Abstract
Dendritic cells (DCs) express multiple Toll-like receptors (TLR) in distinct cellular locations. Herpes simplex viruses (HSV) have been reported to engage both the surface TLR2 and intracellular TLR9 in conventional DCs. However, the contributions of these TLRs in recognition of HSV and the induction of proinflammatory cytokines in DCs remain unclear. Here, we demonstrate that a rare population of HSV, both in laboratory strains and in primary clinical isolates from humans, has the capacity to activate TLR2. This virus population is recognized through both TLR2 and TLR9 for the induction of IL-6 and IL-12 secretion from bone marrow-derived DCs. Further, we describe a previously uncharacterized pathway of viral recognition in which TLR2 and TLR9 are engaged in sequence within the same DC. Live viral infection results in two additional agonists of TLR2 and TLR9. These results indicate that in cells that express multiple TLRs, pathogens that contain multiple pathogen-associated molecular patterns can be detected in an orchestrated sequence and suggest that the innate immune system in DCs is optimized to linking uptake and degradation of pathogens to microbial recognition.
Keywords: cytokines, innate immunity, Toll-like receptor, viral infection, plasmacytoid dendritic cell
Innate recognition of viruses by the mammalian immune system is critical in providing both the immediate antiviral effects, mediated in large part by type I IFNs (1), and in inducing appropriate classes of adaptive immune responses required for clearing viral infection (2, 3). Distinct mechanisms are used to recognize viruses and the viral replication intermediates produced during viral infections. Whereas most cells use the retinoic acid inducible gene-I to recognize certain ssRNA virus infections (4) and melanoma differentiation-associated gene 5 to recognize picornaviruses (5), plasmacytoid dendritic cells (pDCs) use exclusively the Toll-like receptors (TLRs) to recognize ssRNA and dsDNA viruses (4, 6). More recently, a TLR-independent cytosolic recognition of DNA has been shown to play a major role in the induction of type I IFNs (7, 8).
For innate recognition of herpes viruses, at least three pathways have been described. The first pathway involves the detection of viral genomic dsDNA by the TLR9. This pathway is used by the pDCs to recognize Herpes simplex virus (HSV)-1 (9, 10) and HSV-2 (10). TLR9-mediated recognition also represents the predominant pathways in non-pDCs, such as splenic CD11c+ DCs upon HSV-1 infection (9). Further, systemic inoculation of UV-irradiated HSV-2 results in IFNα production in an myeloid differentiation factor (MyD)88 and TLR9-dependent manner (10). Similarly, in vivo recognition of a beta herpes virus, murine cytomegalovirus, has been shown to depend on TLR9 and MyD88 (11, 12). The second type of TLR–herpesvirus interaction occurs when virions engage surface TLR2 on DCs and macrophages. Peritoneal macrophages (13) and microglial cells (14) secrete inflammatory cytokines in response to HSV-1 in a TLR2-dependent manner. The third type of viral recognition results in a MyD88-independent, IFN regulating factor (IRF)-7-dependent pathway. This third pathway has been demonstrated to play a major role during systemic infection with HSV-1, because IFNα secretion and suppression of viral replication depended mostly on IRF7 and to a much lesser extent on MyD88 (6).
Although these three distinct pathways of herpesvirus recognition are known to exist, the relative contributions of these pathways in viral recognition vs. viral pathogenesis are unclear. The ability of HSV-1 to trigger TLR2 has been shown to be responsible for the exacerbation of neonatal herpes encephalitis (13), because neonatal mice deficient in TLR2 secreted less IL-6 and had a higher rate of survival compared with WT mice upon lethal HSV-1 challenge. Further, the ability of HSV-1 and HSV-2 to activate TLR2 has been postulated to play a role in the sepsis-like disease associated with human neonatal herpes infection (15). These studies suggested that inflammatory responses induced by TLR2 engagement by HSV leads to immunopathology in the infected host. However, these studies used laboratory strains of HSV, and the clinical and epidemiological relevance of TLR2-activating HSV-1 and HSV-2, hereby termed HSV-1TLR2* and HSV-2TLR2*, are unknown. Thus, in this study, we examined the occurrence of the HSV-1TLR2* and HSV-2TLR2* in both the laboratory strains and the human clinical isolates from a variety of tissues and mucosal secretions. We show that HSVTLR2* constituted a minor population of HSV found in either the laboratory or the clinical isolates. Further, by plaque purification of the HSV-1TLR2*, we demonstrate that these represent a minor subspecies amongst the non-TLR2 activating HSV-1, or HSV-1TLR2°.
The HSV-1TLR2* in the clinical and laboratory viral isolates can engage both TLR2 and TLR9. However, the relative contributions of these two receptors in viral recognition and cytokine secretion in cells that express both of these receptors are unknown. TLR engagement leads to the activation of the IRF5 via MyD88 and tumor necrosis factor receptor-associated factor 6 (16). This TLR-MyD88-IRF5 axis is required for the production of proinflammatory cytokines, IL-6, IL-12, and TNFα (16). Given that TLR2 functions at the cell surface (17), whereas TLR9 functions in the endosomes (18, 19), if both receptors are involved in viral recognition, it is unknown whether TLR2 and TLR9 induce independent signals or whether the two TLRs synergize or antagonize to produce inflammatory cytokines. Here, we addressed the relative importance of TLR2 and TLR9 in herpesvirus recognition by classical DCs (cDCs) and pDCs.
Results
Only Certain Laboratory Strains of HSV Trigger TLR2 Activation.
