Abstract
A cell-based screen of chemical libraries was carried out to identify small molecules that control the self-renewal of ES cells. A previously uncharacterized heterocycle, SC1, was discovered that allows one to propagate murine ES cells in an undifferentiated, pluripotent state under chemically defined conditions in the absence of feeder cells, serum, and leukemia inhibitory factor. Long-term SC1-expanded murine ES cells can be differentiated into cells of the three primary germ layers in vitro and also can generate chimeric mice and contribute to the germ line in vivo. Biochemical and cellular experiments suggest that SC1 works through dual inhibition of RasGAP and ERK1. Molecules of this kind may not only facilitate practical applications of stem cells in research and therapy, but also provide previously undescribed insights into the complex biology of stem cells.
Keywords: pluripotency, cell-based screen, SC1, ERK1, Ras GTPase-activating protein
In addition to their therapeutic potential, ES cells provide an excellent in vitro system for the study of early development and human diseases (http://stemcells.nih.gov/info/scireport/2001report.htm). Such uses of ES cells, however, require new tools to better understand and control the mechanisms that govern their self-renewal and differentiation. Typically, ES cells are maintained in culture with feeder cells and/or mixtures of exogenous factors. The self-renewal of murine ES (mES) cells largely depends on two key signaling molecules: leukemia inhibitory factor (LIF)/interleukin 6 (IL-6) family members (2) and bone morphogenic protein (BMP) (3, 4). LIF activates STAT signaling through a membrane-bound gp130–LIF receptor complex to promote self-renewal and inhibit mesoderm and endoderm differentiation (2); BMP4 induces expression of Id (inhibitor of differentiation) genes (3) and inhibits MAPK signaling (4) and neuroectoderm differentiation. The combination of BMP4 and LIF can maintain the self-renewal of mES cells in the absence of feeder cells and serum (3). Additionally, the core pluripotency-associated transcriptional regulators, Sox2 (5), Oct4 (6), and Nanog (7, 8), as well as the phosphatidylinositol 3-kinase (PI3K)–AKT signaling pathway (9), are also involved in ES cell self-renewal.
Although significant progress has been made in recent years, we are still far from a complete picture of the dynamic regulatory circuitry that controls the self-renewal of ES cells. Consequently, unbiased cellular screens for small molecules or genes that regulate the self-renewal of ES cells may provide new insights into these processes and also facilitate practical applications of ES cells in research and therapy.
Results and Discussion
High-Throughput Chemical Screen.
To carry out such a screen, an established reporter mES cell line was used, which was derived from heterozygous Oct4-GFP (with the 18-kb Oct4 regulatory region) transgenic OG2 mice (10). OG2-mES cells lose both GFP expression and their compact-colony morphology completely in 4–6 days in the absence of feeder cells and LIF (LIF alone will not sustain self-renewal under feeder-free conditions), affording a robust assay system for self-renewal. Undifferentiated OG2-mES cells were plated into gelatin-coated black 384-well plates at a density of 500 cells per well in ESC-growth media (GM). After overnight incubation, the media was changed to ESC-serum replacement (SR) media and compounds from a library of 50,000 discrete heterocycles (11) were added to each well (5 μM final concentration). After an additional 6 days of incubation, in which media and compound were changed at day 3, cells were analyzed for GFP expression and morphology (with LIF as a “pseudopositive” control). From the primary screen, 28 compounds were identified that maintained colony morphology and GFP expression of OG2-mES cells. Seventeen of these 28 (including a series of pyrimidine derivatives) were shown to maintain the expression of multiple mES cell-specific markers, including SSEA-1, Oct4, and ALP (data not shown).
From this set of compounds, a class of 3,4-dihydropyrimido[4,5-d]pyrimidines was characterized that maintain the undifferentiated phenotype of mES cells in a dose-dependent manner (Scheme 1). A structure-activity-relationship study (Table 1, which is published as supporting information on the PNAS web site) of a second generation focused 3,4-dihydropyrimido[4,5-d]pyrimidine library revealed that: the R1 position can tolerate bulky substituents (e.g., hetero-aromatic substituents, PEG linker), the R2 position tolerates a methyl group well (but not methoxy or hydrogen), and the 3,6-substitution pattern on the phenyl ring at the R3 position is required for activity (e.g., a 5-position methoxy-substitution on the phenyl ring abolishes activity completely). Importantly, an analog SC1, also called pluripotin (Scheme 1), was identified with 10-fold higher activity (EC50 = 1 μM concentration in the ESC-SR media) and relatively low cellular toxicity (>30 μM).
