that cholesterol binding plays an important role in regulating the activity of ion channels to which these PHB-domain proteins bind. Podocin, as we show here, binds to, colocalizes at the slit diaphragm with, and regulates the activity of TRPC6. Our results suggest that these proteins, similar to the proteins associated with MEC-2, may be part of a mechanosensitive protein complex at the slit diaphragm of podocytes. In general, we propose that many of the PHB-domain proteins regulate membrane protein function by binding sterols, perhaps by altering their local lipid environment.
Results
Podocin and MEC-2 Are Cholesterol-Binding Proteins.
Podocin (8), MEC-2 (T.B.H., B.S., R.U.M., and T.B., unpublished data), and several other PHB-domain proteins are found in cholesterol-rich membrane fractions. To test whether Podocin and MEC-2 bind to cholesterol, we expressed both proteins in HEK293T cells and tested for binding of photoactivatable lipids (Fig. 2a). These derivatives attach to associated molecules when they are stimulated by UV light (17). Podocin and MEC-2 bound cholesterol but not phosphatidylcholine (Fig. 2a). Cholesterol binding required the PHB domain, because Podocin lacking this domain (PodocinΔPHB) did not label. Binding to membrane proteins was quite specific, for example, the Ig superfamily member and Podocin-interacting protein Nephrin was not labeled (Fig. 2b). These data were confirmed by using digitonin precipitation (Fig. 7, which is published as supporting information on the PNAS web site).
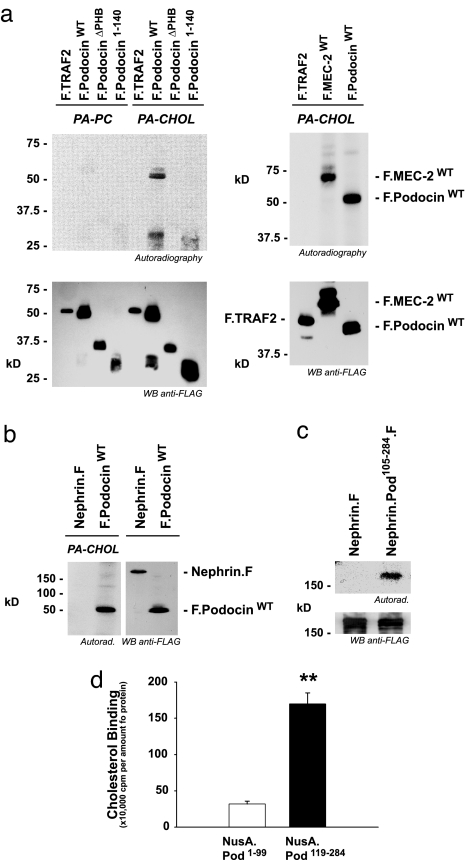
Fig. 2.
Podocin and MEC-2 bind cholesterol. (a) MEC-2 and Podocin expressed in HEK293T cells are labeled with photoactivatable [3H]photocholesterol (PA-CHOL) but not with photoactivatable [3H]phosphatidylcholine (PA-PC) (incubation time, 16 h). (Upper) Photolabeled proteins were resolved by SDS/PAGE and visualized by autoradiography. (Lower) Expression of proteins in the lysates. (b) The Podocin-interacting membrane protein Nephrin does not bind photoactivatable cholesterol. (c) A fusion protein with a C-terminal fusion of the PHB domain and the hydrophobic region of Podocin (amino acids 105–284) with the extracellular and transmembrane domains of Nephrin binds photoactivatable cholesterol, indicating that this domain can convey cholesterol binding. (d) Direct binding of cholesterol to Podocin; in vitro binding of cholesterol. NusA fused to the N-terminal domain of Podocin (amino acids 1–99) or NusA fused to the cholesterol-binding domain of Podocin (amino acids 119–284) was incubated with radioactively labeled cholesterol, washed extensively, and subjected to scintillation counting.
Although a detailed description of the cholesterol-binding sites on Podocin and MEC-2 is outside the scope of this article, we tested the importance of different regions of Podocin for cholesterol binding by fusing them to the extracellular and transmembrane domains of Nephrin. A fusion containing the PHB domain bound cholesterol (data not shown), but more efficient cholesterol binding was observed when the PHB domain and the adjacent N-terminal hydrophobic domain were included (Fig. 2c). To ensure that cholesterol labeling was the result of direct binding and did not occur through passive stochastic attachment of cholesterol at the cell membrane, we produced fragments of Podocin in Escherichia coli and tested their ability to bind [3H]cholesterol in vitro. The PHB domain was sufficient for cholesterol binding, but binding was more efficient when the polypeptide included the PHB domain and the N-terminal adjacent hydrophobic sequence (Fig. 2d). Binding could be competed with excess cold cholesterol (Fig. 8, which is published as supporting information on the PNAS web site).
