Abstract
RPB5 is an essential subunit of eukaryotic and archaeal RNA polymerases. It is a proposed target for transcription activator proteins in eukaryotes, but the mechanism of interaction is not known. We have determined the solution structure of the RPB5 subunit from the thermophilic archeon, Methanobacterium thermoautotrophicum. MtRBP5 contains a four-stranded β-sheet platform supporting two α-helices, one on each side of the β-sheet, resulting in an overall mushroom shape that does not appear to have any structural homologues in the structural database. The position and conservation of charged surface residues suggests possible modes of interaction with other proteins, as well as a rationale for the thermal stability of this protein.
RNA polymerases (RNAPs) are multisubunit enzymes with core components that are conserved among the bacteria, archaea, and eukarya. Both bacteria and archaea contain only one enzyme comprised of 4 and 12 subunits, respectively, whereas eukaryotes have three classes (I–III) comprised of 10–15 subunits. The archaeal RNA polymerases are closely related to the eukaryal enzymes in terms of subunit composition and sequence homology and have become a popular model system for studying all nonprokaryotic RNA polymerases (1, 2). The two largest subunits of the archaeal and eukaryal polymerases (termed RPB1 and RPB2) bear functional and structural homology with the β′ and β subunits of the prototypical bacterial enzyme. The remaining subunits can be divided into two groups. One set is unique to a specific eukaryotic polymerase class (RPB3, RPB4, RPB7, RPB9, and RPB11 for RNAPII) and the other set is shared among the three eukaryotic polymerases (RPB5, RPB6, RPB8, RPB10, and RPB12).
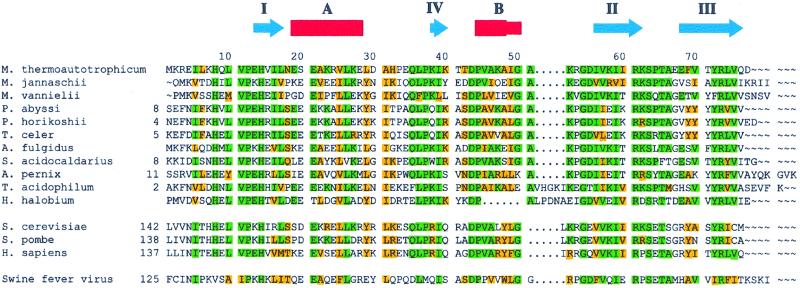
Roles for the RNA polymerase II-specific subunits have been elucidated. RPB3 and RPB11 form a complex that bears structural and functional homology with the α subunit of bacterial RNA polymerases (3, 4). RPB4 and RPB7 form a ssDNA-binding complex, which is essential for transcription initiation (5). RPB9 shares functional and structural homology with TFIIS, a transcription elongation factor (6). The roles for the shared subunits are less clear, but RPB5 has been implicated in the transcription activation process and RPB6 in the initiation of transcription. The structure of RNA polymerase II has been determined at 5 Å resolution by using x-ray crystallography (7). The determination of a high-resolution structure of the complex will likely benefit from ancillary high-resolution structures for many of the smaller subunits. RPB5 is highly conserved among eukarya and archeons. In this study, we have determined the solution structure of the RPB5 homologue from the archeaon Methanobacterium thermoautotrophicum (M.t.). The M.t. subunit (mtRPB5) is smaller than many other RPB5s, suggesting that mtRPB5 contains the minimal functional elements (Fig. 1).
Figure 1.
Alignment of RPB5 from M. thermoautotrophicum (GenBank accession no. O27122) with homologous proteins from the following: Archae: Methanococcus jannaschii (Q58443), Methanococcus vannielii (P41559), Pyrococcus abyssi (CAB49535), Pyrococcus horikoshii (O74019), Thermococcus celer (P31815), Archaeoglobus fulgidus (O28394), Aeropyrum pernix (Q9YAT3), Sulfolobus acidocaldarius (P115210), Thermoplasma acidophilum (Q03588), Halobacterium halobium (P15740). Eukarya: Saccharomyces cerevisiae (NP_009712), Schizosaccharomyces pombe (AF020780), Homo sapiens (CAA11843). Virus: swine fever virus (1097499). Amino acids that are identical and similar in M.t. with at least five other species are highlighted in green and yellow, respectively. The NMR-derived secondary structural elements of mtRPB5 are illustrated above the alignment by using the nomenclature referred to in the text.
