Abstract
During the process of biological nitrogen fixation, the enzyme nitrogenase catalyzes the ATP-dependent reduction of dinitrogen to ammonia. Nitrogenase consists of two component metalloproteins, the iron (Fe) protein and the molybdenum-iron (MoFe) protein; the Fe protein mediates the coupling of ATP hydrolysis to interprotein electron transfer, whereas the active site of the MoFe protein contains the polynuclear FeMo cofactor, a species composed of seven iron atoms, one molybdenum atom, nine sulfur atoms, an interstitial light atom, and one homocitrate molecule. This Perspective provides an overview of biological nitrogen fixation and introduces three contributions to this special feature that address central aspects of the mechanism and assembly of nitrogenase.
Biological nitrogen fixation, as defined by the reduction of dinitrogen to ammonia under physiological conditions, is thermodynamically favorable (1). Even at low intracellular dissolved gas pressure, the reaction has a large negative free energy when reduced ferredoxin (Fdred) serves as the electron donor. Nevertheless, the enzymatic reaction, as catalyzed by the complex metallocluster-containing nitrogenase system, does not proceed without the additional input of substantial quantities of energy in the specific form of ATP hydrolysis. These requirements may be summarized by the following equation:
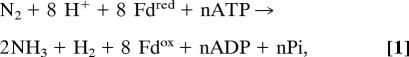 |
where n is the ratio of ATP hydrolyzed per electron transferred. This expression encompasses three central questions relevant to the mechanism of nitrogenase:
What is the role of nucleotide hydrolysis in electron transfer and substrate reduction?
How are substrates bound and reduced at the active site?
How are the nitrogenase metalloclusters assembled and inserted?
These issues are addressed in the contributions to this Nitrogen Fixation Special Feature by the groups of Watt (2); Seefeldt, Dean, and Hoffman (3); and Ribbe, Hodgson, and Hedman (4). The purpose of this Perspective is to provide both a background for these contributions and a summary of our views on the progress made in deciphering the molecular mechanisms of this fascinating process.
Intermolecular Electron Transfer and the Role of ATP Hydrolysis in Nitrogenase
The overall reaction mechanism of biological nitrogen fixation (Eq. 1) may be divided into two parts (5, 6): (i) the control or regulation of electron transfer to the substrate reduction site and (ii) the substrate reduction process itself. What sets nitrogenase apart from essentially all other enzymatically catalyzed redox processes is the number of electrons (eight) that must ultimately be delivered to the substrates each turnover cycle, with the consequent demand for precise timing of the underlying electron transfer events. The first part of this mechanism consists of a cycle involving the ATP-dependent electron transfer between the two protein components of nitrogenase, named Fe protein and MoFe protein, as diagrammatically shown in Scheme 1. The second part of the process, discussed subsequently, involves substrate reduction on the MoFe protein when sufficient cycles of intermolecular electron transfer have occurred.
Scheme 1.
Kinetic scheme depicting ATP-dependent electron transfer between the component proteins of nitrogenase. Av1 and Av2 denote the MoFe protein and Fe protein, respectively, from Azotobacter vinelandii. The superscripts R and Ox denote reduced and oxidized states of Av2, and the superscripts N and N-1 indicate the oxidation levels of Av1 before and after electron transfer from Av2. Fld, flavodoxin.
In this first cycle, the transfer of electrons from the Fe protein to the MoFe protein has an obligatory requirement for ATP hydrolysis by the Fe protein, which occurs only in the complex. Essential features of the electron transfer and ATP hydrolysis processes (Figs. 1 and 2), most notably the structures of the constituent metalloclusters and the nucleotide-binding site, have been defined by high-resolution x-ray crystal structures of the component proteins (7–15). In addition to the individual proteins, structures of complexes between Fe protein and MoFe protein have also been obtained at moderate resolutions (16–19) that correspond to several of the putative intermediates indicated in Scheme 1.
Fig. 1.
Ball-and-stick representation of the nitrogenase metalloclusters. Shown are the Fe-protein [4Fe:4S] cluster (A) and the two clusters of the MoFe protein, FeMo cofactor, and the P cluster (B) assigned to the oxidation state PN (C). The radii of all non-protein atoms have been set to 0.7 Å, and the protein ligands are presented as black bonds. Iron, molybdenum, sulfur, carbon, oxygen, and nitrogen atoms are colored burgundy, orange, yellow, gray, red, and blue, respectively. This figure was generated with the program MolScript (74) from Protein Data Bank entries 2NIP, 1M1N, and 3MIN.
Fig. 2.
Complex of the nitrogenase proteins stabilized by ADP-AIF4−. (Left) ADP-AlF4−-stabilized half-complex between a Fe-protein dimer and an αβ-subunit pair of the MoFe protein. The subunits are depicted as Cα traces with the MoFe α- and β-subunits colored red and blue, respectively, and the individual subunits of each Fe protein colored green and yellow. Non-protein groups are shown in a space-filling representation using the color scheme of Fig. 1, with fluorine and magnesium colored orange and green, respectively. (Right) Transduction pathway coupling the nucleotide and cofactor sites in the nitrogenase complex. This view represents a slice through the complex that includes the ADP-AlF4−, [4Fe:4S]-cluster, P-cluster, and FeMo-cofactor sites. The side chains of Asp-129 of each Fe-protein subunit are depicted as space-filling models to illustrate the locations of these critical residues adjacent to both the nucleotide and cluster sites.
