Abstract
The class II transactivator (CIITA) is the master integrator of expression of MHC class II genes. It interacts with variety of basal transcription factors to initiate and elongate transcription of these genes. Among others, it recruits positive transcription elongation factor b (P-TEFb) to MHC class II promoters. In cells, P-TEFb is found in small active or large inactive complexes. The large complex is composed of P-TEFb, 7SK small nuclear RNA, and hexamethylene bisacetamide-inducible protein 1 (Hexim1). The present study identifies Hexim1 as a potent inhibitor of CIITA-mediated transcription. Not only the exogenously expressed but also IFN-γ-induced CIITA was inhibited by Hexim1. This inhibition did not result from an association between Hexim1 and CIITA but depended on the intact Cyclin T1-binding domain in Hexim1. Importantly, Hexim1 sequestered P-TEFb from CIITA, as documented by binding competition and ChIP assays. Conversely, the depletion of Hexim1 from cells by siRNA increased CIITA-mediated transcription. Thus, modulating ratios between active and inactive P-TEFb complexes is an additional mechanism of regulating transcriptional activators such as CIITA.
Keywords: 7SK small nuclear RNA
MHC class II are glycoproteins presenting exogenous antigens to helper T lymphocytes to initiate adaptive immune responses. Constitutive expression of MHC class II genes is restricted to professional antigen-presenting cells, macrophages, dendritic cells, B cells, medullary thymic epithelial cells, and activated T cells (1). Other somatic cells types lack MHC class II molecules, but their expression can be induced by the exposure to certain cytokines, e.g., IFN-γ (2, 3).
The class II transactivator (CIITA) is the coactivator orchestrating the transcription of MHC class II genes (4). Three different isoforms of CIITA are expressed from three different promoters in certain cell types (5). Whereas the CIITA isoform 1 (CIITA.IF1) is expressed in dendritic cells and macrophages, the CIITA isoform 3 (CIITA.IF3) is present constitutively in B cells but can also be induced by IFN-γ in melanomas, glioblastomas (6–8) and HeLa cells (9). CIITA isoform 4 (CIITA.IF4) is the primary IFN-γ-inducible isoform in nonhematopoietic cells (10).
Promoters of all MHC class II genes contain a specific DNA element, which contains the S, X1/2, and Y boxes. This sequence is first recognized and occupied by RFX and NFY proteins and subsequently by CIITA. CIITA recruits general transcription factors, histone acetyl transferases (p300, CBP, and PCAF), and chromatin remodeling complexes (BRG1) leading to the initiation phase of transcription (11). Early after initiation, RNA polymerase II (RNAPII) is paused by the action of negative transcription elongation factors (NTEF), such as the negative elongation factor and 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB)-sensitivity-inducing factor (12). To overcome this block, CIITA recruits the positive transcription elongation factor b (P-TEFb) to MHC class II promoters. P-TEFb then phosphorylates the C-terminal domain of RNAPII and NTEF, allowing RNAPII to elongate and generate mature mRNA species.
P-TEFb is a heterodimer composed of the cyclin-dependent kinase 9 and one C type cyclin, cyclin T1 (CycT1), cyclin T2, or cyclin K. In cells, P-TEFb is found in two distinct molecular complexes (13, 14). Catalytically active P-TEFb forms a small complex. However, P-TEFb is inactive in a large complex (LC), where it associates with the 7SK small nuclear RNA (7SK snRNA) and hexamethylene bisacetamide (HMBA) inducible protein 1 (Hexim1) (15, 16). In the current view, the binding of 7SK snRNA to the basic region (BR; amino acids 150–177) in Hexim1 leads to the exposure of the CycT1-binding domain (TBD) in its C terminus (17–19). This change allows for the binding between CycT1 and Hexim1 and results in the formation of the LC.
In addition to its role in the inactivation of P-TEFb, Hexim1 can also inhibit the transcriptional activity of a variety of activators, such as the p65 subunit of NF-κB, glucocorticoid receptor and estrogen receptor α (ERα) (20–22). In these cases, Hexim1 binds these activators by its BR and interferes with their function. Moreover, even when Hexim1 binds ERα, this interaction is not sufficient to inhibit completely its activity in cells. Rather, the competition between ERα and Hexim1 for the binding to CycT1 is responsible for decreased ERα-mediated transcription (22).
In this study, we demonstrated that the ability of CIITA to activate transcription of MHC class II genes is inhibited by Hexim1. Indeed, Hexim1 blocked the activity of CIITA by its CycT1-binding domain, even in the absence of its BR. Also, data from in vitro binding experiments and ChIP analyses demonstrated that Hexim1 competes with CIITA for the binding to CycT1. In support of this notion, the depletion of Hexim1 from cells led to the increased activity of CIITA and the induction of CIITA-dependent genes.
Results
Hexim1 Decreases the Transcriptional Activity of CIITA on the HLA-DRA Promoter.