We had previously shown that Podocin homooligomerizes and forms high-molecular-weight complexes by homophilic interactions that require the PHB domain (8). MEC-2 also homooligomerizes (see below) as do several other PHB-domain proteins (18). The size of the complexes suggested that they contain at least 20–50 molecules (Fig. 9a, which is published as supporting information on the PNAS web site). Multimerization, however, does not require cholesterol binding. Limited cholesterol depletion with methyl-β-cyclodextrine (MBCD) of Podocin-expressing cells did not interfere with the formation of high-molecular-weight complexes (Fig. 9b). Thus, Podocin and MEC-2 bind cholesterol, themselves, and other proteins.
Touch Sensitivity Requires Sterol Binding by MEC-2 in C. elegans.
To test the in vivo importance of cholesterol binding, we made use of the requirement for MEC-2 in C. elegans touch sensitivity. Twenty-three mec-2 alleles causing touch insensitivity in C. elegans have missense mutations (9), and we screened most of the resulting proteins for their ability to bind cholesterol, localize to the membrane, multimerize, and interact with associated channels. Cholesterol binding was absent in some mutants and reduced in many others (data not shown). As an example, we have studied the protein MEC-2(P134S) that is produced by the u274 allele. This mutation substitutes a Ser for Pro in the hydrophobic region preceding the PHB domain (Fig. 1b). Worms expressing the mutant allele are completely touch insensitive (2 of 50 animals responded once to five touches). MEC-2(P134S) was not labeled with photoactivatable cholesterol (Fig. 3a), even though it localized to the plasma membrane (data not shown), multimerized (Fig. 3b), and interacted with the MEC-4-related channel αENaC (Fig. 3c). These data suggest that loss of touch sensitivity results from the loss of cholesterol binding of this protein. Furthermore, these data are consistent with a role for MEC-2 in recruiting or maintaining cholesterol in the multiprotein MEC-4 channel complex in vivo, although we cannot exclude that previously localized cholesterol could assist in the association of MEC-2 with the complex.
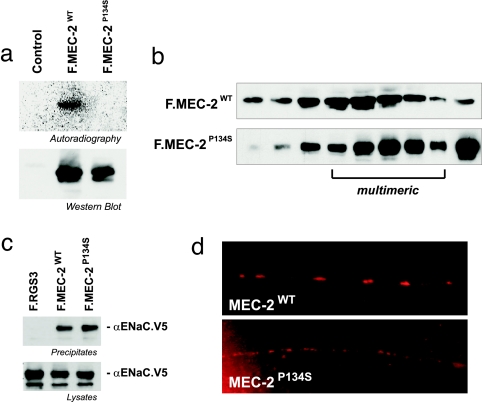
Fig. 3.
MEC-2(P134S) does not bind cholesterol but can bind DEG/ENaC channels and multimerize. (a) Photoaffinity cholesterol labels FLAG-tagged wild-type MEC-2 but not MEC-2(P134S). (Upper) Photolabeled proteins were immunoprecipitated with anti-FLAG antibody, resolved by SDS/PAGE, and visualized by autoradiography. (Lower) The expression of proteins in the lysates. (b) Velocity-gradient centrifugation shows that both wild type and MEC-2(P134S) multimerize. (c) Wild-type MEC-2 and MEC-2(P134S) equivalently coimmunoprecipitate V5-tagged rat αENaC from HEK293T cells (the second transmembrane domain of αENaC can substitute for that of MEC-4 in vivo) (33). The rat channel protein was used because we were unable to express MEC-4 to sufficient levels. This experiment demonstrates that MEC-2 can bind to other DEG/ENaC proteins and that the mutant binding does not depend on cholesterol. (d) Localization of wild-type and mutant MEC-2 in processes of touch-receptor neurons in C. elegans. Both proteins are found in the process (suggesting that both localize to the plasma membrane), but the P134S protein is not found in the characteristic puncta formed by the mechanosensory-channel complex. Proteins were visualized in C. elegans with a MEC-2-specific antibody (9).
To investigate whether the mutant protein localizes correctly in touch-channel puncta of touch neurons in the nematode, we stained wild-type and mutant animals with antibodies directed against MEC-2. The u274 mutation did not prevent the localization of the mutant protein to the plasma membrane (Fig. 3d). The distribution of MEC-2(U247) within the plasma membrane, however, was not wild type, because the protein was not found in puncta (0 of 50 animals) but was more uniformly distributed. These results are consistent with a role for cholesterol binding in the formation of higher-order structures at the independently localized MEC-4 puncta in vivo.
If cholesterol, or a cholesterol derivative (19), is needed for channel function, cholesterol-deprived worms should be touch insensitive. However, when wild-type C. elegans larvae were transferred from normal (13 μM) cholesterol to cholesterol-depleted plates, they produced F1 progeny that arrested as young larvae and that were touch sensitive. Presumably, these arrested larvae were not completely depleted of cholesterol, having, as we show below, sufficient cholesterol for touch sensitivity but not enough for further development.