The Fe protein is a homodimeric protein with a [4Fe:4S] cluster that bridges the two subunits (20). Although traditional in structure (Fig. 1A), this cluster has the unique property of undergoing reversible redox reactions between three oxidation states (21), unlike classical [4Fe:4S] clusters that use only two states (22, 23).
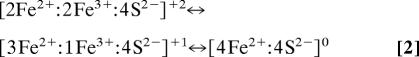 |
Most enzymological studies of nitrogenase have used dithionite as the immediate Fe-protein reductant. Dithionite-reduced Fe protein is well established to exhibit the +1 oxidation state of the cluster and to serve as a single electron donor to the MoFe protein in Scheme 1. As Watt and coworkers discuss in their article (2), if the Fe protein is reduced to the all ferrous 0 state during turnover, each cycle in Scheme 1 could potentially function in the transfer of two electrons to the MoFe protein (24). This could have profound consequences for both doubling the specific activity and providing an ATP/e ratio of 1, rather than the presently accepted value of 2 for a single electron transfer (excluding uncoupling processes). It is this uncertainty that is reflected in the unspecified value for “nATP” indicated in Eq. 1. Watt et al. present evidence that the Azotobacter flavodoxin is a sufficiently strong reductant to produce an all-ferrous state of the Fe protein [intriguingly, a distinct all-ferrous form of this protein has also been described (25)]. If this state is physiologically relevant, then the nitrogenase redox cycle in Scheme 1 could be a two-electron process in vivo, at least when flavodoxin is the Fe-protein electron donor.
Three other properties of Fe protein should be noted for our discussion. First, the protein has the peptide fold and nucleotide-binding domain similar to the G protein family of nucleotide-dependent switch proteins (26). These parallels are reflected through numerous examples of the differential effects of ATP/ADP on Fe-protein properties such as EPR spectra, iron chelation, and reduction potential. Second, ATP hydrolysis coupled to electron transfer is required for substrate reduction. Nucleotide analogs that induce the switch properties of the free Fe protein, nevertheless, do not support substrate reduction with the MoFe protein (27, 28). Third, the Fe protein is the only known functional electron transfer agent for substrate reduction. The metal centers in the MoFe protein can be reversibly oxidized by small electron-transfer proteins such as ferredoxins, flavodoxins, and plastocyanin or agents such as redox dyes (29), but none of these can drive substrate reduction, even if they have a lower E′o than Fe protein. That is, only Fe protein can reduce MoFe protein beyond a level obtained by other reductants.
The heterotetrameric MoFe protein is composed of two copies of the homologous α- and β-subunits, (αβ)2; each αβ pair coordinates two unique metalloclusters, the FeMo cofactor and the P cluster (Fig. 1 B and C), based on iron-sulfur cluster motifs. Because the two sets of clusters are separated by ≈70 Å (30), each αβ pair likely represents the functional unit for electron transfer and substrate reduction. The [8Fe:7S] P cluster (Fig. 1C) is positioned at the α–β-subunit interface along the pseudo twofold axis relating the two subunits (8). Spectroscopic studies indicate that the P cluster is in the all-ferrous state as isolated in dithionite (31). The second MoFe protein metallocluster also contains eight metals (seven Fe and one Mo) and is designated the FeMo cofactor, because, in contrast to the P cluster, it can be extracted from partially denatured protein and inserted into cofactor-deficient MoFe protein (32). The cofactor has several remarkable properties not previously seen in Fe:S cluster chemistry. Whereas iron-sulfur clusters typically have a protein side chain ligand per metal, e.g., the cluster in Fe protein, the FeMo cofactor has only two direct protein ligands for the eight metals. To complete the coordination sphere, there are additional sulfides, an organic acid (homocitric acid), and a core, light element, atom (see discussion below). A plausible oxidation state assignment for the protein-bound FeMo cofactor (33, 34), as isolated in dithionite, is [1Mo4+:4Fe2+,3Fe3+:9S2−]3+.
Potential steps in the cycle depicted in Scheme 1 can be envisioned in terms of the crystal structures of different Fe protein–MoFe protein complexes. For our purposes, we will limit the discussion to complexes of the Azotobacter native proteins (16, 19). Historically, the first complex to be structurally characterized (16) was the putative transition state that was trapped during ATP hydrolysis by AlF4− (35, 36), an analogue for the departing phosphate (Fig. 2). This structure clearly revealed an electron transfer pathway extending from the Fe protein through the P cluster to the FeMo cofactor. Significantly, the Fe protein in this complex is positioned at a separation distance from the P cluster shorter than possible from the simple docking of the two native proteins. Complex formation is facilitated by a structural reorganization of the Fe protein such that the two subunits are folded toward each other by a hinge motion near the [4Fe:4S] cluster. These changes reposition the cluster more toward the Fe-protein surface, which in turn flattens to be complementary to the MoFe protein. Furthermore, this structure highlighted details of the ATP hydrolysis process and the coupling to electron transfer, as shown in Fig. 2. The nucleotide-binding sites are located at the dimer interface of the Fe protein, and the conformational change upon complex formation with the MoFe protein repositions several Fe-protein backbone and side-chain elements to effect catalytic hydrolysis of the nucleotide. Of particular significance is the observation that two catalytic side chains extend across the dimer interface to the opposite subunit, which helps to explain why ATP is not hydrolyzed by Fe protein in the absence of stabilizing MoFe protein.
