Abstract
Eukaryotic cells respond to a variety of DNA insults by triggering a common signal transduction cascade, known as checkpoint response, which temporarily halts cell-cycle progression. Although the main players involved in the cascade have been identified, there is still uncertainty about the nature of the structures that activate these surveillance mechanisms. To understand the role of nucleotide excision repair (NER) in checkpoint activation, we analyzed the UV-induced phosphorylation of the key checkpoint proteins Chk1 and p53, in primary fibroblasts from patients with xeroderma pigmentosum (XP), Cockayne syndrome (CS), trichothiodystrophy (TTD), or UV light-sensitive syndrome. These disorders are due to defects in transcription-coupled NER (TC-NER) and/or global genome NER (GG-NER), the NER subpathways repairing the transcribed strand of active genes or the rest of the genome, respectively. We show here that in G0/G1 and G2/M phases of the cell cycle, triggering of the DNA damage cascade requires recognition and processing of the lesions by the GG-NER. Loss of TC-NER does not affect checkpoint activation. Mutations in XPD, XPB, and in TTDA, encoding subunits of the TFIIH complex, involved in both transcription and NER, impair checkpoint triggering. The only exception is represented by mutations in XPD, resulting in combined features of XP and CS (XP/CS) that lead to activation of the checkpoint cascade after UV radiation. Inhibition of RNA polymerase II transcription significantly reduces the phosphorylation of key checkpoint factors in XP/CS fibroblasts on exposure to UV damage.
Keywords: cell cycle, human fibroblasts, DNA repair, DNA damage response
The nature of the signal that activates DNA damage response pathways after UV radiation is still debated. Some evidence suggests that ssDNA, probably coated by replication protein A (RPA), triggers the checkpoint. In fact recruitment of the ataxia-telangiectasia mutated- and Rad3-related (ATR)-interacting protein (ATRIP) protein kinase complex to sites of DNA damage and ATR-mediated Chk1 activation depends upon RPA-coated ssDNA (1, 2). Others have suggested that checkpoint proteins are able to directly bind UV-damaged DNA without any need for processing of the lesion (3). Moreover, it is still largely unclear whether NER plays any role in activating the checkpoint.
This repair pathway, responsible for removing helix-distorting lesions from DNA, essentially consists of four sequential steps: recognition of the DNA lesion, opening of a bubble around the lesion, incision of DNA upstream and downstream the damage by the endonucleases XPF-ERCC1 and XPG, and finally DNA resynthesis and ligation. Global genome NER (GG-NER) depends on recognition of the damage in nontranscribed DNA by the XPC-RAD23B complex and the UV-damaged DNA-binding activity (UV-DDB), including two subunits (DDB1 and DDB2/XPE). Transcription-coupled NER (TC-NER) is initiated by the arrest of the RNA polymerase at a lesion on the transcribed strand of an active gene, in a process that requires the CSA and CSB proteins. Defects in NER lead to various human inherited syndromes: xeroderma pigmentosum (XP), Cockayne syndrome (CS), trichothiodystrophy (TTD), a mild UV light-sensitive syndrome (UVsS), and a complex pathological phenotype with combined symptoms of XP and CS (XP/CS). Despite sharing cutaneous photosensitivity and genetic heterogeneity, these diseases show distinct clinical features. XP is a cancer-prone disorder, whereas CS and TTD are multisystem diseases characterized by developmental and neurological abnormalities and premature aging. The seven NER-deficient complementation groups identified in XP are defective in both NER subpathways, except for the XP-C and XP-E groups that are specifically defective in GG-NER. Conversely, the genes responsible for CS (CSA and CSB) and a still unidentified gene responsible for four UVsS cases are involved in TC-NER (4, 5).
Some controversies exist in the literature about the role of NER in triggering the checkpoint cascade in response to UV radiation. In yeast, we showed that NER is required to allow checkpoint complexes to bind stably to UV-damaged chromosomes, likely through a physical interaction between NER and checkpoint proteins. Triggering of the cascade seems to require initial processing of the lesions. In fact, a variety of yeast NER mutants fail to activate the G1 and G2 checkpoints after UV irradiation, whereas loss of TC-NER alone does not affect checkpoint activation (6). In agreement with our data, UV-induced H2AX phosphorylation was found defective in resting human primary fibroblasts lacking functional XPA, a DNA damage-binding protein involved in the repair of UV-induced lesions from both active and inactive regions of the genome, but was evident in fibroblasts from CS patients assigned to the CS-B group (7). Similarly, H2AX phosphorylation after UV irradiation within the G1 phase was found to depend on XPA and XPC in human fibroblasts immortalized by transfection with the telomerase gene (8).