We demonstrated the need of cholesterol in C. elegans touch sensitivity in two ways. First, we generated a version of MEC-2 with reduced cholesterol binding by mutating its predicted palmitoylation sites. Substitution of Ala for Cys at amino acids 140 and 174 resulted in the loss of palmitoylation (Fig. 4a) and a reduction of cholesterol binding (Fig. 4b). These alterations did not affect overall protein levels, multimerization, or localization to the plasma membrane (data not shown), but mec-2-null worms expressing the mec-2(C140/174A) gene conditionally depended on cholesterol for touch sensitivity. These animals showed virtually the same touch sensitivity as wild-type animals on plates with normal amounts of cholesterol but reduced touch sensitivity when grown on cholesterol-free plates (Fig. 4 c and d). This defect depended on the cholesterol concentration and could be rescued by substituting lathosterol, ergosterol, and 7-dehydro cholesterol for cholesterol in the growth medium (Fig. 4c). Because touch sensitivity of wild-type animals was not affected by this limited cholesterol depletion, the effects we see with the palmitoylation mutant cannot be attributed to indirect effects on neuronal growth or development.
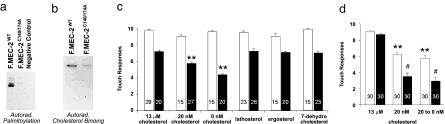
Fig. 4.
Cholesterol dependence of touch sensitivity in C. elegans. Substitution of Ala for Cys in two predicted palmitoylation sites results in the loss of palmitoylation (a) and a reduction of cholesterol binding (b). HEK293T cells were transfected with wild-type MEC-2 or MEC-2(C140/174A) and labeled with [3H]palmitic acid (a) or [3H]photoaffinity cholesterol (b). Equal expression of proteins in the lysates was confirmed on Western blots (data not shown). (c) Touch sensitivity in MEC-2(C140/174A) mutants (black bars) requires cholesterol or its derivatives. Responses of wild-type animals (white bars) are also shown and are not affected by limited cholesterol depletion. mec-2-null worms were transformed with the mec-2(C140/174A) gene and grown on plates with defined sterol concentrations before analysis of touch sensitivity. Depicted is the mean ± SEM (number of animals tested is indicated. ∗∗, P < 0.001 as compared with mec-2(C140/174A) at high cholesterol). (d) Severe cholesterol depletion lowers the sensitivity of wild-type animals (white bars) and mec-2(C140/174A) mutants (black bars). Worms grown on plates containing 20 nM cholesterol for three generations were either maintained on 20 nM cholesterol for another generation or placed on cholesterol-free plates before analysis of response to gentle touch (number of animals tested is indicated; ∗∗, P < 0.001 as compared with wild-type worms on 13 μM cholesterol; #, P < 0.001 as compared with mutants on 13 μM cholesterol).
The second demonstration of cholesterol dependence was seen when we lowered cholesterol levels further by transferring animals grown on minimal (20 nM) cholesterol plates for three generations (we find that the animals arrest their development after approximately four generations) to 0 nM cholesterol plates. Larvae placed on minimal or zero cholesterol never became adults, but arrested in their development. The animals were noticeably more debilitated (many could not move) under these conditions. Nonetheless, wild-type animals that showed normal movement had become relatively insensitive to touch, and animals with the MEC-2 palmitoylation mutations were even less sensitive to touch (Fig. 4d). These data show that touch sensitivity depends on sterols in vivo and suggest that sterols recruited to the MEC-4 channel complex by MEC-2 are needed for its function.
Podocin-Mediated Regulation of TRPC Channel Activity.
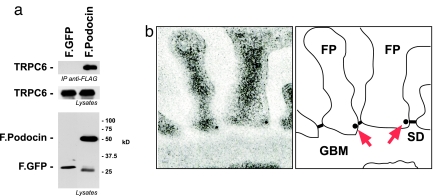
We have found that Podocin, like MEC-2, is associated with ion-channel subunits at the glomerular-slit diaphragm of the kidney. As described above, mutations in the genes encoding Podocin and TRPC6 cause disruption of the kidney filter and focal segmental glomerulosclerosis (15, 16). To test whether these proteins may functionally interact, we first coexpressed tagged versions of the proteins in HEK293T cells and tested for coimmunoprecipitation. Podocin coprecipitated with TRPC6, whereas a control protein did not (Fig. 5a). Similar to MEC-2, which does not influence targeting of the DEG/ENaC ion-channel complex (9), Podocin did not affect TRPC6 localization to the plasma membrane (data not shown). Podocytes express TRPC6 as well as several related TRPC channels (TRPC1, 3, and 4) (Fig. 10a, which is published as supporting information on the PNAS web site). TRPC channels are thought to be heteromeric (20), so it was no surprise that Podocin coprecipitated with these other TRPC channels but not with a control protein (Fig. 10b). Consistent with another study (15), immunofluorescence staining of rat kidney sections confirmed expression of TRPC6 in glomerular podocytes (data not shown). Using immunogold electron microscopy, we localized TRPC6 to the insertion site of the glomerular-slit diaphragm (Fig. 5b), the structure that expresses Podocin (12). Although TRPC6 could be detected in various compartments of the podocyte, immunoreactivity in the secondary processes of the podocyte was clearly confined to the insertion site of the slit diaphragm. Thus, Podocin colocalizes with TRPC6 in vivo.
Fig. 5.
