Abstract
Recent analyses of global pig populations revealed strict correlations between mtDNA phylogenies and geographic locations. An exception was the monophyletic “Pacific clade” (PC) of pigs not previously linked to any specific location. We examined mtDNA sequences of two varieties of Vanuatu sacred pigs, the male pseudohermaphroditic Narave from the island of Malo (n = 9) and the hairless Kapia from the island of Tanna (n = 9), as well as control pigs (n = 21) from the islands of Malo, Tanna, and Epi and compared them with GenBank sequences to determine (i) the distribution of PC and introduced domestic lineages within Vanuatu, (ii) relationship between the Narave and Kapia, and (iii) origin of the PC. All of the Narave share two PC mtDNA sequences, one of which matches the sequence of a Narave collected in 1927, consistent with an unbroken maternal descent of these intersex pigs from the original pigs brought to Vanuatu 3,200 years ago. One-third of the Kapia share a single PC lineage also found in the Narave. The remaining Kapia lineages are associated with recently introduced, globally distributed domestic breeds. The predominant Narave lineage is also shared with two wild boars from Vietnam. These data suggest that PC pigs were recently domesticated within Southeast Asia and dispersed during the human colonization of Remote Oceania associated with the Lapita cultural complex. More extensive sampling of Southeast Asian wild boar diversity may refine the location of Pacific pig domestication and potentially the proximate homeland of the Lapita cultural complex.
Keywords: mtDNA, Pacific, Remote Oceania, Sus scrofa
Recent analyses of mitochondrial DNA (mtDNA) sequences from global wild boar and domestic pig populations describe a tight correlation between geography and phylogeny. The global pig phylogeny is rooted in Southeast Asia, indicating an ancient origin of wild boars in this region followed by dispersal throughout Eurasia. From this dispersed population of wild boars at least six independent domestications of pigs in Europe (n = 2), Asia (n = 2), India (n = 1), and Southeast Asia (n = 1) occurred within the past 9,000 years (1). Of the 686 wild boars and domestic pigs examined, only 2% did not fit a strict phylogeographic pattern (1), mainly reflecting the dispersal of Euro-Asian hybrid domesticates (2) over the last 200 years. Within Asia the correlation between geography and phylogeny is maintained with genetic clines observed within Japan (3), across East Asia (4), and from Southeast Asia through to North Asia (5). In contrast to this general pattern, Pacific Island domestic pigs form a monophyletic “Pacific clade” (PC) with limited diversity and no close associations with other domestic breeds or wild boar populations (1).
Traditional morphology defined a regional species, Sus papuensis in New Guinea, from which other Pacific pigs were thought to be derived. The lack of a close association between the morphologically and genetically distinct PC of pigs and other Asian pig populations suggested that there may have been an additional independent pig domestication from a long resident wild boar population in Wallacea (1). A competing hypothesis views the pig as a recent introduction to the Pacific that was dispersed over the last 3,500 years during the human expansion associated with the Lapita cultural complex (6), implying a recent domestication in Southeast Asia. Neither of these hypotheses is consistent with all of the current data. The first hypothesis predicts that fossil evidence of pigs in Wallacea should exist, but no undisputed evidence of pigs in the Western Pacific before the Lapita expansion has yet been unearthed (7, 8). The second hypothesis predicts that the origin of the PC of pigs should coincide with the putative homeland of the Lapita cultural complex in Taiwan (6, 7, 9, 10). In contrast, wild boars from Taiwan are not closely related to the PC of pigs (1). Thus, the origin of the Pacific pig, the most important domestic animal of the region, has remained unresolved.
The human colonization of Remote Oceania (11) is associated with the dispersal of the pig, dog, chicken, and rat (7). As humans moved farther into the Pacific, the island sizes decreased and the distances between islands increased, creating a series of biogeographical barriers for natural dispersal that increased the importance of human transported landscapes (7). The maintenance of these animals on islands varied, reflecting ecological constraints. On smaller islands of Micronesia and Polynesia in particular the pig disappeared from the archaeological record before European contact, presumably because of its adverse competition with humans for land resources (12, 13). The Pacific rat, Rattus exulans, is thus the most widespread of the introduced animals of the Pacific and as such has been used effectively as a proxy for human settlement and exchange (14–16). Unlike the European introduced rats of distinct species that could not interbreed with the Pacific rat, studies of Pacific pigs, dogs, and chickens are complicated by the introduction of conspecifics within the last few hundred years. The extent to which European introduced pigs interbred with Pacific pigs has not as yet been well studied.