Scheme 1.
Chemical structures of 3,4-dihydropyrimido[4,5-d]pyrimidine scaffold (A) and SC1 (B).
Characterization of SC1's Biological Activity.
Treatment of OG2-mES cells (feeder-cell dependent) or R1-mES cells (feeder-cell independent) grown on gelatin-coated six-well plates (1.6 × 104 cells/cm2) with SC1 either in the ESC-SR media (3 μM) or in chemically defined media without LIF [CDM: ESC-N2B27 (1 μM) or ESC-N2 (300 nM)], maintains their ability to self-renew for >10 passages in an undifferentiated/pluripotent state. SC1-expanded mES cells are positively stained with multiple pluripotency markers, including SSEA-1, Oct4, and ALP (Fig. 1A; see also Fig. 5 A and B, which is published as supporting information on the PNAS web site), and grow as compact colonies, indistinguishable from undifferentiated mES cell colonies grown under feeder cell conditions. SC1-expanded mES cells also express high levels of Oct4, Nanog, and Sox2 as determined by RT-PCR (Figs. 1B and 5C). OG2-mES cells cultured under LIF+BMP4 conditions [LIF+BMP4 treatment in ESC-N2B27 media is a feeder-free culture condition that is used for multiple mES cell lines] (3) have decreased expression of pluripotency markers and exhibit less compact colony morphology compared with OG2-mES cells cultured under feeder cell conditions. In comparison, SC1-expanded OG2-mES cells show an increased number of cells expressing higher levels of pluripotency markers and improved colony morphology, but somewhat slower proliferation rates (Figs. 1C and 5D) and lower plating efficiencies (LIF+BMP4, 85.6 ± 3.6%; SC1+N2B27, 70.1 ± 4.1%). SC1 (in both ESC-SR and ESC-N2B27 media) also maintains OG2-mES cell self-renewal at low cell densities (50 cells/cm2) (Fig. 1 D and E; see Fig. 6, which is published as supporting information on the PNAS web site). Furthermore, the presence of SC1 inhibits differentiation of mES cells induced by either 20% FBS or retinoic acid (RA) (1 μM) treatment (Fig. 1F; see Fig. 7, which is published as supporting information on the PNAS web site). Importantly, SC1-expanded mES cells (>10 passages) form embryoid bodies (EBs) in suspension efficiently and can be selectively induced to differentiate into neural/neuronal (βIII-tubulin and NeuroD1) (12), cardiac muscle (myosin heavy chain and Nkx2.5) (13), and endodermal cells (Sox17 and FOXA2) (14), derivatives of the three primary germ layers, after SC1 depletion (Fig. 1G). And finally, SC1-expanded mES cells can contribute to the germ line and generate healthy chimeric mice in vivo (Fig. 1H; see Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 1.
SC1-expanded mES cells maintain their ability to self-renew under feeder cell/LIF-free conditions. (A and B) Serially passaged, SC1-expanded OG2-mES cells maintain the expression of pluripotency markers by FACS analysis (A) (undifferentiated population: SC1 + SR, 95.9%; DMSO + SR, 1.2%; SC1 + N2B27, 95.7%; DMSO + N2B27, 0.6%; SC1 + N2, 95.8%; DMSO + N2, 0.1%; LIF + BMP4, 87.4%), histocytochemistry (alkaline phosphatase) and by RT-PCR (B) (Nanog, Oct4, and Sox2). mES cells were seeded in gelatin-coated six-well plates at 1.6 × 104 cells/cm2 and maintained with 3 μM SC1 in the ESC-SR media, 1 μM SC1 in the ESC-N2B27 media, or 300 nM SC1 in the ESC-N2 media without LIF or feeder cells. mES cells (seeded at 1.6 × 104 cells/cm2) maintained with 103 units/ml LIF plus 10 ng/ml BMP4 in the ESC-N2B27 media were used as a control. mES cells cultured with DMSO after two passages were used as negative control. Cells were split every 3 days and seeded at the same density (1.6 × 104 cells/cm2 in six-well plates). SC1-expanded mES cells at passage 11 were analyzed with FACS, histocytochemistry, and RT-PCR. (C) Growth curves of OG2-mES cells under different culture conditions. mES cells maintained under the feeder condition were used as a positive control. Cells were split every 3 days, and cell numbers were counted by hemocytometer. Under all conditions, >95% cells