Podocin interacts and colocalizes with TRPC6. (a) Mouse TRPC6 coimmunoprecipitates with FLAG-tagged Podocin (F.Podocin) but not with a control protein (F.GFP). (Top) Coprecipitated TRPC6 channel after immunoprecipitation of Podocin or GFP. (Middle and Bottom) Expression of the proteins in the lysates. (b) TRPC6 is located at the slit diaphragm (SD) of podocyte foot processes (FP) near the glomerular basement membrane (GBM). This localization mimics that of Podocin. Rat kidneys were perfused with ice-cold PBS, fixed in situ, and subjected to immunogold electron microscopy. Arrows indicate the localization of gold particles in the electron micrograph.
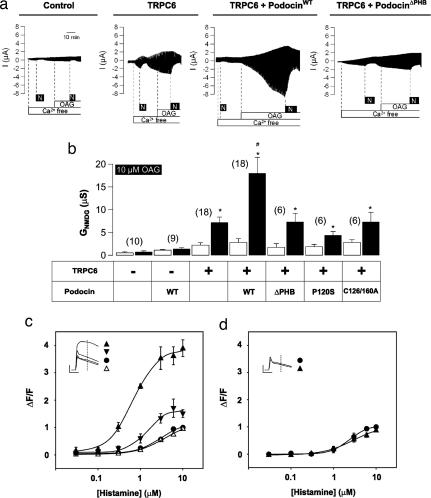
We next tested whether Podocin affected TRPC6 channel activity by examining TRPC6 currents in Xenopus laevis oocytes with and without Podocin. Expression of TRPC6 induced an inward Na+ current in a Ca2+-free bath solution that was further augmented by stimulation with the membrane-permeable diacylglycerol homologue 1-oleoyl-2-acetyl-sn-glycerol (OAG)(Fig. 6a). This increase required the TRPC6 channel; it was not seen in water-injected oocytes. The OAG-induced currents were significantly augmented in oocytes coexpressing TRPC6 and Podocin, but were not increased in oocytes coexpressing TRPC6 and PodocinΔPHB (Fig. 6a). These data show that Podocin interacts with TRPC6 to regulate TRPC6 activity.
Fig. 6.
Activation of TRPC-channel activity by Podocin. (a) Podocin, but not PodocinΔPHB, enhances TRPC6 currents in Xenopus oocytes stimulated with 10 μM OAG. Expression of TRPC6 induces an inward Na+ current in a Ca2+-free bath solution that is further augmented by stimulation with OAG. The OAG-induced currents were significantly augmented in oocytes coexpressing TRPC6 and Podocin but were not increased in oocytes coexpressing TRPC6 and PodocinΔPHB. (b) Podocin increases the effect of OAG (10 μM 1-oleoyl-2-acetyl-sn-glycerol; black bars) on NMDG-sensitive conductance (GNMDG) of TRPC6 channels in Xenopus oocytes, but mutant Podocins do not. Currents in control oocytes (white bars) were not affected. The Podocin mutants used were PodocinΔPHB, PodocinP120S, and PodocinC126/160A (see Fig. 1). The number of oocytes examined is given in parentheses. ∗, P < 0.05 as compared with water-injected oocytes; #, P < 0.05 as compared with TRPC6 coexpressed with PodocinΔPHB. (c) Wild-type, but not mutant, Podocin increases histamine-induced calcium influx (measured as a change in fluorescence, ΔF/F) in HeLa Cx43 cells. Cells were transiently mock transfected (filled circles) or transfected with DNA coding for wild-type Podocin (filled triangles), PodocinΔPHB (open triangles), and PodocinC126/160A (inverted filled triangles) and measured simultaneously in the same experiment by using a FLIPR. Data are means ± SD from three to five independent experiments. Measurements are taken at the shoulder of the response (line in the inset, which shows the calcium responses of the cells challenged with 10 μM histamine). Vertical scale, 104 arbitrary fluorescence units; horizontal scale, 2 min. (d) The stimulation by Podocin is abolished when cells are treated with MBCD to deplete cholesterol.
To test whether the Podocin-mediated activation of TRPC6 also involved cholesterol binding, we coexpressed mutant Podocin defective in cholesterol binding with TRPC6 in oocytes. We quantified the effect of Podocin on the TRPC6 channel currents by replacing Na+ in the extracellular bath solution with impermeable n-methyl- d-glucamine (NMDG) and calculating the NMDG-sensitive conductance (Fig. 6b, GNMDG). Mutation of the proline residue (PodocinP120S) equivalent to MEC-2(P134S) or of the palmitoylation sites (PodocinC126/160A) both resulted in the loss of the OAG-stimulated currents. PodocinP120S did not bind cholesterol, and PodocinC126/160A showed weak cholesterol-binding activity, but both interacted with TRPC6 (not shown).