Within the Pacific, the pig was culturally the most important of the domestic animals. Beyond serving as food, pigs were the main unit of food storage in the humid environment of the tropical Pacific, and by extension, the basis for societal wealth and ecological balance through cycles of competitive giving (17). Throughout the region pigs could and can be used for brideprice, feature importantly as a major ritual item in economic exchanges and as the major ritual sacrificial animal, and are often prominent in origin and subsequent mythology (17–19). Moreover, in the systems of certain Vanuatu cultures, pigs can have souls similar to but different from human souls, and consequently ritual pig manipulation can assist in human spiritual progression. Thus, the importance of pigs in the Pacific reaches its pinnacle in Vanuatu, currently the most culturally diverse region of the world per capita with 113 indigenous languages spoken by ≈200,000 people inhabiting ≈80 islands (20). The origin and maintenance of this extreme cultural diversity among interacting populations within a circumscribed archipelago remains one of the most intriguing aspects of the Pacific.
Within Vanuatu several sacred breeds of pigs with extremely derived morphologies were developed and maintained for cultural purposes. The people of the north and central islands created “tusker” pigs that were imbued with spiritual significance and required for Nimangki, traditional chiefly rank taking. The tusks of boars continuously grow and are sharpened by the opposing upper canines. Avulsion of the upper canines allows the tusks to circularize, completing a full rotation in 6–7 years and a double circle in 10–12 years (18, 21). As the tusks circularize they puncture the cheek and sometimes the jaw, necessitating delicate care that can include the removal of lower jaw back teeth to create space for reentry of the tusk. Tusker boars were often traditionally castrated to reduce their aggression, facilitate their care, and reduce the risks of fights and broken tusks (21).
To attain successively higher chiefly ranks, tusker boars had to be produced and killed. To obtain the highest ranks, in addition to the normal tuskers, special Narave tuskers also had to be produced and killed. The Narave are intersex male boars with variable testis development, hormone production, and spermatogenesis (22, 23) (Fig. 1a). Because Narave often have no external testes, castration is usually not possible. As a result, they are the most difficult tuskers to rear and manage. Early studies suggested that an X-linked mode of inheritance produced this phenotype (23, 24), although more recent studies have identified several autosomal recessive mutations that may be responsible for this condition. For example, mutations in genes that encode steroidogenic enzymes such as 5α-reductase (25) and p450c17 (26) have been shown to cause male pseudohermaphroditism in humans.
Fig. 1.
Narave pigs from Malo and Kapia pigs from Tanna. (a) A Narave tusker with pierced cheeks and two examples of aphallic morphologies with variable testes development. (b) Two Kapia including one wearing the ochre face paint reserved for men of title on Tanna. All photographs were taken by J.K.M.
Our ongoing investigations of steroidogenic production in Narave have revealed a deficient responsiveness to gonadotropin that may result in insufficient steroid production for complete masculinization (D.L.G., unpublished data). Because Narave are incapable of breeding, the intersex morphology is maintained through mothers and sisters of Narave. Typically a mother or a carrier sister (called “soke-rave” or “good-blooded” on Malo) of a Narave will farrow at least one Narave per litter (23, 24). This is the highest rate of intersex animal production of which we are aware.
In the southern islands of Vanuatu Narave are unknown; instead, other sacred breeds of pigs were cultivated. On the island of Tanna, the Kapia, a hairless breed of pig, was developed. In southern Tanna, the Kapia are thought to be the creator Kumwesen's first attempt at humanity and so are culturally viewed as the elder siblings of humans. These pigs can have status equivalent to a “man of title” and are thus able to wear prestige items like male bark cloth belts and ochre face paint (Fig. 1b). Culturally the Kapia are not considered pigs and cannot be exchanged for them but rather are equivalent to humans and sea turtles for the purposes of ceremonial exchanges (27). On Tanna, Kapia represent ≈6% of each generation, but the hairless morphology is not associated with sex-biased inheritance.