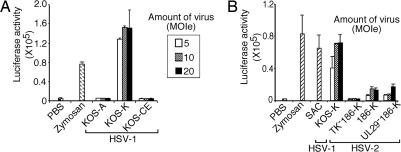
To understand the relative contributions of innate viral recognition through TLR2 and TLR9, we first examined the ability of a number of commonly used laboratory HSV strains to induce activation of NFκB via TLR2. We investigated the ability of the purified HSV-1 and HSV-2 to induce luciferase reporter activity driven by the NFκB promoter by using transiently transfected HEK293T cells. Because very similar results were obtained by using live or UV-inactivated viruses, these forms of viruses were used interchangeably in the luciferase reporter assays. As reported previously (13), HSV-1 KOS strain obtained from David Knipe's laboratory (KOS-K) induced strong activation of TLR2. In contrast, KOS purchased from American Type Culture Collection (KOS-A) or KOS obtained from the laboratory of Gary Cohen and Roselyn Eisenberg (KOS-CE) completely failed to trigger TLR2 activation even at high multiplicity of infection equivalent (MOIe) (Fig. 1A; see Table 1, which is published as supporting information on the PNAS web site). However, not all HSV strains obtained from the Knipe laboratory activated TLR2, as a thymidine kinase (TK) negative mutant, TK− HSV-2 from the same laboratory (TK−186-K) failed to induce NFκB activation via TLR2, whereas the WT 186 and unique long region 29 (UL29)-deficient 186 strains (186-K and UL29−186-K) stimulated low levels of TLR2 activation (Fig. 1B). No other TLR tested (TLR3, TLR4, TLR7, or TLR9) elicited NFκB activation by KOS-K in this reporter cell line (data not shown). These observations prompted us to examine the basis for the difference between the HSV KOS that triggers TLR2 (HSV-1 KOSTLR2*) versus those that do not (HSV-1 KOSTLR2°).
Fig. 1.
Only certain laboratory strains of HSV trigger activation through TLR2. (A) HEK293T cells expressing TLR2, CD14, and NFκB-driven firefly luciferase reporter gene (293T/luc/TLR2/CD14 cells) were cultured with the indicated MOI equivalents (MOIe) of UV-inactivated HSV-1 KOS strains from three different sources (A, ATCC; K, Knipe; CE, Cohen and Eisenberg) or 10 μg/ml zymosan for 7 h, and luciferase activities were measured. (B) 293T/luc/TLR2/CD14 cells were cultured with the indicated MOIe of UV-inactivated HSV-1 or HSV-2 strain, zymosan (10 μg/ml), or 0.02% SAC for 7 h, and luciferase activities were measured. The data are representative of six similar experiments.
A Minor Subspecies of the HSV-1 KOSTLR2* Activates TLR2.
To investigate the molecular basis for the difference in HSV-1 KOSTLR2* and HSV-1 KOSTLR2°, we first prepared genomic DNA from the purified KOS-K and KOS-A and sequenced the whole genome. However, this analysis did not result in meaningful data as we observed sequences representing several subspecies of HSV from bulk cultures of KOS-K and KOS-A (unpublished observations). To obtain pure single clones of viruses, we carried out plaque purification of KOS-K and KOS-A. Sixty-four plaques were picked from the plates inoculated with KOS-K, and pools of 10 plaques were examined for the TLR2-agonist activity. To our surprise, not all pools of KOS-K clones stimulated TLR2 (Fig. 7A, which is published as supporting information on the PNAS web site). Next, some of the individual plaque isolates were examined for their ability to trigger TLR2 activation (Fig. 7 B and C). Of the plaque isolates p11-p20, only p18 had the ability to stimulate TLR2. To compare the abilities of the KOS-K viral clones to trigger TLR2, plaque-purified viruses were propagated and purified, and various MOI of clones were tested for TLR2 activation. At all MOI tested, only the p63 of the plaque isolates p1 and p61–64 had the capacity to stimulate TLR2 activity (Fig. 7D). Thus, these results indicated that KOS-K consisted of a mixture of virus subspecies, most of which failed to activate TLR2. Thus, these results demonstrated that only certain laboratory isolates of KOS possess the ability to trigger TLR2 activation (Table 1), and that those strains consist of a mixture of viral subspecies, of which a minority activates TLR2 (Fig. 7).
Rare Occurrence of HSVTLR2* in Human Clinical Isolates.
The observation that only certain laboratory isolates have the TLR2-activating capacity prompted us to examine the relevance of this type of activity in human clinical isolates. Thus, viruses were propagated from human clinical samples taken from a variety of mucosal and systemic sites from two different hospital sources (Table 2, which is published as supporting information on the PNAS web site). Because of the implications to neonatal herpes (13, 15), we included four samples from the neonates with infection involving the CNS and disseminated HSV-1. A preliminary screening of the primary clinical isolates from Yale New Haven Hospital found no TLR2-agonist activities (Fig. 8, which is published as supporting information on the PNAS web site). To extend this analysis, we examined 32 clinical isolates from various sources (Table 2). Vero cell lysates of certain clinical isolates showed strong activity (Gen A and CNS C), whereas others had modest activity (Ora E, CNS B, CNS D, and CNS F) (Fig. 8 B and C). To confirm these findings with purified viruses, we propagated these clinical isolates and reexamined the purified virions for their TLR2-activating capacity. These analyses revealed that only two isolates (Gen A and CNS C) had the true capacity to activate TLR2 (Fig. 8D). To further examine whether clinical isolates that trigger TLR2 consists of a uniform population or a mixture of subspecies, several plaques were isolated from CNS C. Analysis of the purified plaque-isolated clones of CNS C revealed heterogeneity in the ability of individual clones to trigger TLR2 activation (Fig. 8E). These results indicated that a rare HSV-1 shed by the infected humans activated TLR2, and that such isolates consisted of a mixture of TLR2-activating and non-TLR2-activating subspecies.