Materials and Methods
Sample Preparation.
The sequence encoding the full length protein from M. thermoautotrophicum ΔH was cloned into the pET15b vector (Novagen) as a fusion with an N-terminal hexa-histidine tag and a thrombin cleavage site. The fusion protein was overexpressed in Escherichia coli BL21-Gold (DE3) (Stratagene) cells, which carry an extra plasmid encoding for three rare E. coli tRNAs (AGG, AGA, ATA). The cells were grown at 37°C in M9 minimal media with either 2.5 g/liter of 13C-glucose, 0.7 g/liter 15N-NH4Cl, or both to an OD600 of 0.6 and were induced with isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM. The cells were grown for 5 h more before harvesting. Protein purification was carried out by using standard Ni affinity chromatography. The hexa-histidine tag was cleaved by incubation of the purified protein sample with thrombin overnight in a cutting buffer consisting of 50 mM Tris (pH 7.6), 150 mM NaCl, and 2.5 mM CaCl2 and then was passed through a second Ni column to remove the cleaved tag. The protein was then dialyzed into NMR buffer consisting of 150 mM NaCl and 25 mM phosphate (pH 6.5) and was concentrated by ultrafiltration to approximately 1 mM. Ten percent D2O was added to provide NMR lock signal. For NMR experiments that require a sample dissolved in D2O, the samples were lyophilized and then resuspended in D2O.
NMR Spectroscopy.
NMR spectra were acquired at 25°C by using Varian INOVA 500 and 600 MHz spectrometers equipped with pulse field gradient units and actively shielded triple resonance probes. All NMR data were processed by using nmrpipe software (8) and were analyzed with nmrview 3.0 (9). Backbone resonance assignments were achieved by using 15N-HSQC, HNCACB (10), CBCA(CO)NH (11), CCC-TOCSY (12), and HNCO (13) spectra. HCCH-TOCSY (14), HCC-TOCSY (15), and 15N-edited TOCSY spectra were used to assign sidechain protons. Unambiguous assignment of the two aromatic sidechains (Phe69 and Tyr72) and histidine sidechains was achieved by using a Cβ to aromatic proton correlation sequence (16). All of the backbone and Cβ resonances were assigned, and all of the nonlabile proton sidechains except for those of Met1 and Gln9 were assigned.
Structure Calculations.
Distance restraints were obtained from 15N-edited NOESY and CN-NOESY (17) spectra for the samples in H2O, and two-dimensional homonuclear NOESY and 13C-edited NOESY (18) spectra for the samples in D2O. All NOE mixing times were 150 ms. Dihedral angle restraints were derived from 3JHNHα values obtained via an HNHA (19) experiment. Dihedral angle ranges derived from talos (20) were also implemented when in agreement with the HNHA data. Twenty initial structures were generated by distance geometry and simulated annealing protocols in CNS (21) using a total of 161 (including 128 long range) NOEs, 41 dihedral, and 13 hydrogen bonding restraints. All 20 structures converged to the same general fold.
The 10 lowest energy structures were used as starting structures for further refinement by using aria (22, 23). The 10 lowest energy structures of each aria iteration were used to derive residue-specific assignments of previously unassigned NOEs. A total of 50 structures were refined in the last (ninth) iteration, and the 10 lowest energy were analyzed. A total of 1,430 unambiguous and 1,185 ambiguous distance restraints were obtained. The frequency window tolerance for assigning NOEs was ±0.05 ppm for the proton and ± 0.5 ppm for nitrogen and carbon shifts. The ARIA parameters, p, Tv, and Nv, were as in the work by Nilges et al. (24). The 10 lowest energy structures had no NOE violations greater than 0.5 Å or dihedral angle violations greater than 5°. Residues 1–11 are disordered and were not included in the structural models. The coordinates and restraint tables have been submitted to the Protein Data Bank (ID code 1eik), and the NMR chemical shifts have been deposited in BioMagResBank (accession no. 4678).
Results and Discussion
RPB5 Adopts a Stable Independently Folded Structure.