To further investigate the genetic origins of domesticated pigs in Vanuatu and their relevance to the human expansion into Oceania, we analyzed mtDNA variation in both Narave and Kapia pigs. We collected blood, hair, skin, or tooth samples from 39 pigs from three islands of Vanuatu: Malo (Narave = 9; controls = 8), Tanna (Kapia = 9; controls = 11), and Epi (controls = 2). The control pigs from Malo and Tanna were morphologically normal pigs from the same or a nearby village as the Narave and Kapia. The control pigs from Epi were included because this island is geographically intermediate between Malo and Tanna. DNA was extracted from each sample, and ≈600 bases of the mtDNA control region were sequenced. The resulting data were compared with all pig sequences currently deposited in GenBank to address the following three issues: (i) the distribution of PC and global domesticate lineages within Vanuatu, (ii) the diversity within and between the sacred intersex Narave and hairless Kapia pigs, and (iii) the origin of the PC and its implications for the human settlement of the Pacific.
Results and Discussion
Distributions of PC and Global Domesticate Lineages.
The 39 Vanuatu pigs were represented by eight mtDNA sequences, with lineages 1–3 being associated with the PC and lineages 4–8 being associated with globally distributed domestic (GDD) breeds (Table 1). More than half of the pigs' mtDNA (20/39) belongs to PC lineages, although their frequencies varied among islands. Nearly all of the pigs from Malo (15/17), including all of the Narave, were PC pigs.
Table 1.
Distribution of the eight mtDNA lineages among islands and pig breeds
| Lineage | Island |
GenBank associations | |||
|---|---|---|---|---|---|
| Malo total (Narave/control) | Tanna total (Kapia/control) | Epi | Vanuatu total | ||
| 1 | 6 (5/1) | 3 (3/0) | 9 | PC | |
| 2 | 9 (4/5) | 1 (0/1) | 10 | PC | |
| 3 | 1 (0/1) | 1 | PC | ||
| 4 | 10 (3/7) | 1 | 11 | Jiangquhai, Xiang, Large Black, Landrace | |
| 5 | 3 (2/1) | 1 | 4 | Large White, Creole, Berkshire, Jabugo, Tia Meslan, Yorkshire, Tamworth, Satsuma, Longlia | |
| 6 | 2 (0/2) | 2 | Middle White, Moncai, Tongcheng, Berkshire | ||
| 7 | 1 (1/0) | 1 | Landrace, Berkshire, Tamworth, Duroc, Gloucester Old Spot, British Saddleback | ||
| 8 | 1 (0/1) | 1 | Large Black, Duroc, Berkshire | ||
| Total | 17 (9/8) | 20 (9/11) | 2 | 39 | |
| PC | 88% (100%/75%) | 25% (33%/18%) | 0% | 51% | |
Tanna was historically a major exporter of pigs, providing >8,000 pigs in 1865 alone to sandalwood traders destined for Australia (20). Thus, it is ironic that 75% of the Tanna pigs we sampled, including 66% of the Kapia, have mtDNA lineages associated with recently introduced GDD breeds (Table 1). Lineages 4 (n = 11) and 5 (n = 4) are associated with many GDD breeds (Table 1). Lineages 4 and 5 are found on both Tanna and Epi, accounting for 79% of all of the introduced lineages we observed. Lineage 4 accounts for 58% of the GDD lineages and is found in 5 of the 12 Tanna villages sampled as well as on the island of Epi. Based mainly on their dark coat color that provides added protection from the tropical sun, it was thought that the domestic breeds recently introduced to Vanuatu were Berkshire, Large Black, Hampshire, and Wessex Saddlebacks (21). The five introduced mtDNA lineages we identified are associated with the breeds Berkshire and Large Black (among others), but not Hampshire, and Wessex Saddlebacks (Table 1).
These data highlight extensive introgression of mtDNA lineages associated with GDD on Tanna. Although only two samples were collected on Epi, both of these are GDD lineages that are also found on Tanna, consistent with the large numbers of French plantations on Epi during the colonial era. In contrast, 88% of the pigs on Malo, both Narave and controls, are PC lineages, indicating low levels of introgression of recently introduced lineages.
Diversity Within and Between the Sacred Pseudohermaphroditic Narave and Hairless Kapia Pigs.
All nine of the Narave pigs sampled shared two PC sequences (lineages 1 and 2) (Table 1). The Narave were expected to resist introgression of mtDNA because their sacred intersex morphology predates European contact and is perpetuated maternally. Lineage 2 has not been reported previously, reflecting its local restriction to Vanuatu or, alternatively, the lack of adequate sampling of Southeast Asian and other Pacific pig populations. The nine Narave pigs were collected from four villages (three villages on Malo and one village recently relocated to the neighboring island of Espiritu Santo). Of the three villages where at least two Narave were collected, both lineages 1 and 2 were observed. The distribution of these lineages and the maternal inheritance of the Narave trait suggest that the putative intersex mutation is not recent and predates the divergence of lineages 1 and 2.