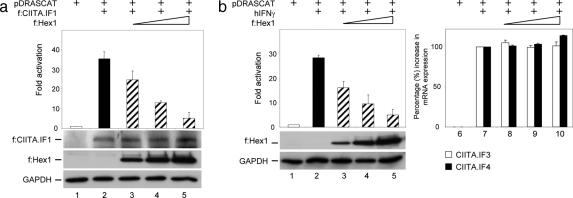
To test the hypothesis that increased levels of Hexim1 lead to the inhibition of CIITA, HeLa cells expressed transiently the target HLA-DRA promoter linked to the chloramphenicol acetyltransferase (CAT) reporter gene (pDRASCAT) and the Flag epitope-tagged CIITA isoform I (f:CIITA.IF1) alone or with increasing amounts of the Flag epitope-tagged Hexim1 (f:Hex1). When CIITA was expressed alone, the CAT activity increased 35-fold (Fig. 1a; compare lanes 1 and 2). However, when the same amount of f:Hex1 was coexpressed, CAT activity decreased to 25-fold (Fig. 1a, lane 3). With increasing amounts of f:Hex1 to CIITA (ratios of 1:5 to 1:10), the activity of CIITA decreased progressively to 5-fold (Fig. 1a, lanes 4 and 5). To exclude the possibility that f:Hex1 led to decreased expression of CIITA, levels of f:CIITA.IF1 and f:Hex1 were assessed by Western blotting (Fig. 1a Top and Middle below the bar graph). As a control, the input levels of GAPDH were also determined by Western blotting (Fig. 1a Bottom). Because the expression of f:Hex1 did not change considerably the expression of f:CIITA.IF1, the decrease in CAT activity was not because of changes in f:CIITA.IF1 levels.
Fig. 1.
Hexim1 decreases the transcriptional activity of CIITA isoforms on the HLA-DRA promoter. (a) Increased levels of Hexim1 block CIITA-mediated transcription on the HLA-DRA promoter. pDRASCAT reporter (0.5 μg) and f:CIITA.IF1 (0.1 μg) were coexpressed with increasing amounts of f:Hex1 (0.1, 0.5, and 1 μg), and CAT assays were performed. CAT activity of the plasmid reporter alone was given as 1 (open bar). The filled bar represents fold activation by CIITA, and hatched bars depict fold activation of CIITA in the presence of increasing amounts of Hexim1. Total amounts of Flag epitope-tagged CIITA.IF1 (f:CIITA.IF1) and Hexim1 (f:Hex1) proteins were assessed by Western blotting (WB) (below the bar graph). Levels of endogenous GAPDH were determined by WB to validate input for each sample (Bottom). Results are representative of three independent CAT assays. Error bars give standard errors of the mean. (b) Hexim1 inhibits transcriptional activity of CIITA induced by hIFN-γ. Expression of CIITA.IF3 and CIITA.IF4 was induced by hIFN-γ (500 units/ml) in HeLa cells expressing pDRASCAT and increasing amounts of f:Hex1 (0.1, 0.5, and 1 μg). CAT assays were done as in a. Protein levels for f:Hex1 and GAPDH were determined by WB (Bottom). Endogenous mRNA for CIITA.IF3 (open bars) and CIITAIF4 (filled bars) were measured by Q-PCR (Right). Samples were normalized to GAPDH mRNA, and values obtained from samples treated only with hIFN-γ were set at 100%. Results are representative of three independent experiments, and error bars give standard errors of the mean.
To determine whether other isoforms of CIITA are also sensitive to Hexim1, the expression of CIITA.IF3 and CIITA.IF4 was induced by human IFN-γ (hIFN-γ) in HeLa cells (9, 23). HeLa cells coexpressed pDRACSAT and increasing amounts of f:Hex1 at the same ratios as in Fig. 1a. Twelve hours later, hIFN-γ was added to the medium (500 units/ml) for another 36 h. hIFN-γ alone increased CAT activity 28-fold (Fig. 1b on the left, compare lanes 1 and 2). However, when increasing amounts of f:Hex1 were coexpressed, CAT activities decreased dramatically to 5-fold (Fig. 1b, compare lanes 3, 4, and 5). Western blotting under the bar graph depicts total amounts of f:Hex1 and GAPDH in HeLa cells (Fig. 1b, below the bar graph). To assure that the expression of f:Hex1 did not interfere with the induction of mRNA levels for CIITA isoforms after IFN-γ treatment, quantitative real-time PCR (Q-PCR) was performed with primers specific for these two isoforms of CIITA (Fig. 1b, on the right). Indeed, levels of mRNA for CIITA.IF3 upon hIFN-γ did not change considerably in the presence of increasing amounts of f:Hex1 and varied from 100% to 105% (Fig. 1b, open bars; compare lane 7 to lanes 8–10). Interestingly, levels of mRNA for CIITA.IF4 went up from 100% to 114% in the presence of the same amounts of f:Hex1 (Fig. 1b, filled bars, lanes 7–10). Relative amounts of these isoforms of CIITA were normalized to the level of GAPDH transcript in each sample. In conclusion, Hexim1 suppresses transcriptional activities of all studied isoforms of CIITA, and this block is downstream of CIITA-mediated transcription.
Hexim1 Lacking BR Blocks the Transcriptional Activity of CIITA.