MtRPB5 was cloned as part of a M. thermoautotrophicum structural proteomics project. Our aim was to determine the feasibility of using NMR to determine the structures of a large number of proteins. A total of 186 small M.t. constructs were expressed in 15N-enriched M9 medium. The recombinant 15N-labeled proteins were purified and “screened” by using 15N-HSQC NMR spectra as a read-out of feasibility of the structure determination by NMR spectroscopy. These results, together with results from circular dichroism spectroscopy, indicated that mtRPB5 adopts a stable, folded structure in NMR buffer (see Materials and Methods) with a mid-point denaturation temperature of 85°C. Equilibrium sedimentation data indicate that mtRPB5 is monomeric.
The Solution Structure of mtRPB5.
The ensemble of structures calculated for mtRPB5 is presented in Fig. 2B, with statistical parameters summarized in Table 1. MtRPB5 is composed of a four-stranded β-sheet that is sandwiched by two helices. The four β-strands comprise residues 13–17 (strand I), which is anti-parallel to the β-hairpin formed by residues 56–62 (strand II) and residues 68–76 (strand III), and residues 38–40 (strand IV), which is parallel to strand III. The first α-helix (referred to as helix A) is nine residues in length (19–28). Helix B is composed of one turn of an α-helix (44–48) followed by another turn of a 3–10 helix (48–50). Residues 1–12 have random coil like backbone chemical shifts and no long- or medium-range NOEs indicative of an unstructured polypeptide chain. A search of the dali (25) database showed no structural similarity to other published three-dimensional structures.
Figure 2.
NMR-derived structure of mtRPB5. (A) Ribbon diagram of a representative structure. (B) The backbone trace of the 10 lowest energy structures superimposed from residue 12 to 77. Drawn in blue are the solvent-inaccessible sidechains that form the hydrophobic core. The β-strands are cyan, and helices are red. Diagrams were created by using molmol (33).
Table 1.
Structural statistics of 10 lowest energy structures of mtRPB5
| Restraint type | Number of restraints |
| Total unambiguous NOE distances | 1,430 |
| Intra-residue | 738 |
| Sequential | 280 |
| Medium range | 143 |
| Long range | 269 |
| Total ambiguous NOES | 1,185 |
| Hydrogen bond constraints | 26 |
| Dihedral angles φ1 | 41 |
| Backbone stereochemistry (residues 12–77) | Percent residues in |
| Most favorable regions | 89% |
| Allowed regions | 7% |
| Generously allowed regions | 3% |
| Non-allowed regions | 0.4% |
| Structure ensemble | Pairwise rms deviation (Å) |
| Backbone (12–77) | 0.59 |
| Heavy atoms (12–77) | 0.94 |
The mtRPB5 structure is stabilized by two hydrophobic cores (Fig. 2B). The first comprises five sidechains: Leu17 at the tip of strand I, Val25, Leu26 from helix A, Ile56 from strand II, and Tyr72 from the strand III. The second core contains sidechains from Ile16 from strand I, Ile39, Val45, and Ala51 from helix B, Val57 from strand II, and Val75 from strand III. These hydrophobic core residues are conserved in archaea and eukarya (Fig. 1), strongly suggesting that these homologous proteins all adopt a tertiary structure very similar to that of mtRPB5.
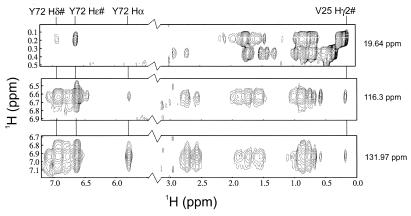
The secondary structure and β-sheet topology observed here for mtRPB5 are similar to those previously determined by NMR for RPB5 from Methanococcus jannaschii (43% sequence identity) (26). Our tertiary structure, however, differs significantly from that reported for M. jannaschii. In mtRPB5, helix A lies on the opposite face of the β-sheet from helix B, whereas in the M. jannaschii structure, helices A and B are on the same side of the β-sheet. The position of helix A in mtRPB5 is supported by several prominent and unambiguous NOEs from Tyr 72 in the β-sheet to Val 25 in helix A (Fig. 3) as well as NOEs from Ile 49 in helix B to His 14 and Ile 16 in strand I (not shown). Tyr72 is the only tyrosine in mtRPB5, and the sequence-specific assignments for these residues are unambiguous.
Figure 3.
Strips from the three-dimensional 13C-edited NOESY in D2O at the carbon planes corresponding to the Cγ2 position of Val 25 (19.6 ppm), and the Cδ (131.9 ppm), and Cɛ (116.3 ppm) of Tyr 72 showing unambiguous NOEs between the Tyr 72 and Val 25.