maintain GFP expression by FACS analysis (Fig. 5D). (D and E) OG2-mES cells can be expanded by SC1 at low cell density (D) and maintain the expression of pluripotency markers after three passages (E) (undifferentiated population: ESC-SR, 90.0%; ESC-N2B27, 97.4%). OG2-mES cells were seeded at a low cell density (50 cells/cm2) and cultured with 3 μM SC1 in ESC-SR media or 1 μM SC1 in ESC-N2B27 media for three passages. Cells were split every 3 days and seeded at the same density (50 cells/cm2). Cell numbers at each passage were counted by hemocytometer. Cells at day 9 were analyzed with FACS. (F) SC1 (1 μM) inhibits differentiation of OG2-mES cells induced by 20% FBS (undifferentiated population: SC1, 89.5%; DMSO, 0.2%) or 1 μM RA (undifferentiated population: SC1, 93.4%; DMSO, 0.4%). OG2-mES cells were seeded at 1.6 × 104 cells/cm2 and cultured with 20% FBS or 1 μM RA in the ESC-N2B27 media with/without 1 μM SC1 for 4 days (FBS) or 2 days (RA) and analyzed with FACS and histocytochemistry. (G) In vitro differentiation potential of SC1-expanded (1 μM concentration in ESC-N2B27 media), passage 11 OG2-mES cells. Neuronal, cardiac muscle, and endodermal differentiation were carried out by using established protocols and cells were stained with antibodies against β III-tubulin, myosin heavy chain, and Sox17, respectively (red, β III-tubulin, myosin heavy chain; green, Sox17; blue, DAPI) and analyzed by RT-PCR. Feeder-cultured OG2-mES cells were used as a control. (H) Germ-line contribution of SC1-expanded (3 μM concentration in ESC-SR media), passage 11 OG2-mES cells. Phase contrast and fluorescent images of female and male gonad (14.5 days postcoitum) and squeezed male gonad tissue clearly show the singular germ cells that express GFP driven by the Oct4 promoter (a marker for ES cells or embryonic germ cells). (SR, ESC-SR media; N2B27, ESC-N2B27 media; N2, ESC-N2 media; ALP, alkaline phosphatase; MHC, myosin heavy chain).
Effects of SC1 on Established Self-Renewal Pathways.
To characterize the mechanism of action of SC1, we initially examined whether SC1 operates through known signaling pathways that control the self-renewal of mES cells. In contrast to LIF, which strongly stimulates STAT3 phosphorylation at tyrosine 705 (2), Western blot analysis indicated that SC1 does not have any effect on STAT activation (Fig. 2A). Knockdown of STAT3 by siRNAs (STAT3 expression was reduced 60% as revealed by Western blot; Fig. 9, which is published as supporting information on the PNAS web site) or inhibition of STAT3 phosphorylation by 50 μM JAK2 inhibitor, AG490, induces differentiation of OG2-mES cells in the presence of LIF and BMP4. However, treatment with either siSTAT3 or AG490 does not block the activity of SC1 (Fig. 9B). These results suggest that SC1 functions through a STAT3-independent mechanism.
Fig. 2.
The effects of SC1 on known self-renewal pathways. (A) Western blot analysis of STAT3 phosphorylation at Tyr-705. OG2-mES cells were cultured in the ESC-N2B27 media overnight and then stimulated with DMSO (negative control), 1 μM SC1, 1 μM SC1− (negative control of SC1), 103 units/ml LIF (positive control), or 1 μM SC1 plus 103 units/ml LIF for 20 min. (B) Quantitative RT-PCR analysis of Id1 gene expression. OG2-mES cells were cultured in the ESC-N2B27 media overnight and then stimulated with DMSO (negative control), 1 μM SC1, or 50 ng/ml BMP4 (positive control) for 45 min. (C) Wnt signaling by Super (8×) TOPflash reporter assay. Stable Super (8×) TOPflash reporter 293T cells were treated with DMSO (negative control), 3 μM SC1, or 2 μM BIO (positive control) for 48 h and assayed for luciferase activity. (D) Transient activation of Nanog expression. OG2-mES cells were treated with 1 μM SC1 in the ESC-N2B27 media, and mRNAs were isolated at 0, 6, and 18 h, separately. In these experiments, OG2-mES cells were seeded at the same density (1.6 × 104 cells/cm2). PD, PD098059; SC1−, negative control of SC1.