In contrast to the Narave, 66% (6/9) of the Kapia have mtDNA lineages associated with GDD (Table 1). The three PC Kapia all share lineage 1, also found in the Narave, indicating a recent derivation of both morphologies from the same ancestral stock. In contrast, the six Kapia sequences inferred to have been recently introduced are represented by three distinct lineages (Tables 1 and 2). The low diversity of PC lineages of the Kapia may reflect either a restricted founding population or the replacement of greater diversity by recent introgression of mtDNA from GDD. Additional sampling of Kapia will be required to distinguish between these alternatives. The inheritance pattern of the hairless morphology remains unclear, but the high rates of mtDNA introgression suggests that it is biparentally inherited and maintained by continuous artificial selection regardless of hybridization.
Table 2.
Sequence variation among Vanuatu pigs and related Southeast Asian wild boars
| Position* | 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 | ||||||
|---|---|---|---|---|---|---|---|
| 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 | |||||||
| 6 6 6 6 6 7 7 7 7 7 7 8 8 8 8 8 8 8 8 8 9 9 9 9 | |||||||
| 1 4 5 7 7 1 3 3 4 5 5 2 2 2 4 4 7 8 9 9 0 2 7 9 | |||||||
| 6 9 7 5 6 4 6 7 1 7 9 2 3 6 0 1 8 7 5 7 9 6 7 5 | |||||||
| Lineage | n | Group | Location | Reference | Source† | GenBank accession nos. | |
| 1 | T C A T T C C G C A C G A C T T G C C C C C A T | 12 | PC | Vanuatu, Vietnam | This study and refs. 1 and 5 | M, A | AY884704, AB053610, DQ994618, DQ994620, DQ994621, DQ994631, DQ994633, DQ994634, DQ994637, DQ994638, DQ994654 |
| 2 | … … … . T … … … … . | 10 | PC | Vanuatu | This study | M | DQ994619, DQ994622, DQ994623, DQ994624, DQ994627, DQ994628, DQ994629, DQ994630, DQ994632, DQ994653 |
| 3 | … … … … … . A … … . | 6 | PC | Polynesia, Papua New Guinea, Vanuatu | This study and refs. 1 and 28 | M, A | DQ444710, AY884821, AY884670, AY884673, DQ994646 |
| 9 | … … … … . T . . A … … . | 1 | PC | Vanuatu | Ref. 1 | A | AY884702 |
| 10 | … … … . T … . . A … … . | 1 | PC | Papua New Guinea | Ref. 1 | A | AY884637 |
| 11 | . T … … … … . . A … … . | 1 | PC | Papua New Guinea | Ref. 1 | A | AY884615 |
| 12 | . T … . . A … … . . A … … . | 1 | PC | Halmahera | Ref. 1 | A | AY884688 |
| 13 | … . . T T A . . T … … … … . | 1 | SEAWB | RyuKyu | Ref. 29 | M | AB015089 |
| 14 | . T … T . A . . T … … … … . | 2 | SEAWB | RyuKyu | Ref. 29 | M | AB015087, D42184 |
| 15 | C T … T . A . . T … … … … . | 2 | SEAWB | RyuKyu | Unpub. data‡ | A | AB050873, AB050874 |
| 16 | … . . T T A T . T … … … … . | 1 | SEAWB | RyuKyu | Ref. 29 | M | AB015090 |
| 17 | . T … T T A . . T … … … … . | 1 | SEAWB | RyuKyu | Ref. 29 | M | AB015088 |
| 18 | C T … T . A . . T … … … . T . . | 1 | SEAWB | RyuKyu | Unpub. data‡ | A | AB050870 |
| 19 | … . . T . A . . T A … . . T … . G . | 3 | SEAWB | Vietnam | Ref. 5 | A | AB053619, AB053618 |
| 20 | … C … A T . T A … . . T … … | 4 | SEAWB | Vietnam | Ref. 5 | A | AB053611 |
| 21 | . T G … . A . . T A … … … … | 1 | SEAWB | Australia | Ref. 30 | M | AY463091 |
| 22 | . T … . . A . . T A … … T T … . | 2 | SEAWB | Myanmar | Ref. 1 | A | AY884623, AY884712 |
| 23 | … . . T . A . G . . G T . C … … . . | 1 | SEAWB | China | Ref. 1 | A | AY884610 |
| 4 | C T . . C . . A T . T A … . A … … C | 11 | GDD | Vanuatu | This study | M | DQ994635, DQ994641, DQ994642, DQ994643, DQ994644, DQ994645, DQ994647, DQ994649, DQ994650, DQ994652, DQ994655 |
| 5 | C T . . C . . A T . T A . . C . A T … … | 4 | GDD | Vanuatu | This study | M | DQ994636, DQ994639, DQ994651, DQ994656 |
| 6 | C T … . T A T . T A … . A T … . . C | 2 | GDD | Vanuatu | This study | M | DQ994625, DQ994626 |
| 7 | . T . . C . . A T . T A . . C . A T … . . C | 1 | GDD | Vanuatu | This study | M | DQ994648 |
| 8 | C T . . C T . A T . T A … . A … T … | 1 | GDD | Vanuatu | This study | M | DQ994640 |
*Position numbering as in ref. 5.