Recently, three structures of complexes have been determined that potentially represent other steps of the cycle (19). Although the Fe protein binds on a single face of the MoFe protein in all these complexes, it can occupy distinct sites on the surface. Furthermore, the Fe protein in each binding mode has a different conformation correlated with the nucleotide state. The complex in which Fe protein has bound the nonhydrolyzable ATP analogue, AMPPCP, is remarkably similar to that from AlF4− trapping in terms of positioning of the [4Fe:4S] cluster over the P cluster, yet the dimer interface of the Fe protein has not closed to the same extent. In the ADP-bound state, the Fe-protein docking site principally involves the MoFe-protein α-subunit, whereas for the nucleotide-free Fe protein, the docking site is primarily on the MoFe-protein β-subunit. The Fe-protein [4Fe:4S] clusters in the latter two structures are now positioned ≈5–8 Å farther away from the P cluster than in the putative transition-state complex.
The structural features of the complexes suggest the outlines of a reaction coordinate that may help in thinking about Scheme 1. In Fig. 3, we have arranged along the abscissa the known protein structures in the order they might be expected in Scheme 1 (the missing structure is Fe protein with ATP). Along one ordinate is the change in hinge angle that describes the opening of the Fe protein relative to the most closed, AlF4−-trapped, putative transition-state model (Fig. 3 Inset). On the other ordinate is the distance between the centroids of the Fe-protein cluster and the P cluster observed in various complexes (for the dissociated proteins, this distance is a lower limit, as indicated by the arrow).
Fig. 3.
Correlation between hinge angle (bars) and the distance between centroids (filled triangles) of the [4Fe:4S] and P cluster of the nitrogenase proteins in a series of states ordered along a potential reaction coordination for nucleotide hydrolysis. (Inset) The hinge angle is defined by the rotation angle about an axis along the dimer interface required to superimpose one subunit of a specified Fe-protein structure onto a subunit of the ADP-AlF4−-stabilized Fe protein, after initially superimposing the other subunits in these structures.
Assuming a correlation between intercofactor distance and electron transfer rate (37, 38), only the AMPPCP complex exhibits a similar close approach as in the putative transition state of the ADP-AlF4− complex; the electron transfer rate in the nucleotide-free or ADP complexes would be several orders of magnitude slower because of their longer distances. However, electron transfer rate governed by shortest distance alone cannot be the overall deciding factor for productive electron transfer because no ATP analogue, including AMPPCP, supports substrate reduction (27, 28, 39). One difference between the structures is the degree of the Fe-protein change as measured by hinge angle; in the AMPPCP complex, the Fe protein remains ≈10–15° more open compared with the Fe protein of the ADP-AlF4− complex. These two structures may begin to separate the two parts of the process that makes Fe protein the unique donor for MoFe protein coupled to the nucleotide hydrolysis. We conjecture that only during hydrolysis does the full subunit closure and reorganization occur, leading to the transition state.
Substrate Reduction
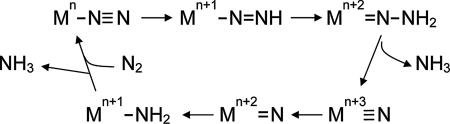
The second major question introduced at the beginning of this Perspective, the substrate reduction mechanism, must start with the redox chemistry of the metals, as shown by model reactions for dinitrogen reduction first suggested by Chatt (40) for molybdenum and realized experimentally by Schrock (41), and more recently for iron by Peters and coworkers (42) (Scheme 2).
Scheme 2.
Generalized Chatt-type mechanism for the reduction of dinitrogen to ammonia catalyzed at a single metal accommodating oxidation states n to n + 3. For the case of M Mo and Fe, n = +3 and +1, respectively.
Mo and Fe, n = +3 and +1, respectively.
As demonstrated by these studies, many of these single-site metal-nitrogen intermediates are feasible for either iron or molybdenum and, in one case (41), capable of reducing dinitrogen triple bonds with catalytic efficiency. Nevertheless, there appears to be a chemical imperative for the decidedly more complex 7Fe:9S:Mo:homocitrate cluster of the FeMo cofactor as the active site for substrate reduction in biological nitrogen fixation. For example, against the burden of acquiring large quantities of a scarce metal, iron, which is highly depleted in the oceanic environment, evolutionary pressures have retained the cofactor-based nitrogenase system for more than a billion years. Even the so-called alternate nitrogenases (43) are believed to be a minor variant on the cofactor theme, where the Mo is replaced by either V or Fe. Furthermore, the cofactor requires the uncommon metabolite, homocitric acid, and requires ≈20 additional proteins for assembly and insertion (44); the mechanism of Mo and homocitrate incorporation during cofactor maturation, and the intriguing role of the Fe protein in this process, is detailed in two reports by Ribbe, Hodgson, and Hedman and coworkers (4, 45) in this special feature. One would imagine that an active site consisting of a simpler one- or two-metal center for nitrogen fixation should been evolutionarily dominant if biologically functional, yet this is not the case.