HSVTLR2* Variants Are Recognized Through TLR2, TLR1, and TLR6 Independently of CD14.
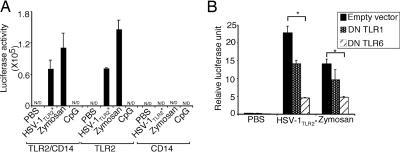
Thus far, we have demonstrated the ability of certain laboratory and clinical isolates of HSV to trigger NFκB activation in HEK293T reporter cells transfected with TLR2. However, the importance of other TLRs that are known to pair with TLR2, such as TLR1 and TLR6 (17), and CD14, a coreceptor involved in TLR2 activation by CMV (20), in HSV-1TLR2* recognition is unknown. To determine the involvement of CD14, HEK293T cells were transfected with TLR2, CD14 or in combination, and the ability of the HSV-1TLR2* to induce NFκB activation was assessed. It is known that HEK293T cells do not express functional levels of CD14 and require exogenous CD14 to respond to LPS (21). Thus, these results indicated that CD14 is not required for HSV-1TLR2* recognition (Fig 2A). Next, we examined the requirement for TLR1 or TLR6 in TLR2-mediated recognition of HSV-1TLR2*. Because HEK293T cells express both TLR1 and TLR6 endogenously (22), we transfected TLR2-expressing HEK293T cells with the dominant negative (DN) TLR1 or DN TLR6. Expression of the DN TLR6, and to a lesser extent, DN TLR1, resulted in a reduction of NFκB activation in cells triggered with HSV-1TLR2*. A similar reduction in the NFκB activation was seen with zymosan (Fig. 2B), a known agonist of TLR2/TLR6 (17). These data suggested that the recognition of HSV-1TLR2* is mostly mediated by TLR2 and TLR6, and to a lesser extent by TLR2/TLR1 complexes independent of CD14.
Fig. 2.
HSVTLR2* variants are recognized through TLR2, TLR1, and TLR6 independently of CD14. (A) HEK293T cells were transfected with TLR2 and/or CD14 plasmid. The cells were stimulated with 5 MOIe of UV-inactivated HSV-1 KOS-K, 10 μg/ml of zymosan, or 5 μg/ml of CpG 1826 for 7 h. Luciferase activity was measured. (B) 293T/luc/TLR2/CD14 cells were transfected with 500 ng of DN TLR1 or DN TLR6 plasmid and Renilla luciferase plasmid. Twenty-four hours later, the cells were stimulated with 5 MOIe of UV-inactivated HSV-1 KOS-K or zymosan for 7 h. Relative luciferase unit was calculated as a ratio of NFκB-dependent firefly luciferase activity to NFκB-independent Renilla luciferase activity. N/D, not detected. ∗, P < 0.05. The data are representative of three similar experiments.
cDCs Use both TLR2 and TLR9 to Recognize the HSVTLR2* Variants.
We thus far have demonstrated that a rare population of HSV-1 and HSV-2 can activate TLR2/TLR6 and TLR2/TLR1 independent of CD14. Key antigen presenting cell types, myeloid DCs, and macrophages, express TLR1, TLR2, TLR6, and TLR9, among other TLRs (3), and are known to respond to viruses including HSV-1 and HSV-2 (9, 13, 23–25). However, it remains controversial through which receptors DCs recognize HSVTLR2*.
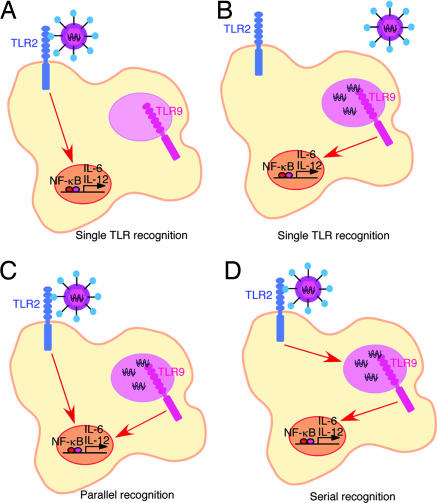
Several potential mechanisms can be postulated for HSVTLR2* recognition by DCs (Fig. 3). The simplest is that in which HSVTLR2* only activates TLR2 or TLR9 in DCs (Fig. 3 A and B). Alternatively, HSVTLR2* might activate both TLR2 and TLR9 in DCs. Within this scenario, there are at least two possibilities, the first is that HSVTLR2* recognition occurs by two separate pathways upon engagement of TLR2 and TLR9 (Fig. 3C). This can occur in the same DC (as drawn), or in separate cells, depending on which receptor happens to be engaged in a given DC. The final possibility is that HSVTLR2* is detected sequentially, first by TLR2 at the cell surface, and subsequently, via TLR9 in the endosome/lysosome compartment in the same DC (Fig. 3D). In this scenario, TLR9 signal depends on the initial activation of the cell through TLR2.
Fig. 3.
Schematic of possible recognition mechanisms of HSVTLR2* by DCs. (A and B) Single TLR-mediated recognition of HSVTLR2* through either TLR2 (A) or TLR9 (B). (C) Parallel recognition of HSVTLR2* via TLR2 and TLR9 in the same cells. (D) Serial recognition of HSVTLR2*, first by the cell surface TLR2 and then by the intracellular TLR9.