†Source: M, modern; A, archaeological sample.
‡T. Watanobe, N. Ishiguro, A. Matsui, and M. Nakano, unpublished data.
The Narave and Kapia are both high-maintenance breeds. As mentioned above, the Narave attain their cultural value through the growth of circular tusks that are fragile and require extensive care and feeding for at least 6 years. This extended care is exacerbated by the often undescended testes and consequent inability to reduce aggression by means of castration. Likewise, the hairlessness of the Kapia leaves them vulnerable to sunburn in the tropics. Thus, both of these sacred pig breeds require more care to maintain than ordinary pigs. From a utilitarian perspective it seems paradoxical that pigs that require more work to maintain are considered of greater value. Narave tuskers for example, are worth twice that of normal boars with the same degree of circularized tusks. Within the traditional systems of Vanuatu, the increased value of the high maintenance Narave and Kapia thus serves as honest reflections of the additional effort required to produce and rear them.
Origin of the PC and Implications for Human Settlement of the Pacific.
To estimate the origin of the PC of pigs, we compared our Vanuatu lineage 1 sequence with all pig sequences currently deposited in GenBank. All pig sequences that were within six substitutions from lineage 1 were retained, and their geographic locations were compiled (Table 2). A network diagram relating our 39 Vanuatu pigs and all of the lineages identified in this search is shown in Fig. 2. Our database and literature search criterion identified an additional 31 pig sequences derived from modern samples, bone and teeth collected within the past 150 years, and archaeological samples resulting in 70 pigs sharing 23 distinct lineages (Table 2 and Fig. 2). All of the pigs identified are either Pacific island pigs (French Polynesia, Hawai'i, Papua New Guinea, and Vanuatu) or wild boars from Southeast Asia (China, Halmahera, Myanmar, Ryukyu Islands, and Vietnam). The exception is a single feral pig from Australia, assumed to have been recently introduced from Southeast Asia (1, 30) but that may have descended from one of the thousands of pigs acquired from Tanna in the 1800s (21). In particular, we found that lineage 1 observed in Narave samples collected from Vanuatu both recently and in 1927 by Baker (23, 24) (Oxford University Museum of Natural History Staff, personal communication) is also shared by two wild boars caught on separate hunting expeditions in northern Vietnam (5).
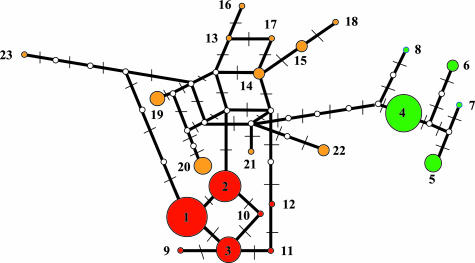
Fig. 2.
Network relationships among the Vanuatu pigs and mtDNA sequences from GenBank within six substitutions of the main Narave sequence (lineage 1). The lineage numbers are described in Tables 1 and 2. The areas of the circles are proportional to the number of individuals sharing a given sequence as listed in Table 2. The hash marks denote the number of inferred substitutions distinguishing each sequence. The three groups are identified by color as follows: red, PC; orange, SEAWBC; green, GDD. The white circles reflect sequences as yet unobserved but presumed to exist among Southeast Asian wild boar populations.