Clearly, detailed analyses of the cofactor structure and chemistry are essential for elucidating the mechanism. Thus, when a 1.16-Å-resolution crystal structure revealed a previously undetected electron density in the cofactor core (19), a multitude of new questions about the enzyme mechanism and potential model analogues were raised. The density corresponding to this interstitial ligand had been obscured in previous structures by a combination of factors derived from resolution-dependent termination effects inherent in Fourier analysis. It is only at resolutions beyond ≈1.3 Å that these effects are minimized and the central atom could be revealed.
What is this central atom? Light elements that best fit the density are C, N, and O; unfortunately, x-ray diffraction is not sufficiently discriminating to distinguish between light elements differing by only one to two electrons such as the C, N, O series, and we are left with this uncertainty in identifying the core atom. Nevertheless, the revelation resolved a thorny question about the iron coordination of cofactor, because this core atom provides the fourth ligand for the trigonally compressed tetrahedral coordination of the six central iron atoms.
Although the identity of this light element cannot be unambiguously established by crystallography, nitrogen was marginally a better fit of the x-ray data and intuitively more satisfying chemically. Subsequently, density functional and other studies have been reported (34, 46–48) incorporating the three alternative light elements, to model the electronic, magnetic, and chemical redox properties of the FeMo cofactor. These studies have found a preference order N > C > O; although not proving the element, it is suggestive of nitrogen as the core atom. However, Seefeldt, Dean, Hoffman, and coworkers (49, 50) have attempted to identify the atom by 14N,15N electron-nuclear double resonance (ENDOR)/electron spin-echo envelope modulation (ESEEM) spectroscopy and have been unable to identify any exchangeable or non-protein-derived N atoms associated with the FeMo cofactor under turnover conditions or in the “as isolated,” resting oxidation state. These studies have concluded that (i) any hypothetical core nitrogen atom does not turn over during catalysis and (ii) the core atom is not a nitrogen. Although we are not being strong advocates for a N core atom, it seems that this spectroscopic approach may not yet be adequate to unambiguously exclude an element such as nitrogen that is spin-coupled in the magnetic core, especially without a clear demonstration of an alternate element. (There is a hint for a natural abundance 13C ENDOR resonance, but its origin is not known and the 14N resonances that are seen have yet to be fully evaluated and associated with any specific species or protein residue, including the cofactor ligand His α442.) For clusters that are electronically complex, multielemental, and redundant of atom type, one component may be masked by unrecognized properties of the system or measurement process, as we found for the core atom in the first place. For example, in the density functional analysis of Lovell et al. (47), the core atom is found to have little spin density (−0.02 of an electron), which would make these spectroscopic features difficult to observe. We would be less restrained and more convinced by the ENDOR/ESEEM results if a positive control were available showing that model compounds, containing an interstitial nitrogen atom spin-coupled as for the cofactor, had signature 14N ESEEM/ENDOR resonances that are missing in the cofactor spectrum.
Identifying the light element will require extraordinary analytical and/or spectroscopic erudition. For example, there are 1,366 N atoms in the protein for every potential one in the cofactor. The numbers are even more unfavorable for O or C. Yet this atom, we suspect, will prove central, and not just geometrically, to understanding dinitrogen, and perhaps all substrate, reduction. The site must certainly provide an electronic bridge that couples the six central iron atoms that likely serve in substrate binding and reduction. In this central location, the ligand contribution to any individual iron could be modulated by redox changes at the other iron sites, allowing incoming substrates to displace the core ligand at one or more iron atoms without significant distortion of the cluster. If the atom were nitrogen, one appealing thought is that the atom might be a remnant after half dinitrogen reduction. Such a notion is less persuasive, however, when the energetics of the cofactor structural rearrangement to expose the site are considered (see refs. 48 and 51). Likewise, fully active MoFe protein can be generated in mutant strains of Azotobacter vinelandii that cannot turn over substrates (52, 53). Hence, it is probable that the core atom is inserted during cluster synthesis, although this does not preclude the atom exchanging during catalysis.
The critical first step in elucidating the nitrogenase mechanism, how substrates or inhibitors bind to the FeMo cofactor, is made more complicated by the plethora of substrates reduced by nitrogenase besides the natural substrates dinitrogen and proton (see refs. 54 and 55). Most substrates and inhibitors have in common a multiple bond (with the notable exception of protons), and the atoms delineating the bond(s) can be C, N, O, and S in various combinations. In model compounds, both end-on and side-on binding modes have been observed, and neither mode has been excluded for the enzyme. In addition, different substrates appear to bind at different net reduction states of the cofactor; that is, a different number of redox cycles in Scheme 1 are required for binding of different substrates.