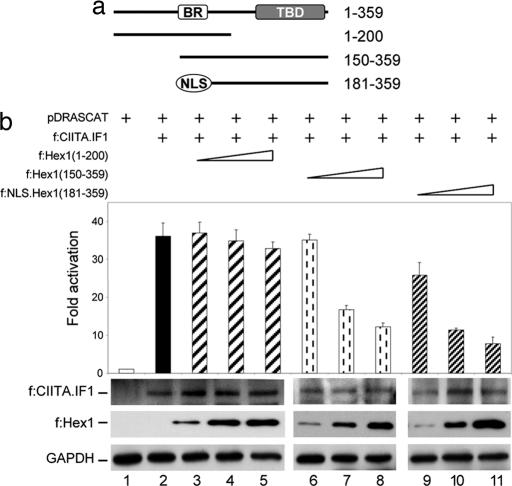
To address the possibility that the binding between the BR of Hexim1 and CIITA is responsible for the inhibition of CIITA, several deletion mutant Flag epitope-tagged Hexim1 proteins were prepared (Fig. 2a). First, the localization of all truncated forms of f:Hex1 was determined by immunocytochemistry performed with αFlag antibodies, and all truncated forms of f:Hex1 demonstrated a nuclear localization as f:Hex1 (Fig. 6 and Supporting Text, which are published as supporting information on the PNAS web site). Then, CAT assays with mutant f:Hex1 proteins were carried out as in Fig. 1a. The mutant f:Hex1(1–200) protein with the BR but no TBD had no effect on CIITA-mediated transcription (Fig. 2b; compare lane 2 with lanes 3–5). In contrast, the mutant f:Hex1(150–359) protein with BR and TBD inhibited CIITA (Fig. 2b, lanes 6–8). Surprisingly, the mutant f:NLS.Hex1(181–359) protein without the BR but containing the TBD inhibited CIITA equivalently (Fig. 2b, lanes 9–11). Levels of f:CIITA.IF1, truncated f:Hex1 proteins, and GAPDH were assessed by Western blotting (Fig. 2b, below the bar graph). Contrary to previous reports, these data suggest an additional mechanism of how Hexim1 modulates transcriptional activity of CIITA, which does not depend on the BR in Hexim1. Indeed, in control experiments where RelA, a subunit of NF-κB, was coexpressed with its target plasmid reporter and increasing amounts of Hexim1 proteins, RelA activity was considerably less inhibited by the mutant f:NLS.Hex1 (181–359) protein than CIITA (Fig. 7 and Supporting Text, which are published as supporting information on the PNAS web site). Thus, the BR of Hexim1 is not necessary for the inhibition of transcriptional activity of CIITA; rather, the intact TBD in Hexim1 is sufficient for this inhibition.
Fig. 2.
The CycT1-binding domain, but not BR, in Hexim1 inhibits CIITA-mediated transcription. (a) Deletion mutant Hexim1 proteins used in CAT assays. Numbers to the right mark boundaries of wild-type and mutant Hexim1 proteins. The white box represents the BR, the gray box marks the TBD, and the white oval depicts the nuclear localization signal (NLS). (b) Hexim1 with the intact TBD represses the activity of CIITA. Experiments were performed as in Fig. 1a, with the same amounts of pDRASCAT (lane 1), f:CIITA.IF1 (lane 2), and deletion mutant Hexim1 proteins. f:Hex1(1–200) (lanes 3–5), f:Hex1(150–359) (lanes 6–8), and f:NLS.Hex1(181–359) (lanes 9–11). Images below the bar graph represent expression of f:CIITA.IF1, truncated Hexim1 proteins, and GAPDH as determined by Western blotting.
Hexim1 Competes with CIITA for the Binding to CycT1 in Vitro.
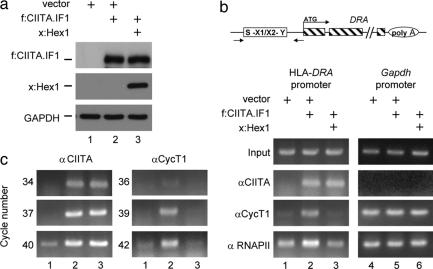
To examine whether Hexim1, instead of binding to CIITA, removes P-TEFb from CIITA to inactivate its activity, we performed in vitro competition assays. The chimera between GST and CycT1 (GST.CycT1) was expressed in Escherichia coli and purified by GST affinity chromatography. f:Hex1 and f:CIITA.IF1 proteins were transcribed and translated by using the rabbit reticulosyte lysate in vitro, and their expressions were determined by Western blotting by using αFlag antibodies. Then, ratios between these proteins were calculated based on densitometric measurements. The GST.CycT1 chimera was incubated with f:CIITA.IF1 and f:Hex1 alone or in combination. f:CIITA.IF1 and f:Hex1 bound the GST.CycT1 chimera protein (Fig. 3, lanes 1 and 2). However, when f:CIITA.IF1 was incubated with a 7-fold excess of f.Hex1, the binding of f:CIITA.IF1 to the GST.CycT1 chimera disappeared almost completely (Fig. 3, lane 3). Lanes 4 and 5 represent input of f:CIITA.IF1 and f:Hex1 for each lane (Fig. 3, lanes 4 and 5). Additionally, f:CIITA.IF1 and f:Hex1 did not bind GST-agarose beads alone (data not presented). Thus, Hexim1 and CIITA compete for the binding to CycT1.
Fig. 3.