The 23 distinct lineages form three groups defined by increasing numbers of substitutions relative to lineage 1. The PC lineages are within 3 substitutions, the “Southeast Asian wild boar clade” (SEAWBC) lineages are within 4–6 substitutions, and the GDD sequences are either 9 or 10 substitutions from lineage 1, respectively (Table 2 and Fig. 2). The PC pigs are characterized relative to the SEAWBC by four transition substitutions at bases 15,714 (100%), 15,737 (86%), 15,878 (86%), and 15,759 (66%), highlighting the diversity of the wild boars currently found in Vietnam (Table 2).
These data indicate that the PC of pigs is nested within a larger clade consisting exclusively of Southeast Asian wild boars. The PC of pigs can be derived from the SEAWBC via at least three mutational pathways (Fig. 2), highlighting the ambiguity of interpreting sequence phylogenies generated over short time periods. The wild boars within this SEAWBC are distributed from Myanmar to the Ryukyu Islands, consistent with the correlation between phylogeny and geography observed in analyses of global pig diversity. The sharing of a single mtDNA sequence (lineage 1) by Narave, Kapia, control pigs, a Narave bone sample collected from Vanuatu in 1927, and also Vietnamese wild boars of extremely large size (5) indicates a recent domestication and size reduction in Southeast Asia followed by transport into the Pacific and culturally mediated diversification. The presence of mtDNA sequences of Vietnamese wild boars within the PC and also throughout the SEAWBC suggests that Southeast Asian pig diversity has not yet been adequately sampled. Although previously examined Taiwanese wild boars do not fall into our SEAWBC, the geographic distribution of this group extends from Myanmar to the Ryukyu Islands and, thus, spans Taiwan, the putative homeland of Austronesian languages and the antecedents of Lapita pottery (Fig. 3) (6, 7, 9, 10).
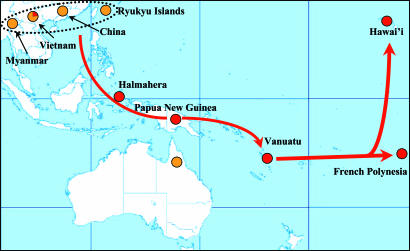
Fig. 3.
Map showing the geographic origins of the PC and SEAWBC sequences examined. The colors are as in Fig. 2. The dotted line encompasses the suggested region PC pig domestication. Note that 22% (2/9) of the Vietnamese wild boars are identical to the main Narave sequence (lineage 1). The red arrows indicate the inferred dispersal of PC pigs with the expansion of the Lapita cultural complex within the last 3,500 years.
The current data are most consistent with a recent domestication of the PC from the SEAWBC followed by its dispersal with the Lapita cultural complex within the last 3,200 years. The spread of the Lapita cultural complex is coincident with the distribution of a group of closely related human mtDNA sequences known by current terminology as Haplogroup B. The proximate origin of Pacific Islander Haplogroup B sequences remains unresolved, with putative origins ranging from Taiwan to Melanesia (31–38). If archaeological remains of pigs in the Pacific do not predate the expansion of the Lapita cultural complex as current data indicate (8), the identification of the location of pig domestication may provide insight on the geographic homeland of the Lapita people. The data presented here suggest that the origin of the Pacific pig may have been in coastal mainland Southeast Asia. Interestingly, Vietnam, where wild boars have mtDNA sequences shared with Vanuatu pigs (Narave, Kapia, and control pigs) is currently one of the few mainland Asian countries where Austronesian languages are still spoken. These languages are found in communities isolated by the recent population expansions of ethnic Han Chinese and Vietnamese. This process of cultural assimilation and absorption has likely characterized much of coastal Southeast Asia (9), obscuring the previously more widespread distribution of human mtDNA Haplogroup B that may have existed 3,500 years ago. Thus, the analyses of pig and wild boar diversity may provide information not available from examining only human variation as has been shown for analyses of the Pacific rat (14–16). Additional sampling of Southeast Asian wild boar diversity is required to further refine the site of domestication of the PC of pigs and potentially the proximate origin of the Lapita cultural complex (Fig. 3).
Our analyses of the genetic diversity of pigs in Vanuatu should be useful to the “Traditional Money Banks in Vanuatu” project funded by the United Nations Educational, Scientific and Cultural Organization (UNESCO) and Japanese Funds-In-Trust for the Preservation and Promotion of the Intangible Cultural Heritage (27). This project seeks to preserve traditional forms of wealth, including pure-blood Narave and Kapia, and integrate these into the local economy. Our data indicate that all of the Narave and the majority of the pigs from Malo have resisted introgression of recently introduced domestic breeds, at least in their maternal lines. Thus, the pigs from Malo may reflect to the greatest extent available the gene pool of the first pigs brought by Lapita colonists to Vanuatu ≈3,200 years ago (7).