Furthermore, quantitative RT-PCR analysis revealed that SC1 treatment does not act through the BMP signaling pathway to increase Id1 gene expression (Fig. 2B). The canonical Wnt signaling pathway recently has been implicated in the self-renewal of mES cells, based on the discovery that a GSK-3β inhibitor, 6-bromoindirubin-3′-oxime (BIO), maintains mES cells in the pluripotent state in the absence of both feeder cells and LIF (15). However, unlike BIO, which activates Wnt/TOPflash reporter activity >200-fold, SC1 does not increase TOPflash reporter activity (Fig. 2C) and does not inhibit GSK-3β activity (data not shown). Thus, it appears that SC1 may function by a previously uncharacterized mechanism. Interestingly, SC1 was found to transiently activate Nanog expression by 7-fold after treatment for 6 h (Fig. 2D). Consistent with this observation, SC1 inhibits the phosphorylation of Ser-315 in p53 (Fig. 10, which is published as supporting information on the PNAS web site), which has been shown to repress Nanog expression (16).
Target Identification.
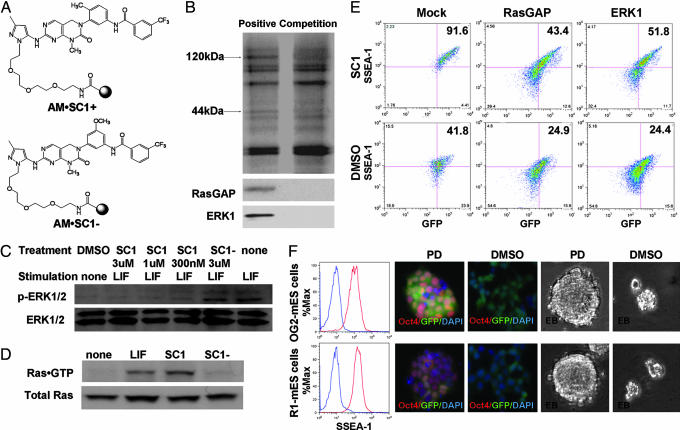
To identify the cellular target(s) of SC1, the compound was linked through the pyrazole N1 position to an agarose affinity matrix via a polyethylene glycol linker based on the above structure-activity-relationship analysis (AM·SC1+) (Fig. 3A). To distinguish specific binding to SC1 versus nonspecific interactions with the affinity matrix, an affinity resin also was synthesized from an inactive analog as a negative control (AM·SC1−). Whole-cell lysates pretreated with the negative affinity matrix were incubated with the positive affinity matrix or positive affinity matrix plus 50 μM SC1 as a competitor. SDS/PAGE analysis revealed that two bands of ≈44 and 120 kDa were bound specifically by the AM·SC1+; addition of SC1 to the lysates during affinity chromatography blocked binding of these two proteins to the affinity matrices (Fig. 3B). These two bands were identified as ERK1 (44 kDa) and Ras GTPase-activating protein (RasGAP, 120 kDa), respectively, by LC/MS and confirmed independently by ERK1/2 and RasGAP Western blot analysis. Direct binding of SC1 to ERK1 or RasGAP (17) was confirmed by surface plasmon resonance with purified enzymes (ERK1: KD = 98 nM; RasGAP: KD = 212 nM; Fig. 11, which is published as supporting information on the PNAS web site). Moreover, SC1 was shown to inhibit ERK1/2 phosphorylation stimulated by LIF in OG2-mES cells (Fig. 3C). In addition, it was demonstrated that SC1 (1 μM) increases the level of Ras·GTP in OG2-mES cells (based on pulldowns by using a GST fusion protein with the Ras-binding domain of Raf-1; Fig. 3D), indicating that SC1 activates Ras by inhibition of RasGAP function.
Fig. 3.