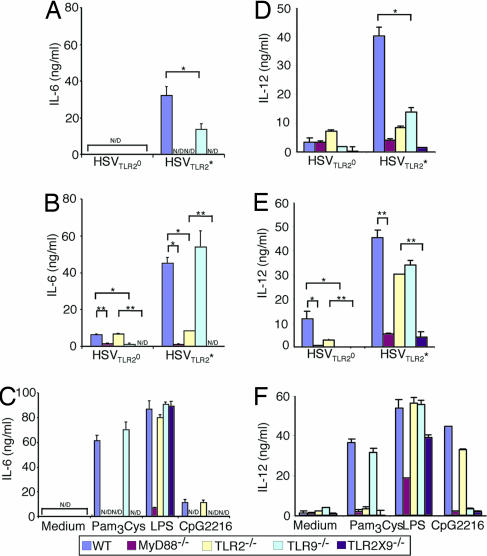
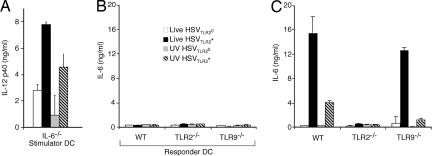
To critically examine the relative roles of TLR2 and TLR9 in HSV recognition, bone marrow (BM) DCs from mice lacking TLR2, TLR9, TLR2 × 9, or MyD88 were stimulated with either UV-inactivated or live HSVTLR2* or HSVTLR2°, and the secretion of a representative proinflammatory cytokines IL-6 and IL-12 were measured (Fig. 4). Consistent with their ability to stimulate TLR2 activation in a reporter system (Fig. 1), UV-irradiated HSVTLR2* induced robust IL-6 and IL-12 secretion that depended mostly on TLR2 and MyD88, whereas little IL-6 or IL-12 was secreted from DCs stimulated with UV-inactivated HSVTLR2° (Fig. 4 A and D). However, to our surprise, IL-6 and IL-12 responses also were reduced significantly in the absence of TLR9 (Fig. 4 A and D). To a control TLR agonist, LPS, TLR2−/−, TLR9−/−, or TLR2 × 9−/− BM DCs secreted WT levels of IL-6 (Fig. 4C) and IL-12 (Fig. 4F). These data indicated that DCs use both TLR2 and TLR9 to recognize HSVTLR2*, but more importantly, a large fraction of the TLR2-dependent recognition of HSVTLR2* also requires TLR9. Thus, a sequential recognition of HSVTLR2* via TLR2 and TLR9 accounts for majority of the TLR2-mediated recognition of UV-irradiated HSVTLR2* virions (Fig. 3D), whereas the rest of the TLR2-dependent recognition is independent of TLR9 in DCs (Fig. 3A). Thus, UV-irradiated HSVTLR2* virions are detected by a combination of the serial TLR2 → TLR9 (Fig. 3D) and single TLR2 (Fig. 3A) recognition mechanisms in DCs.
Fig. 4.
cDCs use both TLR2 and TLR9 to recognize the HSVTLR2* variants. BM DCs from WT or the indicated knockout mice were cultured with 2 MOI of UV-inactivated (A and D), live HSVTLR2* or HSVTLR2° (B and E), or known TLR agonists (C and F) for 24 h. Supernatants were analyzed for IL-6 (A–C) and IL-12 p40 (D–F) by ELISA. N/D, not detected. ∗, P < 0.05; ∗∗. P < 0.01. The data are representative of five similar experiments.
Next, we examined the mechanism of DC recognition of live HSVTLR2* infection. In contrast to the UV-inactivated virions, live HSVTLR2* elicited levels of IL-6 (Fig. 4B) and IL-12 (Fig. 4E) from TLR9−/− BM DC comparable with WT, an observation consistent with a previous study (23). These data suggested that a non-TLR9 MyD88-dependent signals induced by the replicating HSVTLR2* is sufficient to trigger maximum response in DCs. Moreover, TLR2−/− BM DC secreted significant levels of cytokines, particularly IL-12, in response to live HSVTLR2* infection. To determine whether cytokine production from the TLR2−/− DC depends on TLR9, BM DC from TLR2 × 9 double knockout mice were stimulated with the same viruses. The TLR2 × 9−/− DCs secreted minimal to undetectable amounts of IL-6 or IL-12 upon infection with HSVTLR2*, similar to those observed from the MyD88−/− BM DCs. These data indicated that the innate recognition of live HSVTLR2* can be largely accounted for by the combinations of signals from TLR2 and TLR9. Collectively, these data demonstrated that live HSVTLR2* infection of DCs engages two additional types of recognition pathways, TLR2-dependent TLR9-independent (Fig. 3A) and TLR2-independent TLR9-dependent (Fig. 3B), and that deficiency in either receptors can be compensated for by the other. In contrast to the HSVTLR2*, infection of BM DCs with HSVTLR2° resulted in low levels of cytokine secretion, mostly dependent on TLR9. Thus, HSVTLR2° live infection is detected through the TLR2-independent TLR9-dependent pathway (Fig. 3B).
Direct Stimulation of DCs by Viruses Is Required to Induce Cytokine Secretion.
Thus far, our data suggested that a sequential recognition of HSVTLR2* occurs within the same DC through TLR2 followed by TLR9. However, these data also are consistent with the possibility of a positive cross-talk between the responder DCs (recognizing the virus via TLR2) and bystander DCs (activated in a TLR9-dependent manner). To examine the latter possibility, we established a DC-DC coculture system in which the stimulator DCs are treated with HSVTLR2* or HSVTLR2° before addition to the uninfected responder DCs. To ensure that the readout cytokine (IL-6) from the responders are not contaminated by that from the stimulators, we used IL-6−/− DCs as the stimulator cells. Thus, IL-6−/− DCs were incubated with either live or UV-inactivated HSVTLR2* or HSVTLR2° for 24 h, washed thoroughly, UV-irradiated and then were added to the responder DCs. To confirm that the stimulator cells were fully activated by the viral treatment, IL-12p40 secretion from the stimulator cells was measured after the respective treatments (Fig. 5A). These data indicated that the IL-6−/− stimulator DCs responded to the virus infection and secreted cytokine in a manner similar to the WT DCs (Fig. 4). To assess the ability of the stimulator DCs to activate bystander DCs, IL-6 secretion from the responder DC supernatants was measured after 24 h of coculture with the stimulator DCs. Remarkably, the bystander responders failed to secrete IL-6 in response to the virus-infected stimulator cells (Fig. 5B), despite their intact ability to secrete IL-6 after direct virus infection (Fig. 5C). Therefore, these data indicated that bystander activation of the uninfected DCs by other HSV-infected DCs was not observed and that direct recognition of the virus is required to activate DCs to produce inflammatory cytokines.