Others could not detect a strong activation of the ATR-dependent checkpoint in normal human immortalized cell lines, outside the S-phase of the cell cycle (9, 10). XP-C cells, transformed with simian virus 40 were shown to be proficient in the phosphorylation of key ATR targets in response to UV (9). Recent results suggested that XPA-deficient immortalized human cells are defective in activating the DNA damage checkpoint in S-phase, but further processing of UV radiation-induced photoproducts does not seem to be required. Moreover, checkpoint signaling does not seem to be compromised by a defect in only one of the two NER subpathways, GG-NER or TC-NER (10). Despite the available information concerning the first steps of the DNA damage signal transduction cascade triggered by UV irradiation (11), several issues remain open: (i) Can the ATR-mediated checkpoint be activated in nonreplicating G0/G1 or G2 cells? (ii) Is the lesion by itself sufficient to activate the checkpoint, or is the open preincision complex formation required? (iii) Is further processing of the lesion by the NER machinery, which might temporarily lead to ssDNA covered by RPA, needed to activate ATR? (iv) What is the relative contribution of the two NER subpathways, GG-NER and TC-NER, in checkpoint activation?
To address these issues and to understand the role of NER in triggering the signal transduction cascade in response to UV radiation, we analyzed checkpoint activation in primary fibroblasts from several NER-defective patients, representative of distinct clinical, cellular, and molecular alterations. The data we obtained reveal that, in G0/G1 and G2/M phases of the cell cycle, triggering of the DNA damage cascade after UV radiation depends on NER. Both recognition and processing of the UV radiation-induced photoproducts by the GG-NER are required to activate the checkpoint. Conversely, loss of TC-NER does not affect checkpoint activation. In S-phase cells, checkpoint activation does not depend on NER, probably because other kind of lesions, such as exposed single-stranded regions of DNA at stalled replication forks or strand breaks, contribute to trigger the checkpoint cascade. We also found that mutations in XPD, XPB, and TTDA, encoding subunits of the TFIIH complex, involved in both transcription and NER, lead to a failure to activate the checkpoint. Only defects in the XPD gene that result in XP/CS do not impair checkpoint activation, in response to UV radiation. Intriguingly, in XP-D/CS fibroblasts, triggering of the DNA damage cascade depends on ongoing RNA polymerase transcription.
Results and Discussion
With the aim of improving our understanding on the functional relationship between recognition and processing of UV radiation-induced photoproducts by NER and triggering of surveillance mechanisms, we analyzed checkpoint activation in primary fibroblasts from 27 NER-defective patients (Table 1, which is published as supporting information on the PNAS web site). It is important to emphasize that primary human cell lines were chosen for our study because they have intact surveillance mechanisms, compared with certain immortalized cell lines. In particular, fibroblast strains represent an in vitro cell system that still maintains the cell contact inhibition and the cell density-dependent growth typical of the in vivo situation. These features are lost in other cell systems, such as transformed fibroblasts and lymphoblastoid cell lines, and may be important in the cascade of events leading to checkpoint activation.
To estimate possible differences within the cell population, the analysis was performed at the single-cell level by immunofluorescence by using phosphospecific antibodies, recognizing the key ATR targets Chk1 and p53 (anti-P-Chk1-Ser-317 and anti-P-p53-Ser-15).
Checkpoint Activation in G0 and G2/M Fibroblasts Depends on NER.
Nonproliferating fibroblasts from normal donors appeared to be able to activate the checkpoint, because both Chk1 and p53 get phosphorylated after UV irradiation (Fig. 1). Others could not detect a strong ATR-dependent checkpoint activation in immortalized cells that are not replicating their DNA (9, 10). This difference may be due to the analysis of primary cell lines compared with transformed cells. In these immortalized cells, p53 function is compromised, affecting the removal of cyclobutane pyrimidine dimers from genomic DNA, in addition to other pathways (12, 13). Cells from CS patients mutated in either the CSA or CSB gene (14) show no defect in Chk1 and p53 phosphorylation, whereas XP-A fibroblasts (15) are defective in activating the checkpoint after UV irradiation, compared with normal and CS fibroblasts (Fig. 1). In XP-A G2/M-synchronized fibroblasts, the signaling is also drastically compromised (Fig. 2).