Methods
DNA was extracted from pig blood, hair, and skin samples by using the QIAamp DNA Blood Mini Kit (Qiagen Biosciences, Germantown, MD) according to the manufacturer's instructions. DNA was extracted from teeth from two ritually killed pigs from the island of Epi by using a described ancient DNA protocol (39). After extraction the DNA was amplified by PCR using primers L15387 and H16108n as described (1). Approximately 600 bases of the mtDNA control region were sequenced in both directions with the BigDye Terminator Kit on an ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, CA).
We conducted a GenBank Blast search with the main Narave sequence (lineage 1). All sequences within six substitutions of lineage 1 (Table 2) comparing at least 408 bases were used to construct a genetic network (Fluxus-Engineering, fluxus-engineering.com) that was then modified by hand to highlight reticulations (Fig. 2).
Acknowledgments
We thank Chief Vira Joseph of Avunatari Village (Malo), Isi of Nasulnun Village (Santo, recently relocated from Malo), and Joseph Toa (Malo) for kindly providing samples from and stories about their sacred Narave; Chief Tom Numake from Tanna for access to the Kapia pigs and the Crappers, John of the kava crew at the Epi Guest House, Robin for the two boar jaws sampled; and Ralph Regenvanu and the staff of the Vanuatu Kaljoral Senta and Peter Kalsei for lively conversations on the cultural value of Narave over many shells and Ralph M. Garruto for initial chiding. This project was supported by start-up funds from the Research Foundation of Binghamton University (J.K.L.) and by a grant from Templar Sciences (State College, PA) (to J.K.M.).
Abbreviations
- PC
Pacific clade
- GDD
globally distributed domestic
- SEAWBC
Southeast Asian wild boar clade.
Footnotes
References
- 1.Larson G, Dobney K, Albarella U, Fang M, Matisoo-Smith E, Robins J, Lowden S, Finlayson H, Brand T, Willerslev E, et al. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 2.Giuffra E, Kijas JM, Amarger V, Carlborg O, Jeon JT, Andersson L. Genetics. 2000;154:1785–1791. doi: 10.1093/genetics/154.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanobe T, Ishiguro N, Nakano M. Zool Sci. 2003;20:1477–1489. doi: 10.2108/zsj.20.1477. [DOI] [PubMed] [Google Scholar]
- 4.Watanobe T, Ishiguro N, Nakano M, Takamiya H, Matsui A, Hongo H. J Mol Evol. 2002;55:222–231. doi: 10.1007/s00239-002-2320-6. [DOI] [PubMed] [Google Scholar]
- 5.Hongo H, Ishiguro N, Watanobe T, Shigehara N, Anezaki T, Long VT, Binh DV, Tien NT, Nam NH. Zool Sci. 2002;19:1329–1335. doi: 10.2108/zsj.19.1329. [DOI] [PubMed] [Google Scholar]
- 6.Kirch PV. The Lapita Peoples: Ancestors of the Oceanic World. Oxford: Blackwell; 1997. [Google Scholar]
- 7.Kirch PV. On the Road of the Winds: An Archaeological History of the Pacific Islands before European Contact. Berkeley: Univ of California; 2000. [Google Scholar]
- 8.Bellwood P, White P. Science. 2005;309:381. doi: 10.1126/science.309.5733.381a. [DOI] [PubMed] [Google Scholar]
- 9.Bellwood P. Prehistory of the Indo-Malaysian Archipelgo. Honolulu: Univ of Hawai'i Press; 1985. [Google Scholar]
- 10.Diamond JM. Nature. 1988;336:307–308. [Google Scholar]
- 11.Green RC. In: Man and a Half: Essays in Pacific Anthropology and Ethnobotony in Honour of Ralph Bulmer. Pawley A, editor. Auckland: Polynesian Soc; 1991. pp. 491–502. [Google Scholar]
- 12.Intoh M. Man Culture Oceania. 1986;2:1–26. [Google Scholar]