Although the distinctive structure of the FeMo cofactor is generally appreciated, how it differs from conventional [4Fe:4S] clusters can be obscured by the traditional “ball-and-stick” representations often used. For accessibility of substrates and ligands, FeMo cofactor depicted with nominal ionic or van der Waals radii is more appropriate where sulfur atoms have radii around 1.7 Å, more than twice the ionic radii of Fe3+, Fe2+ (0.7 Å) or Mo4+ (0.8 Å) (56, 57) (Fig. 4). The six central iron atoms that lack protein ligands constitute three 4Fe:4S faces dominated by the four sulfur atoms. Ligands approaching a 4Fe:4S face are restricted to the irons by the sulfurs, especially the two axial sulfides (separated by 5 Å), so that van der Waals contact between an approaching dinitrogen and the sulfurs would occur ≈3.4–3.8 Å from an iron. From the direction along the Fe-central ligand vector, dinitrogen could approach somewhat closer (≈2.8 Å), but this distance is still too large to permit direct coordination.
Fig. 4.
Space-filling representation of the FeMo-cofactor (Left) and [4Fe:4S] cluster of aconitase (Right) using chemically appropriate radii for their atoms, including the protein side-chain ligands and homocitric acid. Dinitrogen is included for size comparison. The radii used to generate this figure were as follows: C, 1.5 Å; O, 1.4 Å; interstitial ligand (N3−), 1.4 Å; N in N2, 1.6 Å; S, 1.7 Å; Fe, 0.7 Å; and Mo, 0.8 Å (56, 57).
A comparison to the conventional [4Fe:4S] cluster used for substrate binding and catalysis in aconitase (58) is informative. Both cofactor and aconitase have iron sites that lack protein ligands while remaining coordinated to three sulfides; yet, there are significant differences in the surface geometry of these sites (Fig. 4). For the cofactor, each central iron atom is inside a concave surface of sulfur atoms, whereas in aconitase, the unliganded iron site protrudes from the convex surface of sulfur atoms. Indeed, a solvent molecule coordinates this position in the absence of a substrate for aconitase, indicating that there would be no steric restrictions for a molecule the size of dinitrogen to approach this iron and, hence, a site more in keeping with the bioinorganic models.
The overall impression of the FeMo-cofactor structure is one of steric restriction to the metal sites leading to low chemical reactivity. Both traditional cluster and cofactor metals with protein ligands are sterically blocked by the terminal ligands, but only in the FeMo cofactor are the metals without protein ligands also restricted. What is particularly revealing is the dominance of the sulfur atoms in the overall topology. Restricted access to the metals and low chemical reactivity is contrary to the expectation for the site of dinitrogen reduction. Nevertheless, there is more than conjecture suggesting that FeMo cofactor has unusual chemical reactivity. It has always seemed surprising that the cluster could be extracted intact from the denatured protein without added stabilizing ligands such as thiols, as required for traditional Fe:S clusters (32). The isolated cofactor is unexpectedly stable with only a single iron site bound by excess added thiols, a condition that can lead to decomposition of simpler clusters (59).
Outstanding Mechanistic Issues
Beyond the binding mode, any mechanistic proposal must explain or consider a number of other aspects of the system. Briefly, these are as follow.
Order of Addition of Protons and Electrons.
The reduction vs. proton addition steps are only superficially considered in our representation of substrate reduction (Scheme 2), yet how this occurs has substantial implications. The Seefeldt, Dean, and Hoffman collaboration of coworkers have identified potential intermediates using a combination of mutant proteins, magnetic spectroscopy, and isotopically labeled compounds (60–64). In this special feature (3), they present evidence that for the substrate methyldiazene, only the terminal N atom binds to an iron and that protons are added in an alternating fashion between the two nitrogens with the first proton added to the non-iron-bound nitrogen.
Origin of the Protons.
Proton addition is required for the reduction of all known nitrogenase substrates, but it is not clear whether the protons come from the same donors for all substrates. For example, acetylene reduction is stereospecific (65), but the selectivity changes with substrate concentration (66), suggesting at least a change in mechanism or mode of substrate binding where the proton donor(s) may be different. Both water molecules adjacent to the cofactor and the sulfides of the cluster suggest intriguing possibilities for the proton donors.
Altered or Mutant Proteins.
Altered proteins, generated either by mutagenesis or chemical modification, have been invaluable in elucidating enzyme mechanisms by identifying potential functions of specific amino acid residues. However, altered activity with side-chain changes can be structurally allosteric or compensating, rather than providing unambiguous evidence for direct side chain involvement in the reaction, as experienced with inorganic complexes (67, 68). Ultimately, the use of any modified protein must be justified within the context of the mechanism even when the alteration provides a route to identifying a potential intermediate.
Role of Homocitric Acid.
Homocitric acid is required for dinitrogen reduction (69), but other di- or tricarboxylic acids support some substrate reductions (70), if weakly. As observed with mutations in the protein structure, changes in this ligand are difficult to explain simply by direct effects on a localized site. The cofactor structure and surrounding environment are so complex that changes at one site may be propagated to a distant site to effect the observed altered chemical reactivity expressed in substrate specificity or reduction rates.
Role of Hydrogen Evolution.
In the absence of other substrates, protons are reduced to dihydrogen that can be fully suppressed by most other substrates. In contrast, even at saturating dinitrogen, protons are reduced to dihydrogen (71). In addition, dinitrogen reduction is inhibited by dihydrogen and mediates the formation of HD from D2 (72). A description of the chemical mechanism of dinitrogen reduction cannot be complete without including these unique dihydrogen reactions.