These data suggest that the regulation of TRPC6 by Podocin requires cholesterol binding. We would like to have shown that cholesterol depletion of oocytes abrogated the stimulatory activity of Podocin on TRPC6 currents. However, we cannot efficiently remove cholesterol from oocytes. Therefore, we looked for effects of Podocin on the histamine-stimulated and TRPC channel-dependent increase of calcium in HeLa cells (21), which allow efficient cholesterol depletion (Fig. 6 c and d). Expression of Podocin resulted in a strong increase of the maximal effect to histamine stimulation on transmembrane Ca2+ influx (Fig. 6c). This increase was not found in cells expressing PodocinΔPHB and was strongly attenuated in cells expressing Podocin with mutated palmitoylation sites (Fig. 6c). The weaker effect of the palmitoylation site mutations mirrors that seen with the similar MEC-2 mutant in C. elegans. Consistent with a critical role for Podocin in binding and recruiting cholesterol, limited cholesterol depletion with MBCD abolished the Podocin-dependent stimulation of Ca2+ influx (Fig. 6d). Although treatment of cells with MBCD may have a variety of effects, these data, together with the oocyte experiments, suggest that Podocin-mediated cholesterol recruitment is essential for modulating TRPC-channel function.
Discussion
These data demonstrate that MEC-2 and Podocin bind cholesterol and that this binding regulates ion-channel complexes. Efficient cholesterol binding requires not only the PHB domains but also part of the adjacent hydrophobic domain and covalently attached palmitate chains. Further delineation of the cholesterol-binding domain remains to be elucidated.
These proteins have multiple functions; in addition to binding cholesterol, they multimerize and bind to specific protein targets. Because several PHB-domain proteins have similar hydrophobic and PHB domains to those in Podocin and MEC-2, they may also bind sterols. The binding of cholesterol to Podocin, MEC-2, and similar proteins, especially given the large multimers that these proteins form, could alter the local lipid environment of channel proteins and other targets. For example, MEC-2 localizes to the MEC-4 mechanosensory complex; it is not needed to form the channel complex. In this instance, the recruitment of MEC-2 might change or stabilize the cholesterol content surrounding the complex. The conventional view of cholesterol's function is that it changes the properties of the bilayer, usually by stiffening and/or widening the membrane. The alteration of the lipid environment of the target proteins by the binding of PHB-domain proteins could, thus, alter their structure and function.
Cholesterol and the C. elegans Mechanosensory Complex.
The C. elegans mechanotransduction complex that contains MEC-2 has four other proteins: the DEG/ENaC proteins MEC-4 and MEC-10, which are thought to form the pore of the channel, the paraoxonase-like protein MEC-6, and UNC-24 (9–11, 22). UNC-24 and MEC-6 may also affect the binding or metabolism of lipids associated with the MEC-4/MEC-10 channel. UNC-24 is also a PHB-domain protein, but, unlike MEC-2, it has an additional domain (SCP2, sterol-carrier protein domain 2) that is similar to regions of nonspecific lipid-transport proteins (23). Vertebrates have similar two-domain proteins (e.g., SLP-1) (24). Nonspecific lipid-transport proteins serve as intracellular carriers of cholesterol and other sterols, so the association of a similar domain with a cholesterol-binding PHB domain is suggestive that the two domains could be needed to shuttle cholesterol and other sterols into the plasma membrane. Interestingly, both UNC-24 and SLP-1 are highly enriched in nervous tissue (9, 25).
In contrast to MEC-2 and UNC-24, MEC-6 has a single membrane-spanning domain that puts most of the protein on the extracellular side of the membrane (22). The similarity of MEC-6 to paraoxonases may indicate that it, too, affects the cholesterol content of the membrane, albeit at the outer leaflet of the bilayer, because two of the three vertebrate paraoxonases are secreted and associated with cholesterol-containing high-density lipoprotein particles (the third paraoxonase, PON-2 is, like MEC-6, a widely expressed membrane protein) (26). We speculate that MEC-6, and by analogy, PON-2, may modify or maintain associated lipids on the external side of the lipid bilayer.
Podocin, TRPC6, and Mechanosensation at the Kidney Filtration Barrier.
We show here that Podocin interacts with and regulates the activity of TRPC6. Mutation of TRPC6, like Podocin, causes hereditary nephrotic syndrome in humans (15, 16). Although the TRPC6-associated disease displays a later onset of kidney failure and milder disease than the Podocin disease, the similarity of the defects supports our concept that Podocin modulates TRPC6 function. We have shown that podocytes express several members of the TRPC family and that these bind Podocin. Moreover, various TRPC proteins interact and form heteromultimeric complexes (20). These observations lead us to expect some functional redundancy with other channels of the TRPC family. The more pronounced disease caused by Podocin loss may result from the absence of enhancement of all TRPC channels, not just those containing TRPC6. An intriguing speculation is that Podocin, like MEC-2, may participate in mechanosensation at the kidney filtration barrier. Podocin is part of a multiprotein complex containing the transmembrane proteins Neph1, Neph2, Nephrin, the cytoplasmic adaptor protein CD2AP (3), and, as we show here, the TRP channel TRPC6. Together, these proteins could form a sensor involved in monitoring glomerular pressure or filtration rate. Consistent with this hypothesis, deletion of TRPC6 in mice results in high blood pressure (20).
A Possible Mechanism for Steroid Action at the Membrane.