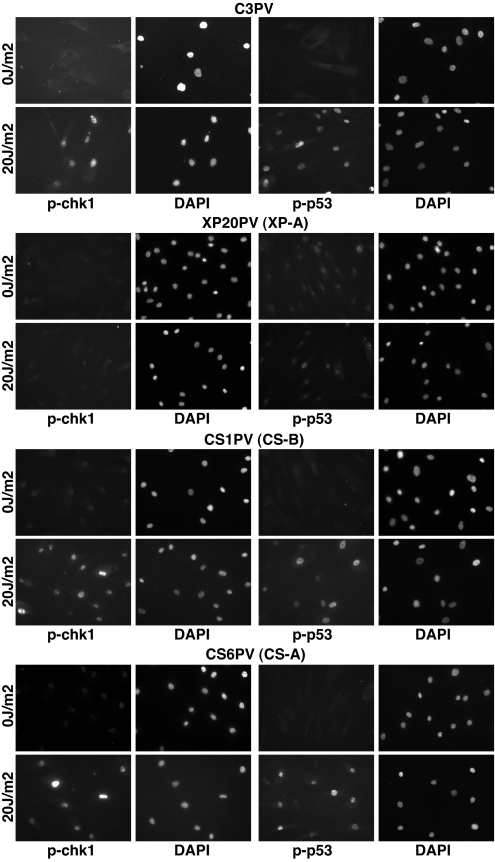
Fig. 1.
XPA-dependent and CSA/CSB-independent checkpoint activation in nonproliferating primary fibroblasts. In normal (C3PV), CS-B (CS1PV), and CS-A (CS6PV) fibroblasts, Chk1-Ser-317 and p53-Ser-15 are phosphorylated 1 h after 20 J/m2 UV-C radiation. Conversely, XP-A (XP20PV) primary fibroblasts are impaired in Chk1 and p53 phosphorylation in response to DNA damage. Cells were held under low serum conditions for 5 days before mock or UV irradiation, and the absence of any cycling cells was monitored by BrdU incorporation for each cell strain analyzed (data not shown). The activation of the DNA damage checkpoint was examined 1 h after UV irradiation, by immunofluorescence by using phosphospecific antibodies, anti-P-Ser-317-Chk1 and anti-P-Ser-15-p53, as described in Materials and Methods.
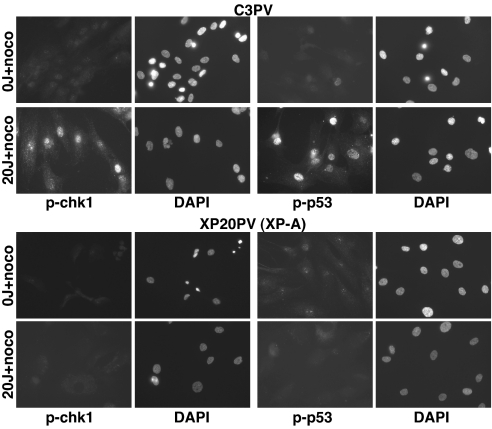
Fig. 2.
XPA-dependent checkpoint activation in G2/M primary fibroblasts. Normal (C3PV) and XP-A (XP20PV) primary fibroblasts were synchronized in G2/M, with nocodazole, before mock or UV irradiation (20 J/m2), and immunofluorescence was performed as described. More than 90% of the cells were in G2/M phases, as determined by flow cytometry analysis (data not shown).
In S-Phase Cells, Checkpoint Activation Does Not Depend on NER.
Asynchronously growing XP-A fibroblasts can activate the checkpoint, but co-staining with either anti-phospho-Chk1 or p53 and anti-BrdU antibodies revealed that phosphorylation of Chk1 and p53 is restricted to S-phase cells (Fig. 3). We noticed that in normal and CS-B proliferating fibroblasts, BrdU-positive cells (i.e., cells in S phase) show a stronger staining with anti-phospho-Chk1 or p53 antibodies than BrdU-negative cells (Fig. 3 and data not shown). The data suggest that, after UV irradiation, in normal fibroblasts the DNA damage checkpoint can be activated in all cell-cycle phases; however, the signal is higher in S phase, when probably other kind of lesions, such as replication forks stalling in front of UV radiation-induced photoproducts or strand breaks, may contribute to enhance the signal activating the checkpoint cascade (16). Thus, the analysis at the single-cell level enabled us to show that mutations in XPA do interfere with UV-induced checkpoint activation in G1 and G2/M phases, but not in S phase, of the cell cycle.
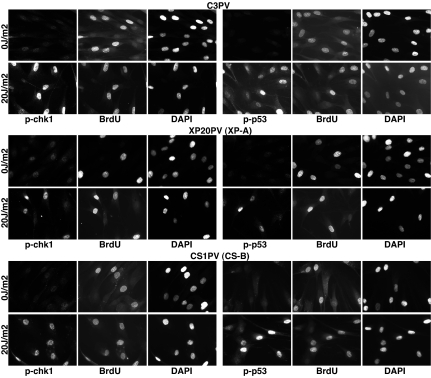
Fig. 3.
Proliferating normal and NER-deficient primary fibroblasts activate the DNA damage checkpoint after UV radiation. Normal (C3PV), XP-A (XP20PV), and CS-B (CS1PV) fibroblasts were pulsed-labeled for 1 h with 50 μM BrdU before mock or UV irradiation (20 J/m2). Cells were fixed 1 h later and coimmunostained with anti-P-Ser-317-Chk1 or anti-P-Ser-15-p53 and anti-BrdU antibodies.