- 13.Kirch PV. In: Australian Archaeologist: Collected Papers in Honour of Jim Allen. Anderson A, Murray T, editors. Canberra: Coombs Academic; 2000. pp. 427–439. [Google Scholar]
- 14.Matisoo-Smith E, Roberts RM, Irwin GJ, Allen JS, Penny D, Lambert DM. Proc Natl Acad Sci USA. 1998;95:15145–15150. doi: 10.1073/pnas.95.25.15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matisoo-Smith E. Hum Biol. 2002;74:489–496. doi: 10.1353/hub.2002.0032. [DOI] [PubMed] [Google Scholar]
- 16.Matisoo-Smith E, Robins JH. Proc Natl Acad Sci USA. 2004;101:9167–9172. doi: 10.1073/pnas.0403120101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knauft BM. From Primitive to Postcolonial in Melanesia and Anthropology. Ann Arbor, MI: Univ of Michigan Press; 2002. [Google Scholar]
- 18.Harrison T. Savage Civilization. New York: Knopf; 1937. [Google Scholar]
- 19.Beckwith M. Hawaiian Mythology. Honolulu: Univ of Hawaii Press; 1976. [Google Scholar]
- 20.Tryon D. In: Arts of Vanuatu. Bonnemaison J, Kaufmann C, Huffman K, Tryon D, editors. Honolulu: Univ of Hawai'i Press; 1996. pp. 170–173. [Google Scholar]
- 21.Weightman B. Agriculture in Vanuatu. Portsmouth, UK: Grosvenor; 1989. [Google Scholar]
- 22.McIntyre JK. Sci New Guinea. 1997;22:137–152. [Google Scholar]
- 23.Baker JR. Br J Exp Biol. 1928;6:56–64. [Google Scholar]
- 24.Baker JR. Man and Animals in the New Hebrides. London: Routledge; 1929. [Google Scholar]
- 25.Imperato-McGinley J, Guerrero L, Gautier T, Peterson RE. Science. 1974;188:1213–1215. doi: 10.1126/science.186.4170.1213. [DOI] [PubMed] [Google Scholar]
- 26.Yanase T. J Steroid Biochem Mol Biol. 1995;53:153–157. doi: 10.1016/0960-0760(95)00029-y. [DOI] [PubMed] [Google Scholar]
- 27.Huffman K. Traditional Money Banks in Vanuatu: Project Summary Report. Port Vila: Vanuatu National Cultural Center; 2005. [Google Scholar]
- 28.Robins JH, Ross HA, Allen MS, Matisoo-Smith E. Nature. 2006;440:7086. doi: 10.1038/nature04770. [DOI] [PubMed] [Google Scholar]
- 29.Okumura N, Ishiguro N, Nakano M, Hirai K, Matsui A, Sahara M. Biochem Genet. 1996;34:179–189. doi: 10.1007/BF02407018. [DOI] [PubMed] [Google Scholar]
- 30.Gongora J, Fleming P, Spencer PB, Mason R, Garkavenko O, Meyer JN, Droegemueller C, Lee JH, Moran C. Mol Phylogenet Evol. 2004;33:339–348. doi: 10.1016/j.ympev.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Lum JK, Rickards O, Ching C, Cann RL. Hum Biol. 1994;66:567–590. [PubMed] [Google Scholar]
- 32.Redd AJ, Takezaki N, Sherry ST, McGarvey ST, Sofro ASM, Stoneking M. Mol Biol Evol. 1995;12:604–615. doi: 10.1093/oxfordjournals.molbev.a040240. [DOI] [PubMed] [Google Scholar]
- 33.Sykes B, Leiboff A, Low-Beer J, Tetzner S, Richards M. Am J Hum Genet. 1995;57:1463–1475. [PMC free article] [PubMed] [Google Scholar]
- 34.Richards M, Oppenheimer S, Sykes B. Am J Hum Genet. 1998;63:1234–1236. doi: 10.1086/302043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lum JK, Cann RL. Am J Phys Anthropol. 2000;113:151–168. doi: 10.1002/1096-8644(200010)113:2<151::AID-AJPA2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 36.Oppenheimer S, Richards M. Sci Prog. 2001;84:157–181. doi: 10.3184/003685001783238989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox MP. Hum Biol. 2005;77:179–188. doi: 10.1353/hub.2005.0037. [DOI] [PubMed] [Google Scholar]
- 38.Trejaut JA, Kivisild T, Loo JH, Lee CL, He CL, Hsu CJ, Lee ZY, Lin M. PloS Biol. 2005;3:e247. doi: 10.1371/journal.pbio.0030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kemp B, Smith DG. Forensic Sci Int. 2005;154:53–61. doi: 10.1016/j.forsciint.2004.11.017. [DOI] [PubMed] [Google Scholar]