Fig. 5.
Direct virus recognition, but not bystander stimulation, induces cytokine secretion from cDCs. IL-6−/− BM DCs (stimulator) were incubated with 2 MOI of live or UV-inactivated HSV-1TLR2* or HSV-1TLR2° for 24 h, and after thorough washing, they either were incubated for additional 24 h for assessment of IL-12 secretion (A) or exposed UV light to prevent de novo viral replication. (B) The stimulator DCs were cocultured with the indicated responder DCs. (C) In parallel, the responder DCs alone were incubated directly with live or UV-inactivated HSV-1TLR2* or HSV-1TLR2° for 24 h, and IL-6 levels were measured. The data are representative of three similar experiments.
pDCs Use TLR9 to Recognize both HSVTLR2* and HSVTLR2°.
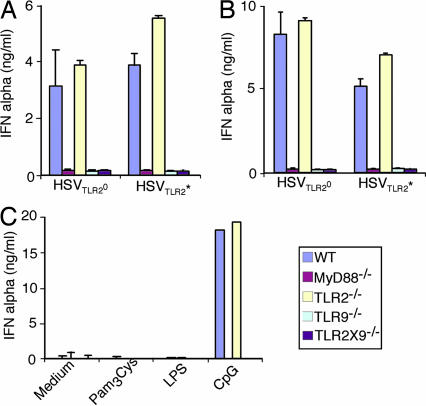
Thus far, the dual receptor-mediated recognition of HSVTLR2* was shown for cDCs in response to live and UV-inactivated viruses. However, IFNα was not detectable from cDCs infected with any of these viruses (data not shown). It is known that pDCs respond to HSV-1 and HSV-2 via TLR9 and secrete robust levels of type I IFNs (9, 10). In an effort to understand innate recognition of HSVTLR2* and HSVTLR2° by pDCs, BM pDCs purified from mice lacking TLR2, TLR9, or both were stimulated with either live or UV-inactivated HSVTLR2* or HSVTLR2°, and IFNα was measured. In contrast to the cDCs, pDCs recognized both HSVTLR2* and HSVTLR2° via TLR9 and not TLR2 resulting in comparable IFNα secretion in response to these viruses (Fig. 6). Further, pDCs failed to respond to either TLR2 or TLR4 agonists, whereas they responded to CpG in a TLR9-dependent manner (Fig. 6C). Thus, unlike cDCs, pDCs recognize both HSVTLR2* and HSVTLR2° via TLR9, regardless of whether the virus is live or UV-inactivated. Further, the ability of an HSV isolate to trigger TLR2 did not alter the recognition of such virus by pDCs via TLR9.
Fig. 6.
pDCs secretion of IFNα depends on TLR9 in response to both HSVTLR2* and HSVTLR2°. BM pDCs from the indicated mice were purified and stimulated with UV-irradiated (A) or live (B) HSVTLR2* or HSVTLR2° at MOI of 2, or with the indicated TLR agonists (C). Supernatants were analyzed for IFNα by ELISA. The data are representative of three similar experiments.
Discussion
The innate immune system is designed to optimally detect a given pathogen through multiple pattern recognition receptors. Most pathogens possess multiple pathogen-associated molecular patterns that are engaged by distinct sets of TLRs. Bacterial surfaces are decorated by LPS, lipopeptide, peptidoglycans, and flagellin which trigger plasma membrane TLRs (2, 26), whereas their genomes activate intracellular TLR9 in the lysosome (27). Further, combinations of pathogen-associated molecular patterns that trigger TLR3 and TLR4 in synergy with TLR7, TLR8, and TLR9 were shown to elicit robust type 1 immunity (28), revealing immunological advantages of engaging multiple TLRs. However, the mechanism of recognition of a live pathogen by a single cell type through multiple TLRs remains unclear. Careful coordination of timing and location of the engagement of the surface TLRs upon phagocytosis and the intracellular TLRs upon uncoating of the outer membranes is expected to be crucial in innate defense against pathogens. Recently, TLR9 and TLR2 were shown to cooperate in the recognition of Mycobacterium tuberculosis, and that additive signals from both TLR9 and TLR2 constituted full recognition in cDCs (29). In the case of M. tuberculosis infection, TLR9 recognition did not depend on the initial engagement of TLR2. Here, we showed that a virus is detected in sequence by TLRs in DCs, first, by the surface TLR2 interacting with the virions, and second, by the intracellular TLR9 recognizing the viral genomic DNA. We showed that such recognition must occur within the same DCs upon direct recognition of the virus and not through activation of bystander DCs. This serial recognition system represents an important mechanism used by DCs to detect HSVTLR2* virions. It is interesting to note that the recognition of a bacterial pathogen (M. Tuberculosis) also requires TLR2 and TLR9, mostly involving parallel TLR2 + TLR9 recognition (29). Upon infection and replication of HSVTLR2*, additional pathways of recognition by cDCs are engaged. First, a robust engagement of the TLR2 likely occurs upon recognition of the newly synthesized viral products leading to TLR2-dependent TLR9-independent activation of cDCs. Second, infection by HSVTLR2* generates a new TLR9 agonist, which is also generated to a lesser extend by infection with HSVTLR2°, which activates cDCs via the TLR2-independent TLR9-dependent pathway. However, activation of TLR2 and TLR9 in DCs can compensate for the deficiency in either receptor. Thus, live HSV infection results in the engagement of additional TLR2 and TLR9, and in the absence of both, little inflammatory cytokines are produced by cDCs in response to HSV infection (Fig. 9, which is published as supporting information on the PNAS web site). Unlike cDCs, pDCs recognize both HSVTLR2* and HSVTLR2° equally through TLR9. Because the pDCs do not express TLR2 (30), this result is not surprising. Further, because experiments involving cDC and pDC were carried out simultaneously by using the same sets of viruses, IFNα levels from the pDCs served as an internal control, as a barometer for the amount of virions present in each well.