To investigate the possibility that the previously reported structure could satisfy our chemical shift assignment and NOE data, we modeled our sequence onto the backbone conformation of mjRPB5 (PDB ID code 1hmj) by using swissmodel (27, 28) and constructed a 5 Å proton-proton contact map for mtRPB5 in this alternative conformation. None of the predicted long range NOEs between helix A and the β-sheet of the mjRPB5 model could be reconciled with our NOE data. We also used this mjRPB5 model as a starting structure for simulated annealing calculations using aria (22–24), which assigns ambiguous NOEs based on a starting model. Using a variety of calculation protocols, we could not reproduce an acceptable ensemble of structures that was consistent with the mjRPB5 model and our chemical shift and NOE assignments.
Surface Features and Protein-Protein Interactions.
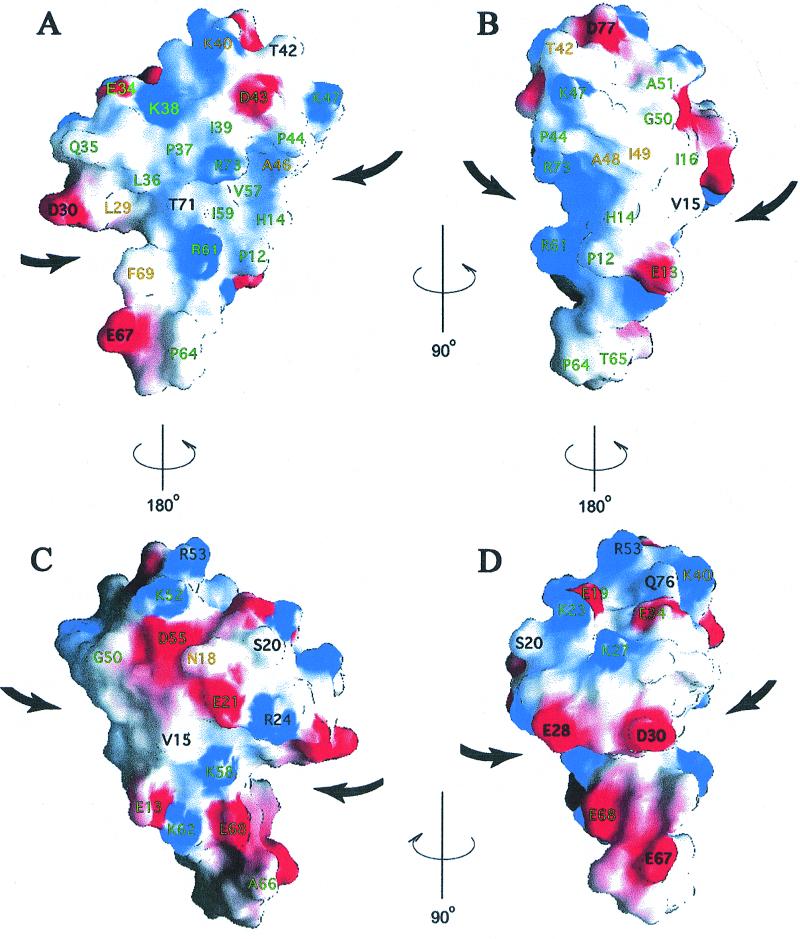
RPB5 is a highly basic protein (calculated pI of 10.1) that resembles a flattened mushroom with the β-hairpin forming the stem and a skewed cap formed mainly by the two helices. There is a groove formed by the “underside” of the mushroom cap surrounding the stem (bold arrows in Fig. 4). This distinctive shape may be involved in specific protein-protein interactions. Approximately half of the total surface area comprises a large, conserved hydrophobic patch with two absolutely conserved Arg residues in the center (Fig. 4 A and B). This surface is a likely candidate for conserved interactions with other polymerase subunits and/or other transcription factors. The presence of the two Arg residues in the middle of this exposed hydrophobic patch at this side of the protein likely prevents the self-aggregation of mtRPB5 even at the high concentrations used for NMR. These conserved Arg residues are also likely to confer a degree of specificity to protein-protein interactions because burial of the positive charges within a multisubunit complex will only be favorable if they are paired with appropriately placed acidic groups.