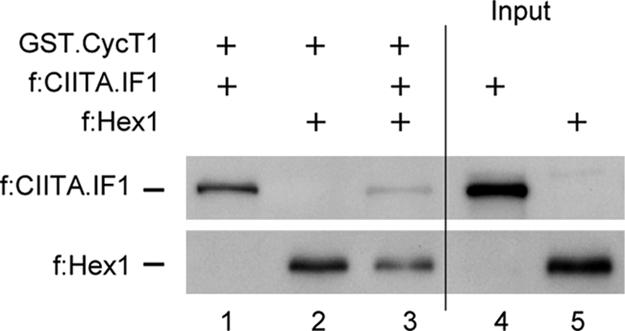
Hexim1 competes with the CIITA for the binding to CycT1 in vitro. The GST.CycT1 chimera was expressed in E. coli and purified from the lysate. f:CIITA.IF1 together with f:Hex1 were synthesized as described in Materials and Methods. f:CIITA.IF1 and f:Hex1 were incubated alone with GST.CycT1 chimera (lanes 1 and 2) or in combination, with a 7-fold excess of f:Hex1 (lane 3). Lanes 4 and 5 represent input for f:CIITA.IF1 and f:Hex1 used in competition assays.
Hexim1 Sequesters P-TEFb from CIITA on Endogenous MHC Class II Promoters.
Thus far, our data indicated clearly that the block in CIITA-mediated transcription was not because of binding of Hexim1 to CIITA but rather the association between CycT1 and Hexim1. To evaluate whether P-TEFb is really removed from CIITA by Hexim1, ChIP assays on an endogenous MHC class II (HLA-DRA) promoter were carried out. First, the f:CIITA.IF1 protein was expressed alone or with the Xpress epitope-tagged Hexim1 (x:Hex1) (ratio 1:1) in HeLa cells. Expression patterns of these proteins were analyzed by Western blotting with αCIITA and αXpress antibodies (Fig. 4a Top and Middle). Also, to ensure that equivalent amounts of lysates were loaded for each sample, levels of GAPDH were determined with αGAPDH antibodies (Fig. 4a Bottom).
Fig. 4.
Hexim1 sequesters P-TEFb from CIITA on the endogenous HLA-DRA promoter. (a) Levels of f:CIITA.IF1 and x:Hex1 proteins in HeLa cells. f:CIITA.IF1 (1 μg) was expressed alone or with x:Hex1 (1 μg) in HeLa cells. The expression of these proteins was determined by Western blotting with αCIITA and αXpress antibodies (Top and Middle). Levels of GAPDH were used to assess inputs of proteins for immunoblotting (Bottom). (b) ChIP assays were carried out with αCIITA, αCycT1, and αRNAPII antibodies. Sequences corresponding to the endogenous HLA-DRA promoter were detected by PCR. Additionally, the promoter for GAPDH gene was used as the negative control. Presented above the analyses is a diagrammatic representation of the HLA-DRA promoter and gene. Arrows below the diagram define positions of primers used in ChIP assays. Antibodies used in ChIP analyses are named next to the gels. (c) All results for PCR analyses were in the linear range of PCR. Amplifications with different numbers of cycles were carried out with αCycT1 and αCIITA immunoprecipitates. Numbers to the left depict actual number of cycles used in PCR.
Next, these proteins were immunoprecipitated with appropriate antibodies, and PCR analyses with primers spanning S-X1/2-Y box of the HLA-DRA promoter and the GAPDH promoter were performed. With αCIITA antibodies, sequences specific for the HLA-DRA promoter were enriched in cells expressing f:CIITA.IF1 alone or in combination with x:Hex1 (Fig. 4b, lanes 2 and 3). In the absence of CIITA.IF1, no CIITA was associated with the DRA promoter (Fig. 4b, lane 1). Importantly, there was no signal detected for the GAPDH promoter with αCIITA antibodies indicating that the association with HLA-DRA promoter is specific. Critically, when antibodies against Cyclin T1 (αCycT1) were used, the HLA-DRA promoter sequence was enriched only in cells expressing f:CIITA.IF1 (Fig. 4b, lane 2). Importantly, no signal for HLA-DRA promoter was observed when f:CIITA.IF1 was co-expressed with x:Hex1 (Fig. 4b, lane 3). Contrary to this observation, GAPDH promoter sequence was occupied by CycT1 irrespective of the expression of f:CIITA.IF1 and x:Hex1 (Fig. 4b, lanes 4–6). Additionally, there was considerably more RNAPII associated with the HLA-DRA promoter when f:CIITA.IF1 was expressed (Fig. 4b, compare lanes 1 and 2). Again, there was no difference in the occupancy of the GAPDH promoter by RNAPII under any condition (Fig. 4b, lanes 4–6). To demonstrate that amplifications were performed in the linear range, PCR with different cycles are presented for immunoprecipitations carried out with αCIITA and αCycT1 antibodies (Fig. 4c Left and Right). We conclude that Hexim1 sequesters P-TEFb away from CIITA on endogenous MHC class II promoters in vivo.
Depletion of Endogenous Hexim1 in HeLa Cells Increases the Activity of CIITA.