Our attempt to study the genetic basis by which HSV engages TLR2 has led to an interesting and somewhat surprising finding. First, the TLR2-activating phenotype was observed in a rare population of laboratory strains and clinical isolates of HSV. Moreover, the viral strains and isolates existed as a collection of subspecies of viral clones, some of which triggered TLR2, whereas the majority did not. Although the mechanism of the existence of diversity among viral clones is unknown, the evolutionary reason for certain HSV to acquire TLR2-activating capacity is of interest. For old viruses such as the members of the herpesviruses that have reached a stable state of parasitism in a variety of hosts ranging from oysters to humans, the advantage of eliciting inflammatory responses via TLR2 only in some infected individual must reflect a design of the viral survival and spreading strategy. It was shown that IL-6-deficient mice are less able to survive ocular HSV-1 challenge compared with WT mice (31). On the other hand, reactivation of the latent HSV-1 was decreased in mice injected with neutralizing antibodies to IL-6 (32). The ability of the HSV-1 to potentiate host survival and virus reactivation via IL-6 may be mediated by the virus' ability to regulate activation of TLR2 in DCs for successful transmission between the hosts. This ability also may manifest in pathology in immunocompromised individuals, but the TLR2-activating phenotype is not required for the inflammatory responses associated with HSV (Table 2).
Phagocytosis and degradation of bacterial and fungal pathogens by DCs and macrophages have been shown to be more efficient when the phagosomes contain TLRs that are engaged by such microbes (33). These phagocytes likely have developed strategies to link degradation and detection of pathogens by placing surface and intracellular TLRs in strategic locations. The sequential detection of HSV described in this study may reflect an evolutionary advantage of linking phagosome maturation with innate detection of microbial genomes by intracellular TLRs that require maturation of the phagosomes. Thus, it is tempting to speculate from our data that TLR2 recognition of virions on the cell surface induces maturation of the endosomes that have taken up the virions, facilitating the subsequent recognition of viral genomic dsDNA by TLR9.
The current study helps to clarify the apparently conflicting results reported in previous studies for the requirement of TLR2 vs. TLR9 in HSV recognition by myeloid DCs (9, 13, 23, 25). Thus, when no dependence on TLR9 was found in macrophages and myeloid DCs, such conditions were created by live HSV-1 infection that likely engaged TLR2 (23). Recognition of HSV-1 KOS has been reported to largely depend on TLR2 when the peritoneal macrophages were infected with HSV-1TLR2* (13). On the other hand, variable levels of TLR2-independent MyD88-dependent secretion of TNFα by the peritoneal macrophages occurred upon HSV-1TLR2* infection because it triggered the replication-dependent TLR9 pathway in these cells (25). Because the induction of the inflammatory cytokines after infection with live HSV-1TLR2* of DCs depends on both TLR2 and to a lesser extent TLR9 (Fig. 9), it would be important to compare susceptibility to infection of the TLR2 × 9 DKO mice to the single TLR KO mice. Such analysis must also take into account the contributions of non-DCs in TLR2-mediated recognition of HSV-1 (14), as well as the importance of type I IFNs secreted by pDCs in a TLR9-dependent manner (Fig. 6).
Conclusion
We have demonstrated that TLR2-activating subspecies are found in both HSV-1 and HSV-2 in the laboratory strains and primary clinical isolates. These viruses engage TLR2 on the cell surface of DCs and induce IL-6 and IL-12 secretion. A significant portion of this TLR2-mediated recognition of virions also requires TLR9 in the lysosome within the same DCs, suggesting the importance of a previously uncharacterized mechanism of sequential recognition of virus via TLR2 → TLR9. These results represent the first demonstration that DCs use two TLRs in a serial manner to detect a pathogen that possesses two distinct pathogen-associated molecular patterns. Understanding of these recognition pathways used by DCs can be used for optimal induction of antiviral immunity as a preventative measure against viral transmission. Further, strategies to treat neonatal herpes and sepsis-like inflammatory symptoms associated with HSV infection in adults must take into account the pathways involving TLR2, TLR9 or both, depending on the phenotype of the virus causing such disease in a given patient.
Methods
Mice.
MyD88−/− (34), TLR2−/− (35), and TLR9−/− (27) mice were previously described and were gifts from S. Akira (Osaka University, Osaka, Japan). TLR2 × 9−/− mice were generated from TLR2−/− × TLR9−/− mice crossed to homozygosity. WT control littermates (F2 generations from 129/svJ × C57BL/6) were purchased from The Jackson Laboratory (Bar Harbor, ME).
Viruses.