Figure 4.
Four views of the surface charge distribution calculated and drawn by using grasp (34), with red representing negative potential (−6.2 kT, full intensity) and blue positive potential (7.6 kT, full intensity). Conserved residues are labeled according to the color scheme in Fig. 1. A is in the same orientation as Fig. 2.
The opposite surface of the molecule (Fig. 4 C and D) is highly charged with a more uniform distribution of positive and negative charges. In this region, we see fewer conserved residues, even among the archaea, suggesting that it may be either solvent-exposed or involved in a species-specific protein-protein interaction. It is interesting to note that Thermoplasma acidophilum has a five-residue insertion between M.t. residues 51 and 52, and four additional residues at the C terminus. Residue 51 is located near C-terminal residue 77 (Fig. 4B), and insertions at residues 51 and 77 would likely introduce an extra lobe at this side of the protein without disrupting the overall fold or existing surface of the protein.
Thermal Stability of mtRPB5.
The sequence and structural features that confer thermo-stable properties to proteins is of great interest in biotechnology. The high thermal stability of mjRPB5 (Tm = 85°C) has been attributed to its high isoleucine content (26). However, mtRPB5 (Tm = 85°C) contains only 10% Ile compared with 17% for mjRPB5, and, therefore, an alternative explanation is more likely. It has been suggested that an increased number of surface ion pairs found in thermophilic proteins may be partly responsible for thermal stability (refs. 29 and 30 and references therein). An increased number of hydrogen bonds and a larger polar surface area, which increases the hydrogen bonding density with water, were also suggested to augment the thermal stability of proteins (29). Vogt et al. (29) define such surface ion pairs as any positively charged sidechain nitrogen atoms in Lys, Arg, and His that are within 4 Å of a negatively charged sidechain oxygen atom of Asp and Glu. For mtRPB5, there are four such pairs: Glu13 to Lys62, Glu19 to Lys23, Glu34 to Lys38, and Glu21 to Arg24. Most of these are conserved in other thermophilic archaeons, but none of these ion pairs are found in the eukaryal or viral RBP5s. The presence of these surface ion pairs likely contributes to the high thermal stability of mtRPB5 and other archaeal subunits. Interestingly, the Glu19 to Lys23 ion pair is conserved in M. jannaschii and Archaeoglobus fulgidus, but the residue types are interchanged.
Comparison with Eukaryotic RPB5s.
The RPB3, RPB11, and RPB10 homologues of archaeal RNAP were shown to form a similar complex to that formed by these subunits in eukaryotic RNAPs (31). This suggests that the quaternary interactions within archaeal RNAPs are similar to those of eukaryotes. RPB5 is an essential subunit in all three eukaryotic RNA polymerases, but the archaeal RPB5s are much smaller than those of eukaryotic RPB5. Mutational studies of the N-terminal region of yeast RPB5, which is not present in the archaeal RPB5s, have implicated this region in interactions with TFIIB (32). Because archaeal RPB5s are highly conserved with the C-terminal regions of the eukaryal subunits, it is likely that the eukaryal proteins are modular with a C-terminal domain that carries out essential polymerase functions and an N-terminal domain that carries out regulatory functions specific to higher organisms. The absence of this N-terminal domain of RPB5 in the archaeal proteins could potentially be caused by an ancient gene fusion event from two separate archaeal proteins. To investigate this possibility, we performed a search of the M.t. genome for ORFs that may have weak homology to the N terminus of eukaryotic RPB5s. We found no candidate ORFs in M.t. that may encode a similar region.
Acknowledgments
We are grateful to Lewis Kay for providing NMR pulse sequences. We thank Sandy Go and Karen Maxwell for technical assistance. This work was supported by grants from the Medical Research Council (MRC) of Canada, the National Cancer Institute of Canada with funds from the Canadian Cancer Society and the Terry Fox Run, and the Ontario Cancer Institute. C.H.A. and A.M.E. are Scientists of the Medical Research Council of Canada.
Abbreviations
- RNAP
RNA polymerase
- M.t.
Methanobacterium thermoautotrophicum
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1eik). Chemical shift assignments have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 4678).