Checkpoint Activation Does Not Require Functional TC-NER but Depends on GG-NER.
We next analyzed a large collection of nonproliferating primary fibroblasts from XP, CS, TTD, and UV light-sensitive syndrome patients to understand whether DNA damage recognition by repair proteins is sufficient for checkpoint activation or further processing of the primary lesion is required and what is the relative contribution of the two NER subpathways, TC-NER and GG-NER. Human fibroblasts were grown in low serum for 5 days, UV-irradiated, and, after 1 h, cells were stained with antibodies anti-phospho-Chk1 and anti-phospho-p53. The nuclear fluorescence intensity of captured images was quantified, and the percentage of positively stained cells whose fluorescence intensity was above a fixed threshold was scored (Fig. 4).
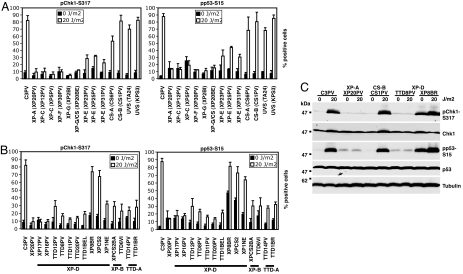
Fig. 4.
Processing of the lesion by GG-NER is required to trigger the signal cascade in response to UV. (A and B) We analyzed checkpoint activation in nonproliferating primary human fibroblasts derived from 27 patients (Table 1), affected by inherited NER-defective syndromes, by immunofluorescence using phosphospecific antibodies, recognizing Chk1 and p53 (anti-P-Chk1-Ser-317 and anti-P-p53-Ser-15). Approximately 300 fibroblasts per cell strain were scored, and the percentage of positively staining cells was measured as described in Materials and Methods. The results represent the mean and SD of three independent experiments. (C) Checkpoint activation was analyzed in nonproliferating primary fibroblasts from a normal donor (C3PV) and four NER-defective patients 1 h after UV radiation (20 J/m2) by immunoblot using the indicated antibodies. In extracts from normal, CS-B (CS1PV), and XP-D/CS (XP8BR) fibroblasts, P-Ser-317-Chk1 and P-Ser-15-p53 strongly increased after DNA damage. XP-A (XP20PV) and XP-D (TTD8PV) fibroblasts are impaired in checkpoint activation.
CS-A and CS-B primary fibroblasts are proficient in checkpoint activation, as well as fibroblasts derived from two UV light-sensitive syndrome patients (Fig. 4A) who exhibit a TC-NER defect but have a proficient GG-NER, like CS cells (5, 17). These findings demonstrate that TC-NER is not necessary for triggering the DNA damage cascade, after UV radiation.
Conversely, a total or partial inability to activate the checkpoint after UV irradiation was observed in XP-C and XP-E primary fibroblasts (Fig. 4A) that are defective, respectively, in the XPC-RAD23B and in the UV-DDB complex, two DNA damage-binding complexes specifically involved in GG-NER in mammals. Although in XP-C cells GG-NER is totally defective, in XP-E only the removal of the most abundant UV radiation-induced photoproduct (the cyclobutane pyrimidine dimer) from nontranscribed DNA is severely impaired (18–21). This different role of XPC and XPE in GG-NER may explain why the defect in checkpoint activation is more severe in XP-C than in XP-E fibroblasts. We showed that a yeast strain exclusively defective in GG-NER (rad7Δ) is proficient in checkpoint activation after UV radiation (6). Rad7 and Rad16 functions in Saccharomyces cerevisiae have many parallels to UV-DDB functions in mammalian cells, even though the two complexes do not show any structural similarity. Both the DDB subunits and Rad7–16 are components of E3 ubiquitin ligase complexes, whose candidate substrate is XPC/Rad4 (4). Mutations in XPE in mammalian cells and in RAD7 in yeast cause similar cellular phenotype; nevertheless, whereas rad7Δ cells are proficient in triggering the checkpoint cascade, XP-E fibroblasts cannot fully activate the checkpoint after UV radiation. A possible explanation of the difference found between yeast and human cells in the involvement of GG-NER in checkpoint activation might be related to the different percentage of transcribed genes relative to the whole genome in the two species. Because a small percentage of mammalian genome is transcribed, given a certain amount of lesions per base pair, it is expected that GG-NER has to deal with many more lesions in the whole genome in human cells compared with yeast cells.
Open Complex Formation and Processing of the Lesion Are Necessary for Checkpoint Activation.
Nonproliferating fibroblasts defective in either XPF or XPG nucleases (22, 23) are unable to phosphorylate Chk1 and p53 after UV irradiation (Fig. 4A). The data are in agreement with the observations we made in yeast, where checkpoint triggering requires the ability to form an opened incision complex in G1 and G2 blocked cells (6).