Substrate Specificity and Binding Sites.
The binding sites for substrates may be defined as much by the redox state of the cofactor as the physical site. The kinetics of mixed substrates are complex and are typically analyzed as competitive, noncompetitive, and uncompetitive depending on the combination of substrates investigated (73). As noted above, the FeMo cofactor has multiple potential binding sites, and it would be difficult to establish kinetic interactions between these potential binding modes. Hence, it would not be surprising if there were more than one detailed mechanism depending on the substrate; although there should be common features, results with one substrate should not uncritically be assigned to all.
Concerning the title question (How many metals does it take to fix N2?), from our perspective, the correct answer for biological nitrogen fixation is 20—the number of unique metals in the FeMo cofactor, P cluster, and Fe protein—because they are all needed to fix dinitrogen, and no one has found a way to simplify this system. This then leads to the corollary question, What are they all doing? The recent progress in this field, exemplified in the contributions to this special feature, suggests that answers to these questions will be forthcoming within the next few years.
Acknowledgments
We thank members of the Howard and Rees groups, past, present, and future, for their contributions to the nitrogenase project in our laboratories. This work was supported in part by National Institutes of Health Grant GM045162.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Alberty RA. J Biol Chem. 1994;269:7099–7102. [PubMed] [Google Scholar]
- 2.Lowery TJ, Wilson PE, Zhang B, Bunker J, Harrison RG, Nyborg AC, Thiriot D, Watt GD. Proc Natl Acad Sci USA. 2006;103:17131–17136. doi: 10.1073/pnas.0603223103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barney BM, Lukoyanov D, Yang T-C, Dean DR, Hoffman BM, Seefeldt LC. Proc Natl Acad Sci USA. 2006;103:17113–17118. doi: 10.1073/pnas.0602130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Y, Corbett MC, Fay AW, Webber JA, Hodgson KO, Hedman B, Ribbe MW. Proc Natl Acad Sci USA. 2006;103:17119–17124. doi: 10.1073/pnas.0602647103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hageman RV, Orme-Johnson WH, Burris RH. Biochemistry. 1980;19:2333–2342. doi: 10.1021/bi00552a009. [DOI] [PubMed] [Google Scholar]
- 6.Thorneley RNF, Lowe DJ. In: Molybdenum Enzymes. Spiro TG, editor. New York: Wiley; 1985. pp. 221–284. [Google Scholar]
- 7.Kim J, Rees D C. Nature. 1992;360:553–560. doi: 10.1038/360553a0. [DOI] [PubMed] [Google Scholar]
- 8.Kim J, Rees DC. Science. 1992;257:1677–1682. doi: 10.1126/science.1529354. [DOI] [PubMed] [Google Scholar]
- 9.Georgiadis MM, Komiya H, Chakrabarti P, Woo D, Kornuc JJ, Rees DC. Science. 1992;257:1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- 10.Bolin JT, Campobasso N, Muchmore SW, Morgan TV, Mortenson LE. Stiefel EI, Coucouvanis D, Newton WE. Molybdenum Enzymes Cofactors and Model Systems. Vol 535. Washington, DC: Am Chem Soc; 1993. ACS Symposium Series; pp. 186–195. [Google Scholar]
- 11.Peters JW, Stowell MHB, Soltis SM, Finnegan MG, Johnson MK, Rees DC. Biochemistry. 1997;36:1181–1187. doi: 10.1021/bi9626665. [DOI] [PubMed] [Google Scholar]
- 12.Schlessman JL, Woo D, Joshua-Tor L, Howard JB, Rees DC. J Mol Biol. 1998;280:669–685. doi: 10.1006/jmbi.1998.1898. [DOI] [PubMed] [Google Scholar]
- 13.Mayer SM, Lawson DM, Gormal CA, Roe SM, Smith BE. J Mol Biol. 1999;292:871–891. doi: 10.1006/jmbi.1999.3107. [DOI] [PubMed] [Google Scholar]
- 14.Jang SB, Seefeldt LC, Peters JW. Biochemistry. 2000;39:14745–14752. doi: 10.1021/bi001705g. [DOI] [PubMed] [Google Scholar]