References
- 1.Gaasterland T. Curr Opin Microbiol. 1999;2:542–547. doi: 10.1016/s1369-5274(99)00014-4. [DOI] [PubMed] [Google Scholar]
- 2.Langer D, Hain J, Thuriaux P, Zillig W. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azuma Y, Yamagishi M, Ueshima R, Ishihama A. Nucleic Acids Res. 1993;19:461–468. doi: 10.1093/nar/19.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakurai H, Miyao T, Ishihama A. Gene. 1996;180:63–67. doi: 10.1016/s0378-1119(96)00406-4. [DOI] [PubMed] [Google Scholar]
- 5.Edwards A M, Kane C M, Young R M, Kornberg R D. J Biol Chem. 1991;266:71–75. [PubMed] [Google Scholar]
- 6.Wang B, Jones D N M, Kaine B P, Weiss M A. Structure (London) 1998;6:555–569. doi: 10.1016/s0969-2126(98)00058-6. [DOI] [PubMed] [Google Scholar]
- 7.Fu J, Gnatt A L, Bushnell D A, Jensen G J, Thompson N E, Burgess R R, David P R, Kornberg R D. Cell. 1999;98:799–810. doi: 10.1016/s0092-8674(00)81514-7. [DOI] [PubMed] [Google Scholar]
- 8.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;66:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 9.Johnson B A, Blevins R A. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 10.Kay L E, Xu G Y, Yamazaki T. J Magn Reson Ser A. 1994;109:129–133. [Google Scholar]
- 11.Grzesiek S, Bax A. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 12.Montelione G T, Lyonns B A, Emerson S D, Tashiro M. J Am Chem Soc. 1992;114:10974–10975. [Google Scholar]
- 13.Muhandiram D R, Kay L E. J Magn Reson Ser B. 1994;103:203–216. [Google Scholar]
- 14.Kay L E, Xu G Y, Singer A U, Muhandiram D R, Forman-Kay J. J Magn Reson Ser B. 1993;101:133–136. [Google Scholar]
- 15.Logan T M, Olejniczak E T, Xu R X, Fesik S W. J Biomol NMR. 1993;3:225–231. doi: 10.1007/BF00178264. [DOI] [PubMed] [Google Scholar]
- 16.Yamasaki T, Forman-Kay J D, Kay L E. J Am Chem Soc. 1993;115:11054. [Google Scholar]
- 17.Pascal S, Muhandiram T, Yamazaki T, Forman-Kay J D, Kay L E. J Magn Reson. 1994;101:197–201. [Google Scholar]
- 18.Ikura M, Bax A, Clore G M, Gronenborn A M. J Am Chem Soc. 1990;112:9020–9022. [Google Scholar]
- 19.Kuboniwa H, Grzesiek S, Delaglio F, Bax A. J Biomol NMR. 1994;4:871–878. doi: 10.1007/BF00398416. [DOI] [PubMed] [Google Scholar]
- 20.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 21.Brunger A T, Adams P D, Clore G M, Delano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges N, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Nilges M, O' Donoghue S. Prog Nucl Magn Reson Spectrosc. 1998;32:107–139. [Google Scholar]
- 23.Linge J P, Nilges M. J Biomol NMR. 1999;13:51–59. doi: 10.1023/a:1008365802830. [DOI] [PubMed] [Google Scholar]
- 24.Nilges M, Macias M, O'Donoghue S, Oschkinat H. J Mol Biol. 1997;269:408–422. doi: 10.1006/jmbi.1997.1044. [DOI] [PubMed] [Google Scholar]
- 25.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 26.Thiru A, Hodach M, Eloranta J J, Kostourou V, Weinzierl R O, Matthews S. J Mol Biol. 1999;287:753–760. doi: 10.1006/jmbi.1999.2638. [DOI] [PubMed] [Google Scholar]
- 27.Peitsch M C. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 28.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 29.Vogt G, Woell S, Argos P. J Mol Biol. 1997;269:631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- 30.Elcock A H. J Mol Biol. 1998;284:489–502. doi: 10.1006/jmbi.1998.2159. [DOI] [PubMed] [Google Scholar]
- 31.Eloranta J J, Kato A, Teng M S, Weinzierl R O J. Nucleic Acids Res. 1998;26:5562–5567. doi: 10.1093/nar/26.24.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyao T, Woychik N A. Proc Natl Acad Sci USA. 1998;95:15281–15286. doi: 10.1073/pnas.95.26.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koradi R, Billeter M, Wuthrich K. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 34.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]