This study highlights a critical role for plasma membrane sterols in modulating ion-channel activity. Our demonstration that Podocin and MEC-2 associate cholesterol with protein complexes suggests that this binding may be a key component of their regulatory effects. This regulation, however, may not be mediated only by sterols; we hypothesize that these proteins could also mediate the action of steroids at the plasma membrane. Although steroid control of transcription through binding to cytoplasmic receptors that localize to the nucleus is well characterized, several “nontranscriptional effects” of steroids, activities that are too rapid to be mediated by transcriptional regulation, have been observed (27). A highly speculative but intriguing hypothesis is that PHB-domain proteins like MEC-2 and Podocin might mediate the nongenomic effects of steroids on ion channels and other membrane proteins. This hypothesis might, in part, explain a curious aspect of Podocin biology. Specifically, although many patients with nephrotic syndrome respond rapidly to treatment with glucocorticoids, individuals lacking Podocin do not. Indeed, the Podocin gene was first cloned in patients with a disease termed steroid-resistant nephrotic syndrome (7). Thus, binding of glucocorticoids to Podocin could be the basis of the therapeutic effects of these compounds.
Materials and Methods
Reagents and Plasmids.
Mouse Podocin cDNA constructs have been described (8, 28). TRPC6 was cloned from a human podocyte cDNA library. MEC-2 cDNA was cloned from a C. elegans ORF AAA87552 (Open Biosystems, Huntsville, AL). Truncations and mutations of Podocin, MEC-2, and TRPC6 were generated by standard cloning procedures. All other constructs have been described (10, 28). Some experiments involving MEC-2 had to be performed with αENaC, a mammalian ENaC protein, instead of MEC-4, because MEC-4 cDNA did not express well in HEK293T cells. Antibodies have been described or were obtained from Sigma, (St. Louis, MO) (anti-FLAG M2), Alomone Labs (Jerusalem, Israel), and Chemicon (Temecula, CA) (anti-TRPC6), and Serotec, Toronto, ON, Canada) (anti-V5). Bacterial vectors for the expression of His-tagged recombinant proteins fused to the C terminus of NusA were kindly provided by Gunter Stier (European Molecular Biology Laboratory, Heidelberg, Germany).
Cell Culture Studies.
Most cell studies used HEK293T cells that were grown in DMEM as described (28). Cholesterol-depleted cells were prepared by growing cells in DMEM with pravastatin (8 μM) for 2 days and then MBCD (5 mM) for 30 min just before the experiment. Immunoprecipitations from HEK293T cells were performed as described (28). Plamitate labeling, the digitonin precipitation assay (29), and photoaffinity labeling (17) were performed as described. Expression and purification of recombinant proteins has been described (30).
PHB protein multimerization was studied by velocity-gradient centrifugation and blue native-gel electrophoresis (31). For preparation of Podocin multimeric complexes, HEK293T cells were lysed in 1 ml of Mes-buffered saline (MBS) in the presence of 1% Triton X-100 and centrifuged for 10 min at 1,000 × g at 4°C. After centrifugation, the supernatant was collected, and SDS was added at a final concentration of 0.1% and incubated for 20 min on ice. Thereafter, the lysate was cleared by centrifugation for 15 min at 100,000 × g. Four milliliters of a discontinuous sucrose gradient (40–5%) was layered on top of a 60% sucrose cushion in an ultracentrifuge tube (Beckman, Fullerton, CA). One milliliter of the cell lysate was adjusted with 1 ml of MBS, added on top of this gradient, and subjected to centrifugation for 16 h at 180,000 × g at 4°C in a Beckman SW-41 rotor. After centrifugation, 2 ml of the supernatant were discarded and 8 fractions (500 μl each) were collected, starting from the top, and analyzed by SDS/PAGE.
In Vitro Cholesterol Interaction.
Podocin truncations were cloned into various bacterial expression vectors and tested for the expression of soluble recombinant fusion proteins. Expression as His-tagged proteins fused to the C terminus of NusA (vectors kindly provided by Gunter Stier) resulted in a large fraction of soluble recombinant Podocin protein that could be affinity-purified on Ni+ columns. Purity of the preparation was confirmed on Coomassie gels. For in vitro cholesterol-interaction assays, 2–20 μg of affinity-purified Podocin protein was bound to 30 μl of Ni+ beads and incubated with 0.1 μCi (1 Ci = 37 GBq) of [3H]cholesterol (Amersham, Piscataway, NJ) complexed with low amounts of MBCD. After binding for 10 min at 37°C, beads were washed extensively and counted in a scintillation counter. To confirm equal loading of the beads, aliquots of the bound protein were run on Coomassie gels. Competition experiments were performed with 1 μg of affinity-purified Podocin protein, varying amounts of [3H]cholesterol, and an ≈100-fold excess of cold cholesterol.
C. elegans Experiments.
C. elegans strains were cultured at 20°C, assayed for touch sensitivity, and prepared for immunofluorescence as described (9, 10). Media for growth on limiting, or no cholesterol, or on other sterols were prepared from chloroform-extracted reagents as described by Matyash et al. (32).
Oocyte Electrophysiology.