HSV-1 KOS (KOS-K), HSV-2 186Syn (186-K), HSV-2 186TKΔKpn (TK−186-K) (36), and HSV-2 5BLacZ (UL29−186-K) (37) were kind gifts of David Knipe (Harvard Medical School, Boston, MA). HSV-1 KOS (KOS-CE) was provided by Gary H. Cohen and Roselyn J. Eisenberg (University of Pennsylvania, Philadelphia, PA). HSV-1 KOS (KOS-S) was provided by Stephen Straus (National Institute of Allergy and Infectious Diseases/National Institutes of Health, Bethesda, MD). HSV-1 clinical isolates (Table 2) were generous gifts from Yale Clinical Virology Laboratory (Marie Landry, Department of Laboratory Medicine) and from University of Pennsylvania as described (38). HSV-1 KOS (KOS-A) and HSV-2 G (HSV-2 G-A) were purchased from American Type Culture Collection. UL-24− KOS (UL24− KOS-C) and TK− KOS (TK− KOS-C) were provided by Donald Coen (Harvard Medical School, Boston, MA). All viral strains were propagated and assayed on Vero cells (39), except UL29−186-K, which were propagated on UL-5 and UL29-expressing Vero cell line, V529 (40). In some cases, the virus was UV-inactivated by exposure to 1 J/cm2 UV light with a Stratalinker UV Cross-linker (Stratagene, La Jolla, CA) as described (41).
Luciferase Assay.
HEK293T cells and HEK293T cells expressing NFκB-driven firefly luciferase reporter gene, TLR2, and CD14 (293T/luc/TLR2/CD14) were generous gifts of Ruslan Medzhitov (Yale University). 293T/luc/TLR2/CD14 cells or HEK293T cells transiently transfected with TLR2 and/or CD14 plasmid were incubated with virus, TLR2 ligands [10 μg/ml zymosan (Molecular Probes, Eugene, OR), 0.02% inactivated Staphylococcus aureus, Cowan's strain (Calbiochem, San Diego, CA), or 200 ng/ml Pam3Cys-4 (InvivoGen, San Diego, CA)], TLR4 ligand (10 μg/ml LPS (List Biological Laboratories, Campbell, CA), or TLR9 ligands [5 μg/ml phosphodiester CpG 1826; TCCATGACGTTCCTGACGTT or CpG 2216; ggGGGACGATCGTCgggggG (synthesized by Invitrogen, Carlsbad, CA)] for 7 h. Cells were lysed and luciferase activity was measured by using Luciferase assay system (Promega, Madison, WI). For measuring relative luciferase activity, 293T/luc/TLR2/CD14 cells were transfected with dominant negative DN TLR1 or DN TLR6 plasmid (42) and Renilla luciferase plasmid and luciferase activity was measured by a Dual-Glo luciferase assay system (Promega).
In Vitro Differentiation and Stimulation of BM DCs.
BM DCs were prepared by the modified protocol as described (43). BM DCs were incubated with the indicated MOI of virus or TLR ligands at 37°C for 24 h. Unless otherwise indicated, KOS-K plaque 63 (p63) and KOS-K plaque 62 (p62) were used as representative HSV-1 KOSTLR2* and HSV-1 KOSTLR2°, respectively. Culture supernatant was collected and IL-6 and IL-12 levels were measured by ELISA with Abs from eBioscience (San Diego, CA) according to manufacturer's instructions. The IFNα levels were measured by ELISA as described (10).
Purification of BM pDCs.
BM was isolated from femurs and tibia, depleted of erythrocytes with ACK lysis buffer (BM cell). The pDCs were purified by using anti-mPDCA-1 microbeads (Miltenyi Biotec Aubum, CA) according to manufacturer's instructions. To further enrich for pDCs, the procedure was repeated twice. The resulting populations of cells were >89% pure by flow cytometric analyses (data not shown).
Bystander DC Stimulation.
BM DCs from IL-6−/− (stimulator) or WT, TLR2−/−, and TLR9−/− (responder) mice were prepared as described above. Stimulator DCs were incubated with 2 MOI of live or UV-inactivated HSV-1TLR2* or HSV-1TLR2° for 24 h, washed three times in PBS, and were either incubated for additional 24 h or exposed to 1 J/cm2 UV light to prevent de novo viral replication. These stimulator DCs (5 × 105 cell per well) were cocultured with responder DCs (5 × 105 cell per well) for 24 h, and IL-6 levels in the supernatants were measured by ELISA. Another group of responder DCs were incubated directly with live or UV-inactivated HSV-1TLR2* or HSV-1TLR2° for 24h.
Supplementary Material
Acknowledgments
We thank the investigators that have provided viruses, particularly, Drs. Gary Cohen, Claude Krummenacher, Roselyn Eisenberg, and Marie Landry; Drs. David Knipe, Claude Krummenacher, and Ruslan Medzhitov for their critical reading of the manuscript; and Jennifer Lund and Melodie Zamora for technical assistance. This work was supported by Public Health Service Grants AI054359 and AI064705 from the National Institutes of Health and the Wyeth Lederle Young Investigator Award (to A.I.).