Mutations in the TFIIH complex, involved in both initiation of basal transcription and opening of a bubble around the lesion in NER, can lead to quite different XP, TTD, or combined XP/CS clinical features (24). The transcriptionally active form of TFIIH is made up of a total of 10 subunits, three of which were found defective in NER syndromes: the helicases XPD and XPB and TTDA, an 8-kDa protein involved in the stabilization of the complex. XPB is essential for TFIIH activity in both transcription and repair, and this probably accounts for the rarity of XP-B patients that show either XP/CS or TTD phenotype. XPD helicase activity is necessary for repair but dispensable for in vitro basal transcription. Thus the XPD gene is rather tolerant of mutations, and XP-D defects have been found in many patients with TTD or with XP and in rare XP/CS patients (24). The XPD gene mutation spectrum in patients is consistent with the hypothesis that the site of the mutation determines the clinical phenotype (25–27). Mutations in p8/TTDA were found in three families with TTD-affected members (28).
We analyzed checkpoint activation after UV irradiation in 10 primary fibroblasts carrying mutations in XPD, two in XPB, and two in TTDA (Fig. 4B; refs. 26–30). With the exception of XP-D cell strains from patients with the combined XP/CS phenotype (XP8BR, XPCS2, and XP1NE), all fibroblasts are totally or partially defective in Chk1 and p53 phosphorylation, 1 h after UV irradiation (between 10% and 30% positively stained XPD, XPB, and TTDA mutated cells were scored, compared with 80–90% positive normal cells, by immunofluorescence with phosphospecific antibodies anti-P-Chk1-Ser-317 and anti-P-p53-Ser-15). The data suggest that TFIIH opening of the DNA bubble around the lesion is required to properly activate the checkpoint in nonproliferating cells after UV radiation. It is interesting to notice that such a defect in checkpoint activation can be found both in fibroblasts from patients with XP features (e.g., XP16PV and XP17PV) and in fibroblasts from patients with TTD features (e.g., TTD11PV and TTD20PV). TTD-A cells also show a defect in checkpoint activation (Fig. 4B; TTD14PV and TTD1BR), in agreement with the recently demonstrated specific requirement of p8/TTD-A in NER, where it stimulates the opening of the DNA around the damage and XPA recruitment (31). TFIIH deficiency can thus lead to a similar defect in checkpoint activation in both XP and TTD patients, despite their extremely different clinical features and incidence of skin cancer. These findings support the hypothesis that the transcriptional defect present in TTD may somehow prevent in these patients the full development of a precarcinogenic lesion into a proper cancer (32).
Triggering of the checkpoint cascade in resting fibroblasts from a normal donor and from four patients representative of NER-defective syndromes, XP, CS, TTD, and XP/CS, was also analyzed by immunoblot by using phosphospecific antibodies, anti-P-Chk1-Ser-317 and anti-P-p53-Ser-15 (Fig. 4C). In agreement with the immunofluorescence data, whereas CS-B resting fibroblasts are proficient in checkpoint activation, mutations in XPA and XPD genes leading to XP and TTD clinical features, respectively, severely impair phosphorylation of checkpoint factors. The total amount of p53 protein does not increase after DNA damage, possibly because we analyzed triggering of the checkpoint cascade 1 h after UV irradiation, whereas others reported an increase in p53 only at later time points after UV radiation (15). A low level of p53 phosphorylation could be observed both in mock and UV-treated XP20PV and TTD8PV fibroblasts, probably because of unrepaired endogenous DNA damage that accumulated in normal growing conditions. UV radiation did not further increase the amount of P-Ser-15-p53 in these cell strains.
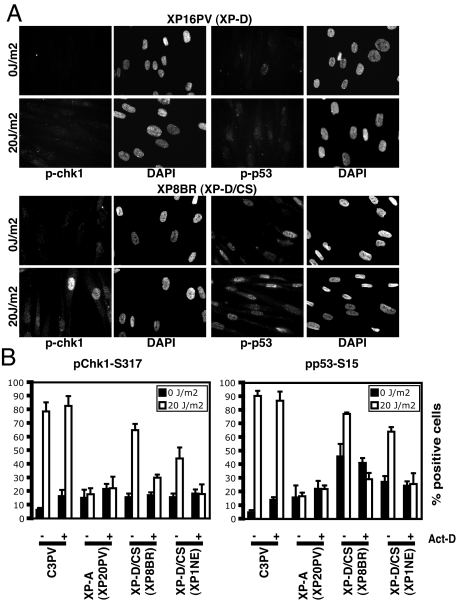
Transcription-Associated Checkpoint Activation in XP-D Fibroblasts from XP/CS Patients.