- 15.Einsle O, Tezcan FA, Andrade SLA, Schmid B, Yoshida M, Howard JB, Rees DC. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 16.Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. Nature. 1997;387:370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- 17.Schmid B, Einsle O, Chiu H-J, Willing A, Yoshida M, Rees DC, Howard JB. Biochemistry. 2002;41:15557–15565. doi: 10.1021/bi026642b. [DOI] [PubMed] [Google Scholar]
- 18.Chiu H-J, Peters JW, Lanzilotta WN, Ryle MJ, Seefeldt LC, Howard JB, Rees DC. Biochemistry. 2001;40:641–650. doi: 10.1021/bi001645e. [DOI] [PubMed] [Google Scholar]
- 19.Tezcan FA, Kaiser JT, Mustafi D, Walton MY, Howard JB, Rees DC. Science. 2005;309:1377–1380. doi: 10.1126/science.1115653. [DOI] [PubMed] [Google Scholar]
- 20.Hausinger RP, Howard J. J Biol Chem. 1983;258:13486–13492. [PubMed] [Google Scholar]
- 21.Watt GD, Reddy KRN. J Inorg Biochem. 1994;53:281–294. [Google Scholar]
- 22.Carter CW, Kraut J, Freer ST, Alden RA, Sieker LC, Adman E, Jensen LH. Proc Natl Acad Sci USA. 1972;69:3526–3529. doi: 10.1073/pnas.69.12.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berg JM, Holm RH. Spiro TG. Iron-Sulfur Proteins. New York: Wiley; 1982. pp. 1–66. [Google Scholar]
- 24.Erickson JA, Nyborg AC, Johnson JL, Truscott SM, Gunn A, Nordmeyer FR, Watt GD. Biochemistry. 1999;38:14279–14285. doi: 10.1021/bi991389+. [DOI] [PubMed] [Google Scholar]
- 25.Angove HC, Yoo SJ, Burgess BK, Münck E. J Am Chem Soc. 1997;119:8730–8731. [Google Scholar]
- 26.Howard JB, Rees DC. Annu Rev Biochem. 1994;63:235–264. doi: 10.1146/annurev.bi.63.070194.001315. [DOI] [PubMed] [Google Scholar]
- 27.Weston MF, Kotake S, Davis LC. Arch Biochem Biophys. 1983;225:809–817. doi: 10.1016/0003-9861(83)90093-0. [DOI] [PubMed] [Google Scholar]
- 28.Deits TL, Howard JB. J Biol Chem. 1989;264:6619–6628. [PubMed] [Google Scholar]
- 29.Watt GD, Wang Z-C. Biochemistry. 1989;28:1844–1850. [Google Scholar]
- 30.Bolin JT, Ronco AE, Mortenson LE, Morgan TV, Williamson M, Xuong N-h. Gresshoff PM, Roth LE, Stacey G, Newton WE. Nitrogen Fixation: Achievements and Objectives. New York: Chapman & Hall; 1990. pp. 117–124. [Google Scholar]
- 31.McLean PA, Papaefthymiou V, Orme-Johnson WH, Münck E. J Biol Chem. 1987;262:12900–12903. [PubMed] [Google Scholar]
- 32.Shah VK, Brill WJ. Proc Natl Acad Sci USA. 1977;74:3249–3253. doi: 10.1073/pnas.74.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo SJ, Angove HC, Papaefthymiou V, Burgess BK, Münck E. J Am Chem Soc. 2000;122:4926–4936. [Google Scholar]
- 34.Vrajmasu V, Münck E, Bominaar EL. Inorg Chem. 2003;42:5974–5988. doi: 10.1021/ic0301371. [DOI] [PubMed] [Google Scholar]
- 35.Renner KA, Howard JB. Biochemistry. 1996;35:5353–5358. doi: 10.1021/bi960441o. [DOI] [PubMed] [Google Scholar]
- 36.Duyvis MG, Wassink H, Haaker H. FEBS Lett. 1996;380:233–236. doi: 10.1016/0014-5793(96)00019-1. [DOI] [PubMed] [Google Scholar]
- 37.Page CC, Moser CC, Dutton PL. Curr Opin Chem Biol. 2003;7:551–556. doi: 10.1016/j.cbpa.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Gray HB, Winkler JR. Q Rev Biophys. 2003;36:341–372. doi: 10.1017/s0033583503003913. [DOI] [PubMed] [Google Scholar]
- 39.Thorneley RNF, Ashby GA, Julius C, Hunter JL, Webb MR. Biochem J. 1991;277:735–741. doi: 10.1042/bj2770735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatt J, Dilworth JR, Richards RL. Chem Rev. 1978;78:589–625. [Google Scholar]
- 41.Yandulov DV, Schrock RR. Science. 2003;301:76–78. doi: 10.1126/science.1085326. [DOI] [PubMed] [Google Scholar]
- 42.Betley TA, Peters JC. J Am Chem Soc. 2004;126:6252–6254. doi: 10.1021/ja048713v. [DOI] [PubMed] [Google Scholar]
- 43.Eady RR. Chem Rev. 1996;96:3013–3030. doi: 10.1021/cr950057h. [DOI] [PubMed] [Google Scholar]
- 44.Dos Santos PC, Dean DR, Hu Y, Ribbe MW. Chem Rev. 2004;104:1159–1173. doi: 10.1021/cr020608l. [DOI] [PubMed] [Google Scholar]
- 45.Hu Y, Corbett MC, Fay AW, Webber JA, Hodgson KO, Hedman B, Ribbe MW. Proc Natl Acad Sci USA. 2006;103:17125–17130. doi: 10.1073/pnas.0602651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hinnemann B, Nørskov JK. J Am Chem Soc. 2003;125:1466–1467. doi: 10.1021/ja029041g. [DOI] [PubMed] [Google Scholar]