Abbreviations
- BM
bone marrow
- cDC
classical dendritic cell
- DN
dominant negative
- HSV
Herpes simplex virus
- IRF
interferon regulatory factor
- MOI
multiplicity of infection
- MOIe
multiplicity of infection equivalent
- MyD88
myeloid differentiation factor 88
- PAMP
pathogen-associated molecular pattern
- pDC
plasmacytoid dendritic cell
- TK
thymidine kinase
- TLR
Toll-like receptor
- UL
unique long region.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Katze MG, He Y, Gale M., Jr Nat Rev Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 2.Akira S. Curr Opin Immunol. 2003;15:5–11. doi: 10.1016/s0952-7915(02)00013-4. [DOI] [PubMed] [Google Scholar]
- 3.Iwasaki A, Medzhitov R. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 4.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 6.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 7.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 8.Stetson DB, Medzhitov R. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Krug A, Luker GD, Barchet W, Leib DA, Akira S, Colonna M. Blood. 2004;103:1433–1437. doi: 10.1182/blood-2003-08-2674. [DOI] [PubMed] [Google Scholar]
- 10.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. J Exp Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delale T, Paquin A, Asselin-Paturel C, Dalod M, Brizard G, Bates EE, Kastner P, Chan S, Akira S, Vicari A, et al. J Immunol. 2005;175:6723–6732. doi: 10.4049/jimmunol.175.10.6723. [DOI] [PubMed] [Google Scholar]
- 12.Tabeta K, Georgel P, Janssen E, Du X, Hoebe K, Crozat K, Mudd S, Shamel L, Sovath S, Goode J, et al. Proc Natl Acad Sci USA. 2004;101:3516–3521. doi: 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurt-Jones EA, Chan M, Zhou S, Wang J, Reed G, Bronson R, Arnold MM, Knipe DM, Finberg RW. Proc Natl Acad Sci USA. 2004;101:1315–1320. doi: 10.1073/pnas.0308057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravalli RN, Hu S, Rowen TN, Palmquist JM, Lokensgard JR. J Immunol. 2005;175:4189–4193. doi: 10.4049/jimmunol.175.7.4189. [DOI] [PubMed] [Google Scholar]
- 15.Kurt-Jones EA, Belko J, Yu C, Newburger PE, Wang J, Chan M, Knipe DM, Finberg RW. J Infect Dis. 2005;191:746–748. doi: 10.1086/427339. [DOI] [PubMed] [Google Scholar]
- 16.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 17.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. Proc Natl Acad Sci USA. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rutz M, Metzger J, Gellert T, Luppa P, Lipford GB, Wagner H, Bauer S. Eur J Immunol. 2004;34:2541–2550. doi: 10.1002/eji.200425218. [DOI] [PubMed] [Google Scholar]
- 19.Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 20.Compton T, Kurt-Jones EA, Boehme KW, Belko J, Latz E, Golenbock DT, Finberg RW. J Virol. 2003;77:4588–4596. doi: 10.1128/JVI.77.8.4588-4596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. J Biol Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 22.Kirschning CJ, Wesche H, Merrill Ayres T, Rothe M. J Exp Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochrein H, Schlatter B, O'Keeffe M, Wagner C, Schmitz F, Schiemann M, Bauer S, Suter M, Wagner H. Proc Natl Acad Sci USA. 2004;101:11416–11421. doi: 10.1073/pnas.0403555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malmgaard L, Melchjorsen J, Bowie AG, Mogensen SC, Paludan SR. J Immunol. 2004;173:6890–6898. doi: 10.4049/jimmunol.173.11.6890. [DOI] [PubMed] [Google Scholar]
- 25.Mansur DS, Kroon EG, Nogueira ML, Arantes RM, Rodrigues SC, Akira S, Gazzinelli RT, Campos MA. Am J Pathol. 2005;166:1419–1426. doi: 10.1016/S0002-9440(10)62359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medzhitov R. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 27.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 28.Napolitani G, Rinaldi A, Bertoni F, Sallusto F, Lanzavecchia A. Nat Immunol. 2005;6:769–776. doi: 10.1038/ni1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A. J Exp Med. 2005;202:1715–1724. doi: 10.1084/jem.20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu YJ. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 31.LeBlanc RA, Pesnicak L, Cabral ES, Godleski M, Straus SE. J Virol. 1999;73:8145–8151. doi: 10.1128/jvi.73.10.8145-8151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kriesel JD, Gebhardt BM, Hill JM, Maulden SA, Hwang IP, Clinch TE, Cao X, Spruance SL, Araneo BA. J Infect Dis. 1997;175:821–827. doi: 10.1086/513977. [DOI] [PubMed] [Google Scholar]
- 33.Blander JM, Medzhitov R. Science. 2004;304:1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 34.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 36.Jones CA, Taylor TJ, Knipe DM. Virology. 2000;278:137–150. doi: 10.1006/viro.2000.0628. [DOI] [PubMed] [Google Scholar]
- 37.Da Costa XJ, Bourne N, Stanberry LR, Knipe DM. Virology. 1997;232:1–12. doi: 10.1006/viro.1997.8564. [DOI] [PubMed] [Google Scholar]
- 38.Krummenacher C, Baribaud F, Ponce de Leon M, Baribaud I, Whitbeck JC, Xu R, Cohen GH, Eisenberg RJ. Virology. 2004;322:286–299. doi: 10.1016/j.virol.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Gao M, Bouchey J, Curtin K, Knipe DM. Virology. 1988;163:319–329. doi: 10.1016/0042-6822(88)90272-3. [DOI] [PubMed] [Google Scholar]
- 40.Da Costa X, Kramer MF, Zhu J, Brockman MA, Knipe DM. J Virol. 2000;74:7963–7971. doi: 10.1128/jvi.74.17.7963-7971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eloranta ML, Alm GV. Scand J Immunol. 1999;49:391–394. doi: 10.1046/j.1365-3083.1999.00514.x. [DOI] [PubMed] [Google Scholar]
- 42.Alexopoulou L, Thomas V, Schnare M, Lobet Y, Anguita J, Schoen RT, Medzhitov R, Fikrig E, Flavell RA. Nat Med. 2002;8:878–884. doi: 10.1038/nm732. [DOI] [PubMed] [Google Scholar]
- 43.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.