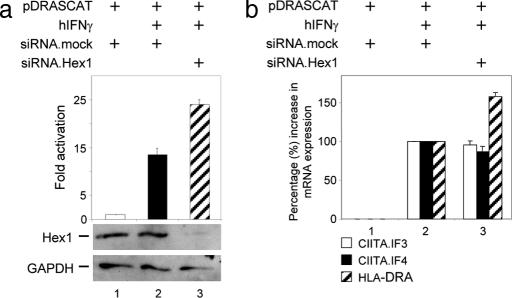
Because the transcriptional activity of CIITA depends mainly on active P-TEFb, the depletion of Hexim1 from cells should increase its activity. To address this hypothesis, the expression of endogenous Hexim1 was decreased by siRNA against Hexim1 (siRNA.Hex1), and CAT assays were performed as in Fig. 1b. The mock siRNA (siRNA.mock) (Fig. 5a, lanes 1 and 2) or specific siRNA.Hex1 (Fig. 5a, lane 3) were coexpressed with pDRASCAT in HeLa cells and after 12 h, IFN-γ was administered for an additional 24 h. When siRNA.mock was used, CIITA activity induced by IFN-γ increased 16-fold (Fig. 5a, compare lanes 1 and 2). Critically, when levels of Hexim1 were reduced extensively by siRNA.Hex1 (Fig. 5a Top), CIITA activity was increased by 60% in comparison to siRNA.mock (Fig. 5a, compare lanes 2 and 3). Western blotting with αHexim1 antibodies revealed that Hexim1 was almost completely abolished when siRNA.Hex1 was used (Fig. 5a Top, lane 3), but there was no effect of siRNA.mock on the expression of Hexim1 (Fig. 5a, compare lanes 1 and 2). To exclude the possibility that the actual increase in CAT activity after IFN-γ administration was because of the enhanced expression of mRNA for CIITA.IF3 and CIITA.IF4, Q-PCR was performed as in Fig. 1b. Surprisingly, transcripts for CIITA.IF3 and CIITAIF4 decreased to 95% and 87% in siRNA.Hex1-treated cells compared with siRNA.mock-treated cells (Fig. 5b, open and filled bars; compare lanes 2 and 3). Additionally, the abundance of endogenous MHC class II transcripts (HLA-DRA) was measured by Q-PCR. Reduced levels of endogenous Hexim1 in HeLa cells resulted in consistently greater expression of MHC class II genes, in average ≈58% (Fig. 5b, hatched bars; compare lanes 2 and 3), which nicely correlates with observed 60% activation on the artificial reporter plasmid (Fig. 5a). These data confirmed our premise that P-TEFb liberated from Hexim1 can be more efficiently recruited and consequently used by CIITA to promote MHC class II transcription in cells.
Fig. 5.
Depletion of endogenous Hexim1 increases the activity of CIITA and transcription of its dependent gene after IFN-γ stimulation. (a) Decreased levels of endogenous Hexim1 elevate transcription from a CIITA-dependent plasmid reporter. HeLa cells were transfected with either siRNA.mock (lanes 1 and 2) or siRNA.Hex1 (lane 3) and pDRASCAT. The next day, hIFN-γ (500 units/ml) was added to the medium (lanes 2 and 3). After an additional 24 h, CAT assays were performed as in Fig. 1b. Amounts of endogenous Hex1 and GAPDH after siRNA treatment were assessed by immunoblotting with αHexim1 and αGAPDH antibodies (below the bar graph). (b) Decreased levels of the endogenous Hexim1 protein augment transcription of CIITA-dependent genes in HeLa cells. HeLa cells were transfected with either siRNA.mock (lanes 1 and 2) or siRNA.Hex1 (lane 3). Subsequently, total RNA was isolated, and endogenous mRNA species for CIITA.IF3 (open bars), CIITA IF4 (filled bars), and HLA-DRA (hatched bars) were measured by Q-PCR. Samples were normalized to GAPDH mRNA, and the value obtained from the sample treated only with hIFN-γ was set to 100%. Results are representative of three independent experiments, and error bars give standard errors of the mean.
Discussion
In this study, we demonstrated that Hexim1 is a potent inhibitor of CIITA-mediated transcription. Surprisingly, the TBD but not the BR in Hexim1 was sufficient for this inhibition. Additionally, Hexim1 sequestered P-TEFb away from CIITA bound to the HLA-DRA promoter. Finally, the depletion of Hexim1 by siRNA led to the increased activity of CIITA and consequently higher expression of MHC class II genes. We are proposing an additional mechanism of how Hexim1 can block certain activators. Instead of binding to the activator via its BR, Hexim1 sequesters P-TEFb away and inhibits its function.
To elucidate whether Hexim1 can also inhibit the activity of CIITA, which requires P-TEFb for its function, we examined first transcription from the HLA-DRA promoter in the absence or presence of Hexim1. Indeed, Hexim1 blocked not only the exogenously expressed CIITA.IF1 but also IFN-γ-induced CIITA.IF3 and CIITA.IF4 (Fig. 1). Because Spilianakis et al. (9) also detected CIITA.IF3 in HeLa cells treated with IFN-γ, our induction of CIITA.IF3 does not contradict previously published data on CIITA.IF3 in melanomas and glioblastomas (6–8). Thus, results in Fig. 1 illustrate clearly that Hexim1 possesses the ability to block the activity of all CIITA isoforms.