- 47.Lovell T, Liu T, Case DA, Noodleman L. J Am Chem Soc. 2003;125:8377–8383. doi: 10.1021/ja0301572. [DOI] [PubMed] [Google Scholar]
- 48.Dance I. Chem Commun. 2003:324–325. doi: 10.1039/b211036a. [DOI] [PubMed] [Google Scholar]
- 49.Lee H-I, Benton PMC, Laryukhin M, Igarashi RY, Dean DR, Seefeldt LC, Hoffman BM. J Am Chem Soc. 2003;125:5604–5605. doi: 10.1021/ja034383n. [DOI] [PubMed] [Google Scholar]
- 50.Yang TC, Maeser NK, Laryukhin M, Lee HI, Dean DR, Seefeldt LC, Hoffman BM. J Am Chem Soc. 2005;127:12804–12805. doi: 10.1021/ja0552489. [DOI] [PubMed] [Google Scholar]
- 51.Hinnemann B, Nørskov JK. J Am Chem Soc. 2004;126:3920–3927. doi: 10.1021/ja037792s. [DOI] [PubMed] [Google Scholar]
- 52.Jacobson MR, Cash VL, Weiss MC, Laird NF, Newton WE, Dean DR. Mol Gen Genet. 1989;219:49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- 53.Wolle D, Dean DR, Howard JB. Science. 1992;258:992–995. doi: 10.1126/science.1359643. [DOI] [PubMed] [Google Scholar]
- 54.Burgess BK. In: Molybdenum Enzymes. Spiro TG, editor. New York: Wiley Interscience; 1985. pp. 161–219. [Google Scholar]
- 55.Seefeldt LC, Rasche ME, Ensign SA. Biochemistry. 1995;34:5382–5389. doi: 10.1021/bi00016a009. [DOI] [PubMed] [Google Scholar]
- 56.Bondi A. J Phys Chem. 1964;68:441–451. [Google Scholar]
- 57.Cotton FA, Wilkinson G, Murillo CA, Bochmann M. Advanced Inorganic Chemistry. New York: Wiley Interscience; 1999. [Google Scholar]
- 58.Beinert H, Kennedy MC, Stout CD. Chem Rev. 1996;96:2335–2373. doi: 10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- 59.Conradson SD, Burgess BK, Holm RH. J Biol Chem. 1988;263:13743–13749. [PubMed] [Google Scholar]
- 60.Lee H-I, Igarashi R, Laryukhin M, Doan PE, Dos Santos PC, Dean DR, Seefeldt LC, Hoffman BM. J Am Chem Soc. 2004;126:9563–9569. doi: 10.1021/ja048714n. [DOI] [PubMed] [Google Scholar]
- 61.Barney BM, Yang TC, Igarashi RY, Dos Santos PC, Laryukhin M, Lee HI, Hoffman BM, Dean DR, Seefeldt LC. J Am Chem Soc. 2005;127:14960–14961. doi: 10.1021/ja0539342. [DOI] [PubMed] [Google Scholar]
- 62.Barney BM, Laryukhin M, Igarashi RY, Lee HI, Dos Santos PC, Yang TC, Hoffman BM, Dean DR, Seefeldt LC. Biochemistry. 2005;44:8030–8037. doi: 10.1021/bi0504409. [DOI] [PubMed] [Google Scholar]
- 63.Igarashi RY, Laryukhin M, Dos Santos PC, Lee HI, Dean DR, Seefeldt LC, Hoffman BM. J Am Chem Soc. 2005;127:6231–6241. doi: 10.1021/ja043596p. [DOI] [PubMed] [Google Scholar]
- 64.Dos Santos PC, Igarashi RY, Lee HI, Hoffman BM, Seefeldt LC, Dean DR. Acc Chem Res. 2005;38:208–214. doi: 10.1021/ar040050z. [DOI] [PubMed] [Google Scholar]
- 65.Dilworth MJ. Biochim Biophys Acta. 1966;127:285–294. doi: 10.1016/0304-4165(66)90383-7. [DOI] [PubMed] [Google Scholar]
- 66.Han JH, Newton WE. Biochemistry. 2004;43:2947–2956. doi: 10.1021/bi035247y. [DOI] [PubMed] [Google Scholar]
- 67.Pool JA, Lobkovsky E, Chirik PJ. Nature. 2004;427:527–529. doi: 10.1038/nature02274. [DOI] [PubMed] [Google Scholar]
- 68.Fryzuk MD. Nature. 2004;427:498–499. doi: 10.1038/427498a. [DOI] [PubMed] [Google Scholar]
- 69.Hoover TR, Robertson AD, Cerny RL, Hayes RN, Imperial J, Shah VK, Ludden PW. Nature. 1987;329:855–857. doi: 10.1038/329855a0. [DOI] [PubMed] [Google Scholar]
- 70.Imperial J, Hoover TR, Madden MS, Ludden PW, Shah VK. Biochemistry. 1989;28:7796–7799. doi: 10.1021/bi00445a040. [DOI] [PubMed] [Google Scholar]
- 71.Simpson FB, Burris RH. Science. 1984;224:1095–1096. doi: 10.1126/science.6585956. [DOI] [PubMed] [Google Scholar]
- 72.Burgess BK, Wherland S, Newton WE, Stiefel EI. Biochemistry. 1981;20:5140–5146. doi: 10.1021/bi00521a007. [DOI] [PubMed] [Google Scholar]
- 73.Burris RH. In: A Treatise on Dinitrogen Fixation. Hardy RWF, Bottomley F, Burns RC, editors. New York: Wiley; 1979. pp. 569–604. [Google Scholar]
- 74.Kraulis PJ. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]