X. laevis oocytes were isolated from adult frogs (Kähler, Hamburg, Germany), dispersed and defolliculated by a 45-min treatment with collagenase (type A; Boehringer, Ingelheim, Germany), rinsed, and kept at 18°C in ND96 buffer: 96 mmol/liter NaCl, mmol/liter KCI 2, 1.8 mmol/liter CaCl2, 1 mmol/liter MgCl2, 5 mmol/liter Hepes, 2.5 mmol/liter sodium pyruvate, pH 7.55), supplemented with theophylline (0.5 mmol/l) and gentamycin (5 mg/l). cRNAs (1–10 ng) for TRPC6, Podocin and Podocin-ΔPHB were transcribed in vitro from cDNAs by using the T7 promoter and polymerase (Promega, Madison, WI) and injected into oocytes after dissolving in 47 nl of double-distilled water (Nanoliter Injector; World Precision Instruments, Berlin, Germany). Water-injected oocytes served as controls. Two to four days after injection, oocytes were impaled with two electrodes (Clark Brothers Instrument, Shelby Township, MI) that had resistances of <1 MΩ when filled with 2.7 mol/liter KCI. By using two bath electrodes and a virtual-ground headstage, the voltage drop across Rserial was effectively zero. Membrane currents were measured by voltage clamping of the oocytes (oocyte clamp amplifier OC725C; Warner Instruments, Hamden, CT) in intervals from −60 to +40 mV, each 1 s. Conductances were calculated according to Ohm's law. Na+ conductances were determined by replacing Na+ by N-methyl-d-glucamine (GNMDG) in a Ca2+-free bath solution, before and after stimulation with 10 μM dioctanoyl glycerol (Sigma-Aldrich, Munich, Germany). During the whole experiment, the bath was continuously perfused at a rate of 5–10 ml/min. All experiments were conducted at room temperature (22°C).
Ca2+-FLIPR Assay.
HeLa-Cx43 cells were loaded with 4 μM FLUO-4/AM and 0.04% Pluronic F-127 (both from Molecular Probes, Eugene, OR) in HBS but with 20 mM Hepes and 2.5 mM probenecid, and assays were performed as described (21).
Statistical Analysis.
Data are expressed as mean ± SEM of n experiments. Statistical evaluation was performed by using Student's t test or ANOVA for repeated measures, followed by a Bonferroni test as posttest (SigmaPlot; Jandel Scientific, San Rafael, CA, and Instat2, GraphPad Software, San Diego, CA). Values of P < 0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank Christina Engel, Stefanie Keller, Petra Dämisch, Charlotte Meyer, and John Byun for technical assistance; Gunter Stier for providing plasmids; and members of the Benzing and Chalfie laboratories for helpful discussions. This study was supported by the Deutsche Forschungsgemeinschaft (T.B.H., T.B., and G.W.) and the National Institutes of Health (M.C.).
Abbreviations
- MBCD
methyl-β-cyclodextrine
- NMDG
n-methyl-d-glucamine
- OAG
oleoyl-2-acetyl-sn-glycerol
- PHB
prohibitin.
Note Added in Proof.
Spassova et al. (34) find that TRPC6 channels can be mechanically gated, a finding that supports our hypothesis that TRPC6 acts as a mechanosensor in the mammalian kidney.
Footnotes
Conflict of interest statement: Columbia University has filed a provisional patent application based on this research.
References
- 1.Ponting CP, Schultz J, Milpetz F, Bork P. Nucleic Acids Res. 1999;27:229–232. doi: 10.1093/nar/27.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrow IC, Parton RG. Traffic. 2005;6:725–740. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 3.Benzing T. J Am Soc Nephrol. 2004;15:1382–1391. doi: 10.1097/01.asn.0000130167.30769.55. [DOI] [PubMed] [Google Scholar]
- 4.Ernstrom GG, Chalfie M. Annu Rev Genet. 2002;36:411–453. doi: 10.1146/annurev.genet.36.061802.101708. [DOI] [PubMed] [Google Scholar]
- 5.Stewart GW. Curr Opin Hematol. 2004;11:244–250. doi: 10.1097/01.moh.0000132240.20671.33. [DOI] [PubMed] [Google Scholar]
- 6.Huang M, Gu G, Ferguson EL, Chalfie M. Nature. 1995;378:292–295. doi: 10.1038/378292a0. [DOI] [PubMed] [Google Scholar]
- 7.Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, Dahan K, Gubler MC, Niaudet P, Antignac C. Nat Genet. 2000;24:349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 8.Huber TB, Simons M, Hartleben B, Sernetz L, Schmidts M, Gundlach E, Saleem MA, Walz G, Benzing T. Hum Mol Genet. 2003;12:3397–3405. doi: 10.1093/hmg/ddg360. [DOI] [PubMed] [Google Scholar]