Next, we wanted to identify the mechanism of Hexim1-mediated repression. Because BR in Hexim1 is necessary for the inhibition of p65, ERα, and glucocorticoid receptors, we were interested in whether the same is true for CIITA (20–22). Surprisingly, the mutant f:NLS.Hex1(181–359) protein lacking the BR still inhibited the activity of CIITA to the same extent as the mutant f:Hex1 (150–359) protein. To validate that the inhibition of CIITA was not because of its binding to the mutant f:NLS.Hex1 (181–359) protein, we also performed binding assays in vitro. Indeed, the mutant GST.Hex1(180–359) chimera was not able to bind CIITA.IF1 but could still bind CycT1 (data not shown). These findings suggested that Hexim1 represses transcription by a mechanism, which is independent of its association with the activator. Indeed, we were able to dissect the capacity of Hexim1 to sequester P-TEFb from CIITA in vitro and in vivo (Figs. 3 and 4).
Our data can be extrapolated to the control of MHC class II transcription during development. Hexim1 not only inhibits MHC class II transcription but also is important for the differentiation of neural and hematopoietic cells (24–26). Because HMBA induces Hexim1 (20, 27), it also differentiates a variety of cells (28, 29). Importantly, HMBA differentiates Raji cells and decreases their expression of the HLA-DRA gene (30). Additionally, Hexim1 could also influence later stages of differentiation of mature B cells to plasma cells. During this transition, MHC class II expression is extinguished, in part, by the B lymphocyte-induced maturation protein I (BLIMP-I) (31). In this scenario, Hexim1 might enhance this inhibition. Indeed, Hexim1 inhibited the induction of MHC class II by IFN-γ in HeLa cells. However, no differences were observed for MHC class I expression in these cells (data not shown). Thus, HMBA or Hexim1 could be used to decrease antigen processing and presentation in a variety of diseases where MHC class II determinants are expressed inappropriately.
The inhibition of activators by the sequestration of active P-TEFb into the inactive complex with Hexim1 and 7SK small nuclear RNA is an attractive global mechanism of transcriptional repression. Principally, any changes between inactive and active form of P-TEFb should impact cellular homeostasis. In fact, when P-TEFb is released from Hexim1 in cardiac myocytes, it results in cardiac hypertrophy (32–34). The liberation of P-TEFb most likely affects a certain cluster of genes that depend on P-TEFb for their transcription (Fig. 5b). Indeed, we observed increased CIITA-mediated transcription when Hexim1 was depleted with siRNA (Fig. 5). This finding reflects different sensitivities for effects of Hexim1 on regulated and housekeeping genes.
Our observation extends already established mechanisms of repression by Hexim1, where its BR binds transcriptional activators and blocks their activity (20–22). In addition, our data reveal that Hexim1 can inhibit transcriptional activators in trans, by decoying P-TEFb away from them, which presents a conceptually previously undescribed mechanism for the inhibition of transcription by Hexim1. Nevertheless, more studies are needed to understand pathways orchestrating the transition between active and inactive P-TEFb complexes in a physiological context.
Materials and Methods
Plasmid Construction.
Plasmid reporter pDRASCAT bearing HLA-DRA promoter in front of CAT reporter gene was described (35). Plasmid coding for Flag epitope-tagged CIITA.IF1 (f:CIITA.IF1) was described (36). f:Hex1 and f:Hex1 (1–200) were generously provided by H. Tanaka (20). Plasmids coding for f:Hex1 (150–359) and f:NLS.Hex1 (181–359) were prepared by the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) (37). In the case of f:NLS.Hex1 (181–359), cDNA coding for the SV-40 NLS was then inserted into HindIII-EcoRI sites of the pFLAG-CMV-2 vector. The Xpress epitope-tagged Hexim1 (x:Hex1) is described in ref. 18. The Flag epitope-tagged Hexim1 (f:Hex1) protein, which was used in in vitro competition studies, was generated by mutating the Xpress epitope-tag to the Flag epitope-tag in x:Hex1. The plasmid encoding the fusion protein between GST and Cyclin T1 (GST.CycT1) was described (38).
Cell Culture.
HeLa cells (purchased from American Type Culture Collection, Manassas, VA) were grown at 37°C with 5% atmosphere of CO2 in DMEM containing 10% FCS, 100 mM l-glutamine, and 50 μg each of penicillin and streptomycin per milliliter.
Transient Transfections and CAT Assays.
HeLa cells were seeded into 60-mm dishes ≈24 h before transfection. The next day, cells were cotransfected with pDRASCAT (0.5 μg) and different plasmid effectors with FuGENE6 reagent (Roche Applied Science, Indianapolis, IN). The ratios of plasmids encoding activators (f:CIITA.IF1) vs. repressors (f:Hex1) were 1:1; 1:5, and 1:10, respectively. Amounts of DNA used for transfections were balanced to the total 1.6 μg with appropriate empty vectors. At 36 h after transfection, cells were harvested, and CAT enzymatic assays were performed as described (39). The activity of reporter plasmid alone was set to 1. Fold transactivation represents the ratio between the CIITA-activated transcription and the activity of the reporter plasmid alone. Error bars give standard errors of the mean. Three independent transfections were performed for each experiment.