Affinity chromatography and target confirmation. (A) Structures of the positive affinity matrix (AM·SC1+) and the negative affinity matrix (AM·SC1−). (B) Silver staining and Western blot analysis of proteins retained by affinity supports. (C) SC1 inhibits autophosphorylation of ERK1/2 at Thr-202/Tyr-204. OG2-mES cells were cultured in the ESC-N2B27 media overnight and then stimulated with 103 units/ml LIF for 15 min in the presence or absence of SC1 or SC1−. (D) SC1 activates Ras activity. OG2-mES cells were treated with 1 μM SC1, 103 units/ml LIF, or 1 μM SC1− in the ESC-N2B27 media for 6 h, and whole-cell lysates were analyzed with the Ras Activation Assay Kit. (E) Overexpression of ERK1 and RasGAP. OG2-mES cells (seeded at 1.6 × 104 cells/cm2) transiently transfected (transfection efficiency is ≈50%) with ERK1 or RasGAP were cultured in the ESC-N2B27 media with/without 1 μM SC1 for 3 days and analyzed with FACS (undifferentiated population: mock, 41.8 ± 1.9%; ERK1, 24.9 ± 2.5%; RasGAP, 24.4 ± 2.9%; Mock + SC1, 91.6 ± 3.6%; ERK1 + SC1, 43.4 ± 2.1%; RasGAP + SC1, 51.8 ± 2.0%). Empty vector was used as a negative control. (F) OG2-mES cells and R1-mES cells infected with lentivirus of shRasGAP were purified and cultured in the ESC-N2B27 media with 40 μM PD098059 for >10 passages. Stable cell lines of GFPhigh population were seeded at 1.6 × 104 cells/cm2 and cultured with/without PD098059 (40 μM) in the ESC-N2B27 media without feeder cells, LIF, and serum. Cells were split every 3 days. Cells with PD098059 treatment at passage 11, and cells without PD098059 treatment at passage 6 were analyzed with FACS (red line, with PD098059; blue line, without PD098059), immunocytochemistry (red, Oct4; green, GFP; blue, DAPI), and EB formation.
To confirm that SC1's effects on self-renewal of mES cells are mediated by its interaction with ERK1 and RasGAP, additional genetic and pharmacological studies were performed. ERK1 was transiently overexpressed for 3 days in OG2-mES cells cultured under ESC-N2B27 CDM conditions; FACS analysis revealed that ectopic expression of ERK1 induced differentiation of mES cells both in the presence and absence of SC1 and that SC1 can partially rescue ERK1-induced differentiation (Fig. 3E). Consistent with this result, it is known that MEK inhibitor PD098059 (40 μM) partially blocks differentiation of mES cells by inhibiting ERK phosphorylation under feeder-free conditions (18). However, PD098059 itself is not sufficient to maintain mES cells in the pluripotent state: ES cells cultured in ESC-N2B27 media with 40 μM PD098059 cannot be passaged more than three times. Similarly, transient ectopic expression of RasGAP induced differentiation in mES cells; again, RasGAP-induced differentiation can be partially rescued by SC1 (Fig. 3E). To further show that SC1's activity is mediated through its combined effects on ERK1 and RasGAP, mES cells (both OG2 and R1 lines) stably expressing a short hairpin RNA (shRNA) against RasGAP (19) were generated by using a lentivirus-based expression vector pLVTHM (which also expresses a GFP reporter) (20). Stable cell lines of GFPhigh population were isolated by cell sorting and confirmed to have RasGAP expression reduced >90% (Western blot; Fig. 12, which is published as supporting information on the PNAS web site). Treatment of these cells with PD098059 (40 μM) maintained them in the undifferentiated/pluripotent state under basal CDM conditions (i.e., without feeder cells, LIF, and serum) for >10 passages. These cells also continue to proliferate (Fig. 13A, which is published as supporting information on the PNAS web site), express multiple mES cell specific markers, and can form normal EBs in suspension culture (Figs. 3F and 13B). In contrast, control cells engineered with the empty pLVTHM vector completely differentiated within two passages under the same basal CDM conditions, whereas those expressing the shRNA but not treated with PD098059 lose both the ESC-like colony morphology and Oct4 expression after five passages and eventually failed to form regular EBs in the suspension culture (Fig. 3F). These experiments collectively suggest that SC1 is a dual function small molecule inhibitor of both ERK1 and RasGAP and that simultaneous inhibition of both protein activities is required and sufficient for SC1's effects on mES cells. This result underscores a potential advantage (and complexity) in the use of small molecules in cell-based phenotypic and reporter screens in that compounds can modulate more than one target to achieve a desired biological effect.
Mechanism of Action.
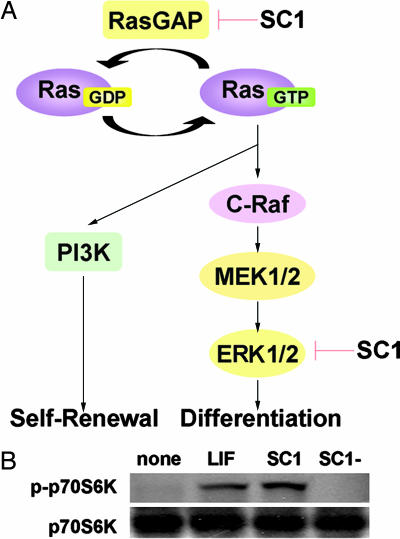
The ERK signaling pathway has been shown to control a vast array of cellular processes, including cell division, proliferation, and differentiation (21). A number of experiments suggest that ERK activation is involved in the differentiation of mES cells. For example, sustained ERK1/2 activation leads to neural differentiation (22), and evidence suggests that BMP promotes LIF-dependent self-renewal by inhibiting ERK and p38 (4). Consequently, inhibition of ERK1/2 by SC1 would be expected to contribute to the self-renewal of mES cells by blocking ERK-dependent differentiation. However, as demonstrated previously, treatment of mES cells with the MEK inhibitor PD098059 alone is not sufficient to sustain mES cell self-renewal, suggesting that modulation of the Ras pathway by SC1 also is required for the self-renewal of mES cells.