- 9.Zhang S, Arnadottir J, Keller C, Caldwell GA, Yao CA, Chalfie M. Curr Biol. 2004;14:1888–1896. doi: 10.1016/j.cub.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 10.Goodman MB, Ernstrom GG, Chelur DS, O'Hagan R, Yao CA, Chalfie M. Nature. 2002;415:1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- 11.O'Hagan R, Chalfie M, Goodman MB. Nat Neurosci. 2005;8:43–50. doi: 10.1038/nn1362. [DOI] [PubMed] [Google Scholar]
- 12.Roselli S, Gribouval O, Boute N, Sich M, Benessy F, Attie T, Gubler MC, Antignac C. Am J Pathol. 2002;160:131–139. doi: 10.1016/S0002-9440(10)64357-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kestila M, Lenkkeri U, Mannikko M, Lamerdin J, McCready P, Putaala H, Ruotsalainen V, Morita T, Nissinen M, Herva R, et al. Mol Cell. 1998;1:575–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 14.Shih NY, Li J, Karpitskii V, Nguyen A, Dustin ML, Kanagawa O, Miner JH, Shaw AS. Science. 1999;286:312–315. doi: 10.1126/science.286.5438.312. [DOI] [PubMed] [Google Scholar]
- 15.Reiser J, Polu KR, Moller CC, Kenlan P, Altintas MM, Wei C, Faul C, Herbert S, Villegas I, Avila-Casado C, et al. Nat Genet. 2005;37:739–744. doi: 10.1038/ng1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winn MP, Conlon PJ, Lynn KL, Farrington MK, Creazzo T, Hawkins AF, Daskalakis N, Kwan SY, Ebersviller S, Burchette JL, et al. Science. 2005;308:1801–1804. doi: 10.1126/science.1106215. [DOI] [PubMed] [Google Scholar]
- 17.Thiele C, Hannah MJ, Fahrenholz F, Huttner WB. Nat Cell Biol. 2000;2:42–49. doi: 10.1038/71366. [DOI] [PubMed] [Google Scholar]
- 18.Umlauf E, Mairhofer M, Prohaska R. J Biol Chem. 2006;281:23349–23356. doi: 10.1074/jbc.M513720200. [DOI] [PubMed] [Google Scholar]
- 19.Chitwood DJ. Crit Rev Biochem Mol Biol. 1999;34:273–284. doi: 10.1080/10409239991209309. [DOI] [PubMed] [Google Scholar]
- 20.Freichel M, Vennekens R, Olausson J, Stolz S, Philipp SE, Weissgerber P, Flockerzi V. J Physiol. 2005;567:59–66. doi: 10.1113/jphysiol.2005.092999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirokova E, Schmiedeberg K, Bedner P, Niessen H, Willecke K, Raguse JD, Meyerhof W, Krautwurst D. J Biol Chem. 2005;280:11807–11815. doi: 10.1074/jbc.M411508200. [DOI] [PubMed] [Google Scholar]
- 22.Chelur DS, Ernstrom GG, Goodman MB, Yao CA, Chen L, O'Hagan R, Chalfie M. Nature. 2002;420:669–673. doi: 10.1038/nature01205. [DOI] [PubMed] [Google Scholar]
- 23.Barnes TM, Jin Y, Horvitz HR, Ruvkun G, Hekimi S. J Neurochem. 1996;67:46–57. doi: 10.1046/j.1471-4159.1996.67010046.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Morrow JS. J Biol Chem. 2000;275:8062–8071. doi: 10.1074/jbc.275.11.8062. [DOI] [PubMed] [Google Scholar]
- 25.Seidel G, Prohaska R. Gene. 1998;225:23–29. doi: 10.1016/s0378-1119(98)00532-0. [DOI] [PubMed] [Google Scholar]
- 26.Getz GS, Reardon CA. Curr Opin Lipidol. 2004;15:261–267. doi: 10.1097/00041433-200406000-00005. [DOI] [PubMed] [Google Scholar]
- 27.Losel RM, Falkenstein E, Feuring M, Schultz A, Tillmann HC, Rossol-Haseroth K, Wehling M. Physiol Rev. 2003;83:965–1016. doi: 10.1152/physrev.00003.2003. [DOI] [PubMed] [Google Scholar]
- 28.Huber TB, Kottgen M, Schilling B, Walz G, Benzing T. J Biol Chem. 2001;276:41543–41546. doi: 10.1074/jbc.C100452200. [DOI] [PubMed] [Google Scholar]
- 29.Charrin S, Manie S, Thiele C, Billard M, Gerlier D, Boucheix C, Rubinstein E. Eur J Immunol. 2003;33:2479–2489. doi: 10.1002/eji.200323884. [DOI] [PubMed] [Google Scholar]
- 30.Benzing T, Brandes R, Sellin L, Schermer B, Lecker S, Walz G, Kim E. Nat Med. 1999;5:913–918. doi: 10.1038/11354. [DOI] [PubMed] [Google Scholar]
- 31.Schagger H, von Jagow G. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 32.Matyash V, Entchev EV, Mende F, Wilsch-Brauninger M, Thiele C, Schmidt AW, Knolker HJ, Ward S, Kurzchalia TV. PLoS Biol. 2004;2:e280. doi: 10.1371/journal.pbio.0020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong K, Driscoll M. Nature. 1994;367:470–473. doi: 10.1038/367470a0. [DOI] [PubMed] [Google Scholar]
- 34.Spassova MA, Hewavitharana T, Xu W, Soboloff J, Gill DL. Proc Natl Acad Sci USA. 2006;103:16586–16591. doi: 10.1073/pnas.0606894103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.