Contrary to all of the other TFIIH mutant cells that we analyzed, fibroblasts from patients XP8BR, XPCS2, and XP1NE, with a combined XP-D/CS phenotype of different degrees of clinical severity, are able to activate the checkpoint after UV irradiation (Figs. 4 B and C and 5). It was shown that in cells from these patients breaks are introduced into cellular DNA on exposure to UV damage, but these breaks are not at the sites of the damage, and it has been proposed that they are introduced erroneously by the NER machinery at sites of transcription initiation (33). Interestingly, triggering of the checkpoint cascade that can be observed after UV irradiation in these cells is associated with transcription. In fact, inhibition of RNA polymerase II transcription by actinomycin D significantly reduced Chk1 and p53 phosphorylation after UV irradiation in XP-D/CS fibroblasts, but it did not affect checkpoint activation in either normal or XP-A cells (Fig. 5B). We also analyzed checkpoint activation in extracts from resting XP8BR fibroblasts by immunoblot (Fig. 4C). Both Chk1-Ser-317 and p53-Ser-15 were already phosphorylated in mock-treated cells. A high percentage of positive cells were also scored among XP-D/CS mock-treated fibroblasts (Fig. 4B). We suppose that the checkpoint might be partially activated by oxidative damage that accumulates in normal growth conditions in XP-D/CS fibroblasts. In fact, DNA breaks were shown to be also generated by the introduction of plasmids harboring oxidative damage in XP-D/CS cells (33). After UV irradiation, a clear increase in the amount of phosphorylation of these key checkpoint factors could be observed.
Fig. 5.
XP-D/CS fibroblasts can phosphorylate Chk1 and p53 after UV damage, and checkpoint activation is associated with transcription. (A) Although in XP-D fibroblasts from the XP patient XP16PV phosphorylation of Chk1-Ser-317 and p53-Ser-15 cannot be detected 1 h after UV radiation (20 J/m2), XP/CS fibroblasts can activate the checkpoint in the same conditions. (B) The indicated nonproliferating cell strains were either mock-treated or UV irradiated, with or without adding actinomycin D (0.5 μg/ml) to the medium for 2 h, before irradiation. The percentage of positively staining cells was measured, and the results represent the mean and SD of three experiments.
In summary, in the present work, the analysis of checkpoint activation after UV irradiation in a large collection of primary fibroblasts shows that in nonproliferating cells, NER recognition and processing of the lesion are required to trigger the signal transduction cascade that leads to the phosphorylation of key checkpoint proteins, such as Chk1 and p53. Functional GG-NER is necessary and sufficient to activate the checkpoint response, whereas TC-NER may be dispensable. In XP-D fibroblasts from XP/CS patients, despite the fact that UV lesions are not processed by NER, the checkpoint might be activated by DNA breaks that occur at sites of transcription initiation.
Materials and Methods
Cell Culture.
Primary human fibroblasts from normal and repair-deficient individuals (Table 1) were cultured in Ham's F10 medium containing 15% FCS (Euroclone, Milan, Italy) and kept at 37°C in a humidified atmosphere with 5% CO2. To bring the cells into a nonproliferating state, cells were held under low serum conditions for 5 days (0.5% FCS), and the absence of any S-phase cells was monitored by BrdU incorporation for each cell strain analyzed (data not shown). Cells were synchronized in G2/M, adding 0.3 μM nocodazole (Sigma-Aldrich, St. Louis, MO) to Ham's F10 medium containing 15% FCS for 20 h. To detect cells that were replicating their DNA, 50 μM BrdU (Sigma) was added to the medium of exponentially growing fibroblasts for 1 h before UV radiation.
UV Irradiation.
For UV irradiation, medium was removed, and cells were washed once with PBS and then irradiated with a Vilber Lourmat (Marne-la-Valée) 12-W lamp (predominantly 254 nm) at a dose of 20 J/m2 (dose rate of ≈1 J/m2 per s). Subsequently, the medium was added back to the cells and the cells returned to culture conditions for 1 h.
Transcription Inhibition.
Fibroblasts held under low-serum conditions for 5 days were treated with actinomycin D (Sigma-Aldrich) at a concentration of 0.5 μg/ml for 2 h before irradiation. The transcription inhibitor was also added during the postirradiation 1-h incubation period.
Antibodies.
Phosphospecific antibodies anti-P-Ser-317-Chk1 and anti-P-Ser-15-p53, anti-Chk1, and anti α/β tubulin antibodies (rabbit polyclonal) were purchased from Cell Signaling Technology (Beverly, MA). Anti-p53 antibodies were purchased from Active Motif (Carlsbad, CA). Alexa Fluor 555 goat anti-rabbit secondary antibodies were from Molecular Probes (Eugene, OR). FITC-anti-BrdU antibodies were from Beckton Dickinson (Franklin Lakes, NJ).