The Ras family of small GTPases are monomeric guanine nucleotide-binding proteins that serve as molecular switches to regulate cellular growth, morphogenesis, and mobility (23). RasGAP modulates Ras signaling by stimulating the GTP-hydrolysis activity of Ras to form the inactive Ras·GDP complex (24). By inhibiting RasGAP, SC1 activates signaling by Ras or a Ras-like GTPase that, in turn, may enhance self-renewal through its downstream signaling pathways. Indeed, overexpression of ERas promotes mES cells self-renewal through the activation of PI3K-AKT signaling and enhanced cell growth (25). SC1 activates phosphorylation of p70S6K (Fig. 4B), a mitogen-stimulated serine/threonine kinase regulated by PI3K (26), suggesting that SC1 also may mediate some of its effects through PI3K signaling. Thus, it appears the SC1 acts through two targets: by inhibiting RasGAP SC1 blocks RasGAPs differentiation-inducing activity and activates self-renewal pathways, while at the same time blocking ERK-dependent differentiation. The self-renewal of ES cells can be largely regarded as a combined phenotypic outcome of cellular proliferation together with inhibition of differentiation and cell death. ES cells may have the intrinsic ability to self-renew, and, therefore, inhibition of differentiation may be sufficient to maintain the undifferentiated state. The identification and characterization of SC1's mechanism of action underscores such basic requirements for the self-renewal of stem cells.
Fig. 4.
Mechanism of action. (A) Proposed mechanism of action of SC1. (B) SC1 activates p70S6K phosphorylation. OG2-mES cells were treated with 103 units/ml LIF, 1 μM SC1, or 1 μM SC1− in the ESC-N2B27 media for 10 h.
Conclusion
In summary, we have identified a previously undescribed small molecule, SC1, from a phenotypic cellular screen that can maintain the self-renewal of mES cells in the absence of feeder cells and exogenous factors. This molecule appears to act by inhibition of ERK1- and RasGAP-dependent signaling pathways. In addition, this molecule represents a previously undescribed class of dual inhibitor of a protein kinase and a small GTPase-activating protein. We are exploring the effects of SC1 on human ES cells and adult stem cells. This and other such molecules are likely to provide insights into the molecular mechanism that control stem cell fate and ultimately may be useful to in vivo stem cell biology and therapy.
Materials and Methods
Culture Media. ESC-GM.
Knockout DMEM (GIBCO, Carlsbad, CA) was supplemented with 15% knockout Serum Replacement (GIBCO)/1× nonessential amino acids (GIBCO)/2 mM l-glutamine (GIBCO)/0.1 mM 2-mercaptoethanol (GIBCO)/103 units/ml LIF (Chemicon).
ES-SR.
Knockout DMEM (GIBCO) was supplemented with 15% knockout Serum Replacement (GIBCO)/1× nonessential amino acid (GIBCO)/2 mM l-glutamine (GIBCO)/0.1 mM 2-mercaptoethanol (GIBCO).
ESC-N2B27 (Completely Serum-Free Condition).
DMEM/F12 (GIBCO) was supplemented with 0.5× N2 (GIBCO)/0.5× B27 (without vitamin A; GIBCO)/50 μg/ml BSA fraction V (GIBCO), 1× nonessential amino acids (GIBCO)/2 mM l-glutamine (GIBCO)/0.1 mM 2-mercaptoethanol (GIBCO).
ESC-N2 (Completely Serum-Free Condition).
DMEM/F12 was supplemented with 1× N2 (GIBCO)/50 μg/ml BSA fraction V (GIBCO)/1× nonessential amino acids (GIBCO)/2 mM l-glutamine (GIBCO)/0.1 mM 2-mercaptoethanol (GIBCO).
mEB-GM.