For induction of CIITA isoforms by hIFN-γ, HeLa cells were cotransfected with pDRASCAT (0.5 μg) and f:Hex1 (0.1, 0.5, and 1 μg) plasmids with FuGENE6 reagent. At 12 h after the transfection, new medium with hIFN-γ (500 units/ml; Roche Applied Science) was administered, cells were grown under standard conditions for 36 h, and then CAT enzymatic assays were performed.
Q-PCR.
Total RNA was prepared with TRIzol reagent (Invitrogen, Carlsbad, CA). Synthesis of cDNA from RNA was performed with M-MLV reverse transcriptase (Invitrogen). Q-PCR was done with the Mx3005P QPCR System (Stratagene). PCR conditions for all reactions included an initial 10-min denaturation step at 95°C, followed by 42 cycles of 30 sec at 95°C, 30 sec at 61°C, and 30 sec at 72°C. Primers for different isoforms of CIITA and HLA-DRA can be obtained upon request. The sequences of primers for the GAPDH gene have been described (9). Presented values from Q-PCR were calculated on the basis of standard curves generated for each gene. Samples were normalized by dividing the number of copies of CIITA.IF3, CIITA.IF4, and MHC class II mRNA by the number of copies of GAPDH mRNA.
Western Blotting.
To determine the expression of proteins used in CAT assays, one-fourth of lysates used for CAT assay were mixed with double-strength Laemmli sample buffer and boiled for 5 min. Samples were then subjected to 10% SDS/PAGE, electrotransferred onto Hybond-P membrane (Amersham Biosciences, Piscataway, NJ), immunodetected by using mouse monoclonal αFlag M2 antibodies (F3165; Sigma-Aldrich, St. Louis, MO), and rabbit polyclonal αCIITA amino acid 1–333 (Rockland, Gilbertsville, PA), followed by incubation with the appropriate secondary antibody, and visualized by Western Lightning chemiluminescence Reagent Plus (Perkin-Elmer Life Sciences, Boston, MA). To determine the amounts of expressed proteins used for immunoprecipitation in ChIP assays, mouse monoclonal αCIITA (sc-13556; Santa Cruz Biotechnology, Santa Cruz, CA), and mouse monoclonal αXpress antibodies (Invitrogen) were used. When endogenous Hexim1 was depleted by specific siRNA, its level was determined by usage of rabbit polyclonal αHexim1 antibody generated against Hexim1 epitope LHRQQERAPLSKFGD (Antibody Solutions, Mountain View, CA). Mouse monoclonal αGAPDH antibodies (Ambion, Austin, TX) were used to detect total protein in each sample.
In Vitro Competition Assays.
The GST.CycT1 fusion protein was produced and purified as described (38). Proteins f:Hex1 and f:CIITA.IF1 were transcribed and translated in vitro by using the TNT-T7 Coupled Reticulocyte Lysate System (Promega, Madison, WI). Each binding reaction was performed in 250 μl of binding buffer [20 mM Hepes (pH 7.9)/0.5% Igepal CA-630/1% Triton X-100/0.7% 2-mercaptoethanol/0.2% BSA/150 mM KCl) for 2 h at 4°C. After binding, GST-agarose beads with bound proteins were extensively washed with binding buffer at 4°C. Samples were then mixed with double-strength Laemmli sample buffer, boiled for 5 min, and finally subjected to SDS/PAGE, followed by Western blotting with mouse monoclonal αFlag M2 antibody.
ChIP Assay.
HeLa cells (5 × 106) were used for each ChIP assay. They were cotransfected with 1 μg of f:CIITA.IF1 alone or in combination with 1 μg of x:Hex1. Chromatin was prepared, and protein–DNA complexes were immunoprecipitated with the following antibodies: rabbit polyclonal αCIITA amino acid 1–333 (Rockland) and goat polyclonal αCycT1 and polyclonal rabbit αRNAPII (sc-8127, sc-899; Santa Cruz Biotechnology) antibodies at 4°C overnight (40). Elution of immunocomplexes from beads, reverse cross-linking, phenol-chloroform protein extraction, and DNA precipitation by ethanol were done as in ref. 40. Finally, 2 μl of DNA was used with appropriate primer sets, and PCR products, taken at various cycle numbers, were separated on 1.2% agarose gel and visualized with ethidium bromide. All PCRs were carried out at cycles where amplification was in the linear range. Sequences of primers used in for PCRs: HLA-DRA promoter: DRA.sense, 5′-GCCAAAATTCAGACAATCTCCATGGC-3′; DRA.antisense, 5′-CCCAATTACTCTTTGGCCAATCAGAAAAATATTTTG-3′. Primers for GAPDH promoter were used from ref. 9.
Knockdown of Hexim1 Using siRNA and CAT Reporter Assays.
Freshly grown HeLa cells, at 60% confluence in a six-well plate, were transfected with a 0.5 pM concentration of either siRNA.Hex1 or siRNA.mock per well by using 5 μl of Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. After 4 h, the medium was removed, and cells were washed with PBS and transfected with pDRASCAT plasmid reporter (0.5 μg) with FuGENE6 (Roche Applied Sciences). After 2 h, the medium was removed, cells were washed with PBS, and fresh DMEM with 10% FBS was supplied. At 12 h after the transfection, medium with hIFN-γ (500 units/ml) was supplied, and cells were cultured under standard conditions for 24 h. RNA oligonucleotides, based on publication (41), can be obtained upon request.