Immunofluorescence Microscopy.
For the detection of phosphorylated Chk1-Ser-316 and p53-Ser-15, cells were grown onto sterile coverslips. After being rinsed in PBS, they were fixed in 2% paraformaldehyde in PBS for 10 min at room temperature. After being rinsed twice in PBS, cells were permeabilized in 0.2% Triton X-100 in PBS for 2 min at 4°C. This was followed by another rinse in PBS. Cells were then incubated for 1 h at room temperature in 5% Normal Goat Serum (Sigma-Aldrich) in PBS. Primary antibody (anti-P-Ser-317-Chk1 or anti-P-Ser-15-p53) incubations were performed for 2 h at room temperature at 1:100 dilutions in PBS supplemented with 2% BSA (Sigma-Aldrich) and followed by extensive washing in PBS. Incubations with Alexa Fluor 555 goat anti-rabbit secondary antibodies were performed for 1 h at room temperature at 1:500 dilution in 2% BSA in PBS. Nuclei were counterstained with 10 μg/ml DAPI (Sigma-Aldrich) in PBS for 2 min at room temperature. After extensive washing in PBS, coverslips were mounted in Vectashield (Vector Laboratories). For detection of incorporated BrdU, immunofluorescence was performed essentially as described above, except that after incubation with secondary antibodies, cells were fixed again with 2% paraformaldehyde in PBS, washed, treated with 4N HCl 10 min, and incubated 1 h with FITC-anti-BrdU antibodies 1:40. Images were taken with Olympus (Melville, NY) BX61 microscope analySIS software. Quantification of fluorescence intensity was performed on blind captured images by using standardized capture settings; image processing and quantification were performed with the ImageJ 1.36b software (Wayne Rasband, National Institutes of Health, Bethesda, MD). Cells were considered positive when the fluorescence value was above a threshold of 108 on a 0–255 grayscale. The error bars represent the standard deviation of the mean. A minimum of three independent experiments was carried out where error bars are shown.
Western Blotting.
Protein extracts and immunoblots were performed as suggested by Cell Signaling Technology. Briefly, cells were lysed in 1× SDS sample buffer (62.5 mM Tris·HCl, pH 6.8/2% wt/vol SDS/10% glycerol/50 mM DTT/0.01% wt/vol bromophenol blue), sonicated 10 sec, and heated to 95°C for 5 min, and equal amounts of total protein extract were loaded onto 10% SDS/PAGE gels. The blots were incubated overnight with polyclonal antibodies anti-P-Ser-317-Chk1, anti-Chk1, anti-P-Ser-15-p53, and anti-α/β tubulin (Cell Signaling Technology) and with anti-p53 (Active Motif) at 1:1,000 dilutions.
Supplementary Material
Acknowledgments
We thank Pietro Transidico, Imaging Unit, FIRC Institute of Molecular Oncology Foundation (Milan, Italy) for support in the quantification of the immunofluorescence images and all of the members of our laboratory for helpful discussions. F.M. was supported by International Centre for Genetic Engineering and Biotechnology funding. This work was supported by grants from Associazione Italiana Ricerca sul Cancro, Consorzio Interuniversitario per le Biotecnologie, Ministero dell' Università e della Ricerca, Fondazione Cariplo, and the European Union FP6 Integrated Project DNA repair. The financial support of Telethon-Italy Grant GGP030406 (to M.M.F.) is gratefully acknowledged.