DMEM (GIBCO) was supplemented with 15% ES-qualified FBS (GIBCO)/0.5× l-glutamine solution (GIBCO)/1× nonessential amino acids (GIBCO).
Expansion of mES Cells in the Presence of SC1.
mES cells were seeded in gelatin-coated six-well plates at 1.6 × 104 cells/cm2 and maintained with 3 μM SC1 in ESC-SR media, 1 μM SC1 in ESC-N2B27 media, or 300 nM SC1 in ESC-N2 media without LIF or feeder cells. mES cells maintained with 103 units/ml LIF plus 10 ng/ml BMP4 in ESC-N2B27 media were used as a positive control. mES cells treated with DMSO in ESC-SR media, ESC-N2B27 media, or ESC-N2 media for two passages were used as negative controls. Cells were split every 3 days and seeded at same density (1.6 × 104 cells/cm2). mES cells at passage 11 were analyzed with FACS, immunocytochemistry, histocytochemistry, and RT-PCR.
Expansion of mES Cells in the Presence of SC1 at Low Cell Density.
OG2-mES cells were seeded at a low cell density (50 cells/cm2) and cultured with 3 μM SC1 in ESC-SR media or 1 μM SC1 in ESC-N2B27 media for three passages. Cells were split every 3 days and seeded at the same density (50 cells/cm2). Cell numbers at each passage were counted by hemocytometer. The cells at day 9 were analyzed with FACS.
EB Formation and in Vitro Differentiation.
OG2-mES cells cultured with 1 μM SC1 in ESC-N2B27 media for 10 passages were trypsinized and plated on ultra-low attachment dishes in mEB-GM. To induce neuronal differentiation, 1 μM RA was added. After 4 days in culture, EBs were seeded on laminin-coated plates and grown in neuronal differentiation basal media [consisting of DMEM/F-12 (GIBCO)/0.5× N2 (GIBCO)/0.5× B27 (GIBCO)/50 μg/ml BSA fraction V (GIBCO)] for an additional 4 days. To induce cardiac myogenic differentiation, 4-day EBs (without RA treatment) were seeded on gelatin-coated plates and grown in DMEM (GIBCO) plus 10% FBS (GIBCO) for an additional 6 days. To induce endodermal differentiation, mES cells were grown on gelatin-coated plates in the ESC-GM (without a feeder layer) for 4 days and then treated with Activin A (100 ng/ml) in RPMI medium 1640 (GIBCO) supplemented with 1× Glutamax (GIBCO) and 10% FBS (GIBCO) for an additional 6 days. Media was changed every 3 days.
Flow Cytometry Analysis.
Cells were trypsinized and blocked with 4% FBS in DPBS for 10 min on ice. Cells were then resuspended in primary antibody staining buffer [mouse anti-SSEA-1 (1:100; Chemicon, Temecula, CA) in DPBS with 0.4% FBS] and incubated for 40 min on ice. After washing twice with DPBS, cells were resuspended in secondary antibody staining buffer [CyIII-conjugated anti-mouse (1:500; Jackson ImmunoResearch, West Grove, PA) in DPBS with 0.4% FBS] and incubated for 40 min on ice. After washing twice with DPBS, cells were used for FACS analysis. Data are acquired with BD LSRII System (BD Science, Franklin Lakes, NJ) and analyzed with FLOWJO software (Tree Star, San Carlos, CA).
Affinity Chromatography.
Cells were lysed with PY buffer (pH 7.2; 60 mM β-glycerophosphate/15 mM nitrophenyl phosphate/25 mM Mops/15 mM EGTA/15 mM magnesium chloride/2 mM DTT/1 mM sodium vanadate/1 mM sodium fluoride/1 mM sodium phenylphosphate/100 μM benzamidine) plus 0.1% Nonidet P-40 for 10 min on ice and analyzed by affinity chromatography with methods described in ref. 1. Whole-cell lysates were pretreated with the control affinity matrix at 4°C for 1.5 h and the negative affinity matrix at 4°C for 1 h. After washing three times, samples were incubated with the positive affinity matrix or positive affinity matrix plus 50 μM SC1 as a competitor at 4°C for 1 h. After heat shock, samples were loaded and separated on a 4–20% Tris-Glycine SDS/PAGE (Invitrogen, Carlsbad, CA) stained with a Silver Stain Plus Kit (Bio-Rad, Hercules, CA). The bands were cut, destained, and analyzed with LC/MS. For Western blot analysis, samples were electroblotted on a nitrocellulose membrane; the membrane was blocked for 1 h at room temperature with 5% nonfat milk in DPBS and immunoblotted overnight at 4°C with rabbit anti-RasGAP (1:1,000; Alexis, San Diego, CA) and rabbit anti-ERK1/2 (1:1,000; Cell Signaling Technology, Beverly, MA). The membrane was then washed, incubated with anti-rabbit peroxidase-conjugated affinity-purified secondary antibody (1:1,000, Pierce, Rockford, IL) at room temperature for 1 h, and developed by SuperSignal chemiluminescence (Pierce).