Abbreviations
- NER
nucleotide excision repair
- TC-NER
transcription-coupled NER
- GG-NER
global genome NER
- XP
xeroderma pigmentosum
- TTD
trichothiodystrophy
- CS
Cockayne syndrome
- XP/CS
a pathological phenotype with combined symptoms of XP and CS
- ATR
ataxia-telangiectasia mutated- and Rad3-related.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Zou L, Elledge SJ. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 2.Cortez D. Genes Dev. 2005;19:1007–1012. doi: 10.1101/gad.1316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang G, Sancar A. Mol Cell Biol. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedberg EC, Walker GC, Siede W, Wood RD, 1Schultz RA, Ellenberger T. Washington, DC: Am Soc Microbiol Press; 2005. DNA Repair and Mutagenesis. [Google Scholar]
- 5.Spivak G. Mutat Res. 2005;577:162–169. doi: 10.1016/j.mrfmmm.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 6.Giannattasio M, Lazzaro F, Longhese MP, Plevani P, Muzi-Falconi M. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 8.Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. Proc Natl Acad Sci USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward IM, Minn K, Chen J. J Biol Chem. 2004;279:9677–9680. doi: 10.1074/jbc.C300554200. [DOI] [PubMed] [Google Scholar]
- 10.Bomgarden RD, Lupardus PJ, Soni DV, Yee MC, Ford JM, Cimprich KA. EMBO J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou BBS, Elledge SJ. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 12.Bowman KK, Sicard DM, Ford JM, Hanawalt PC. Mol Carcinog. 2000;29:17–24. [PubMed] [Google Scholar]
- 13.Koberle B, Roginskaya V, Wood RD. DNA Repair (Amst) 2006;5:641–648. doi: 10.1016/j.dnarep.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Colella S, Nardo T, Mallery D, Borrone C, Ricci R, Ruffa G, Lehmann AR, Stefanini M. Hum Mol Genet. 1999;8:935–941. doi: 10.1093/hmg/8.5.935. [DOI] [PubMed] [Google Scholar]
- 15.Conforti G, Nardo T, D'Incalci M, Stefanini M. Oncogene. 2000;19:2714–2720. doi: 10.1038/sj.onc.1203583. [DOI] [PubMed] [Google Scholar]
- 16.Garinis GA, Mitchell JR, Moorhouse MJ, Hanada K, de Waard H, Vandeputte D, Jans J, Brand K, Smid M, et al. EMBO J. 2005;24:3952–3962. doi: 10.1038/sj.emboj.7600849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh T, Ono T, Yamaizumi M. Mutat Res. 1994;314:233–248. doi: 10.1016/0921-8777(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 18.Chavanne F, Broughton BC, Pietra D, Nardo T, Browitt A, Lehmann AR, Stefanini M. Cancer Res. 2000;60:1974–1982. [PubMed] [Google Scholar]
- 19.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JHJ. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 20.Rapic-Otrin V, Navazza V, Nardo T, Botta E, McLenigan M, Bisi DC, Levine AS, Stefanini M. Hum Mol Genet. 2003;12:1507–1522. doi: 10.1093/hmg/ddg174. [DOI] [PubMed] [Google Scholar]
- 21.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Proc Natl Acad Sci USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lalle P, Nouspikel T, Constantinou A, Thorel F, Clarkson SG. J Invest Dermatol. 2002;118:344–351. doi: 10.1046/j.0022-202x.2001.01673.x. [DOI] [PubMed] [Google Scholar]
- 23.Okinaka RT, Perez-Castro AV, Sena A, Laubscher K, Strniste GF, Park MS, Hernandez R, MacInnes MA, Kraemer KH. Mutat Res. 1997;385:107–114. doi: 10.1016/s0921-8777(97)00031-1. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann AR. Genes Dev. 2001;15:15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- 25.Stefanini M. In: DNA Repair and Human Disease. Balajee AS, editor. Austin, TX: Landes Bioscience; 2006. pp. 30–46. [Google Scholar]
- 26.Taylor EM, Broughton BC, Botta E, Stefanini M, Sarasin A, Jaspers NGJ, Fawcett H, Harcourt SA, Arlett CF, Lehmann AR. Proc Natl Acad Sci USA. 1997;94:8658–8663. doi: 10.1073/pnas.94.16.8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Botta E, Nardo T, Broughton BC, Marinoni S, Lehmann AR, Stefanini M. Am J Hum Genet. 1998;63:1036–1048. doi: 10.1086/302063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giglia-Mari G, Coin F, Ranish JA, Hoogstraten D, Theil A, Wijgers N, Jaspers NG, Raams A, Argentini M, van der Spek PJ, et al. Nat Genet. 2004;36:714–719. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 29.Stefanini M, Lagomarsini P, Giliani S, Nardo T, Botta E, Peserico A, Kleijer WJ, Lehmann AR, Sarasin A. Carcinogenesis. 1993;14:1101–1105. doi: 10.1093/carcin/14.6.1101. [DOI] [PubMed] [Google Scholar]
- 30.Vermeulen W, Scott RJ, Rodgers S, Muller HJ, Cole J, Arlett CF, Kleijer WJ, Bootsma D, Hoeijmakers JHJ, Weeda G. Am J Hum Genet. 1994;54:191–200. [PMC free article] [PubMed] [Google Scholar]
- 31.Coin F, Proietti De Santis L, Nardo T, Zlobinskaya O, Stefanini M, Egly JM. Mol Cell. 2006;21:215–226. doi: 10.1016/j.molcel.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 32.Berneburg M, Clingen PH, Harcourt SA, Lowe JE, Taylor EM, Green MH, Krutmann J, Arlett CF, Lehmann AR. Cancer Res. 2000;60:431–438. [PubMed] [Google Scholar]
- 33.Theron T, Fousteri MI, Volker M, Harries LW, Botta E, Stefanini M, Fujimoto M, Andressoo JO, Mitchell J, Jaspers NG, et al. Mol Cell Biol. 2005;25:8368–8378. doi: 10.1128/MCB.25.18.8368-8378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.