Lentiviral Infection and Long-Term Passage.
mES cells (both OG2 and R1 lines) were infected with lentivirus generated from the expression vector pLVTHM-shRasGAP. After infection, mES cells were maintained under feeder conditions for 6 days (split every 3 days). Stable cell lines of the GFPhigh population isolated by cell sorting were seeded in six-well plates at 1.6 × 104 cells/cm2 and cultured with/without PD098059 (40 μM) in ESC-N2B27 media without feeder cells, LIF, and serum for >10 passages.
Supporting Information.
Detailed materials and methods are presented as Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Prof. Alfred Wittinghofer (Max Planck Institute of Molecular Physiology, Dortmund, Germany) for providing plasmids of RasGAP for protein expression, Prof. Didier Trono (Swiss Institute of Technology, Lausanne, Switzerland) for providing pLVTHM vector, and Dr. Wen Xiong for helpful discussion. This work is supported by Novartis Research Foundation. This work is manuscript 18028-CH of the Scripps Research Institute.
Abbreviations
- BMP
bone morphogenic protein
- EB
embryoid body
- GM
growth media
- LIF
leukemia inhibitory factor
- mES cells
murine ES cells
- PI3K
phosphatidylinositol 3-kinase
- RA
retinoic acid
- RasGAP
Ras GTPase-activating protein
- SR
serum replacement.
Footnotes
The authors declare no conflict of interest.
References
- 1.Ding S, Wu TY, Brinker A, Peters EC, Hur W, Gray NS, Schultz PG. Proc Natl Acad Sci USA. 2003;100:7632–7637. doi: 10.1073/pnas.0732087100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niwa H, Burdon T, Chambers I, Smith A. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ying QL, Nichols J, Chambers I, Smith A. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 4.Qi X, Li TG, Hao J, Hu J, Wang J, Simmons H, Miura S, Mishina Y, Zhao GQ. Proc Natl Acad Sci USA. 2004;101:6027–6032. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 7.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 8.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 9.Paling NR, Wheadon H, Bone HK, Welham MJ. J Biol Chem. 2004;279:48063–48070. doi: 10.1074/jbc.M406467200. [DOI] [PubMed] [Google Scholar]
- 10.Ohbo K, Yoshida S, Ohmura M, Ohneda O, Ogawa T, Tsuchiya H, Kuwana T, Kehler J, Abe K, Scholer HR, Suda T. Dev Biol. 2003;258:209–225. doi: 10.1016/s0012-1606(03)00111-8. [DOI] [PubMed] [Google Scholar]
- 11.Ding S, Gray NS, Wu X, Ding Q, Schultz PG. J Am Chem Soc. 2002;124:1594–1596. doi: 10.1021/ja0170302. [DOI] [PubMed] [Google Scholar]
- 12.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 13.Maltsev VA, Rohwedel J, Hescheler J, Wobus AM. Mech Dev. 1993;44:41–50. doi: 10.1016/0925-4773(93)90015-p. [DOI] [PubMed] [Google Scholar]
- 14.D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 15.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 16.Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 17.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 18.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Dev Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- 19.Kunath T, Gish G, Lickert H, Jones N, Pawson T, Rossant J. Nat Biotechnol. 2003;21:559–561. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- 20.Wiznerowicz M, Trono D. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson GL, Lapadat R. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Li L. Dev Biol. 2005;284:1–11. doi: 10.1016/j.ydbio.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Gamblin SJ, Smerdon SJ. Curr Opin Struct Biol. 1998;8:195–201. doi: 10.1016/s0959-440x(98)80038-9. [DOI] [PubMed] [Google Scholar]
- 24.Egan SE, Giddings BW, Brooks MW, Buday L, Sizeland AM, Weinberg RA. Nature. 1993;363:45–51. doi: 10.1038/363045a0. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi K, Mitsui K, Yamanaka S. Nature. 2003;423:541–545. doi: 10.1038/nature01646. [DOI] [PubMed] [Google Scholar]
- 26.Alessi DR, Kozlowski MT, Weng QP, Morrice N, Avruch J. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.