Supplementary Material
Acknowledgments
We thank Marek Gajdusek for expert secretarial assistance, members of the Peterlin laboratory for discussion and helpful comments on the manuscript, and Dr. Tanaka (University of Tokyo, Tokyo, Japan) for reagents. This work was supported by grants from the National Institutes of Health.
Abbreviations
- CIITA
class II transactivator
- HMBA
hexamethylene bisacetamide
- Hexim1
HMBA inducible protein 1
- P-TEFb
positive transcription elongation factor b
- BR
basic region
- TBD
cyclin T1-binding domain
- RNAPII
RNA polymerase II
- CycT1
cyclin T1
- ERα
estrogen receptor α
- f:Hex1
Flag epitope-tagged Hexim1
- f:CIITA.IF1
Flag epitope-tagged CIITA isoform I
- hIFN-γ
human IFN-γ
- x:Hex1
Xpress epitope-tagged Hexim1
- siRNA.Hex1
siRNA against Hexim1
- siRNA.mock
mock siRNA
- CAT
chloramphenicol acetyltransferase
- Q-PCR
quantitative real-time PCR.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Ting JP, Trowsdale J. Cell. 2002;109(Suppl):S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 2.Harton JA, Ting JP. Mol Cell Biol. 2000;20:6185–6194. doi: 10.1128/mcb.20.17.6185-6194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pattenden SG, Klose R, Karaskov E, Bremner R. EMBO J. 2002;21:1978–1986. doi: 10.1093/emboj/21.8.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reith W, Mach B. Annu Rev Immunol. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 5.Muhlethaler-Mottet A, Otten LA, Steimle V, Mach B. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deffrennes V, Vedrenne J, Stolzenberg MC, Piskurich J, Barbieri G, Ting JP, Charron D, Alcaide-Loridan C. J Immunol. 2001;167:98–106. doi: 10.4049/jimmunol.167.1.98. [DOI] [PubMed] [Google Scholar]
- 7.Goodwin BL, Xi H, Tejiram R, Eason DD, Ghosh N, Wright KL, Nagarajan U, Boss JM, Blanck G. Cell Growth Differ. 2001;12:327–335. [PubMed] [Google Scholar]
- 8.Piskurich JF, Linhoff MW, Wang Y, Ting JP. Mol Cell Biol. 1999;19:431–440. doi: 10.1128/mcb.19.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spilianakis C, Kretsovali A, Agalioti T, Makatounakis T, Thanos D, Papamatheakis J. EMBO J. 2003;22:5125–5136. doi: 10.1093/emboj/cdg496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldburger JM, Suter T, Fontana A, Acha-Orbea H, Reith W. J Exp Med. 2001;194:393–406. doi: 10.1084/jem.194.4.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drozina G, Kohoutek J, Jabrane-Ferrat N, Peterlin BM. Curr Top Microbiol Immunol. 2005;290:147–170. doi: 10.1007/3-540-26363-2_7. [DOI] [PubMed] [Google Scholar]
- 12.Sims RJ III, Belotserkovskaya R, Reinberg D. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen VT, Kiss T, Michels AA, Bensaude O. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Zhu Q, Luo K, Zhou Q. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 15.Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. Mol Cell Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 17.Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM. EMBO J. 2005;24:4291–4303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blazek D, Barboric M, Kohoutek J, Oven I, Peterlin BM. Nucleic Acids Res. 2005;33:7000–7010. doi: 10.1093/nar/gki997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulte A, Czudnochowski N, Barboric M, Schonichen A, Blazek D, Peterlin BM, Geyer M. J Biol Chem. 2005;280:24968–24977. doi: 10.1074/jbc.M501431200. [DOI] [PubMed] [Google Scholar]
- 20.Ouchida R, Kusuhara M, Shimizu N, Hisada T, Makino Y, Morimoto C, Handa H, Ohsuzu F, Tanaka H. Genes Cells. 2003;8:95–107. doi: 10.1046/j.1365-2443.2003.00618.x. [DOI] [PubMed] [Google Scholar]
- 21.Shimizu N, Ouchida R, Yoshikawa N, Hisada T, Watanabe H, Okamoto K, Kusuhara M, Handa H, Morimoto C, Tanaka H. Proc Natl Acad Sci USA. 2005;102:8555–8560. doi: 10.1073/pnas.0409863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wittmann BM, Fujinaga K, Deng H, Ogba N, Montano MM. Oncogene. 2005;24:5576–5588. doi: 10.1038/sj.onc.1208728. [DOI] [PubMed] [Google Scholar]
- 23.Greer SF, Zika E, Conti B, Zhu XS, Ting JP. Nat Immunol. 2003;4:1074–1082. doi: 10.1038/ni985. [DOI] [PubMed] [Google Scholar]
- 24.De Falco G, Bellan C, D'Amuri A, Angeloni G, Leucci E, Giordano A, Leoncini L. Cancer Biol Ther. 2005;4:277–281. doi: 10.4161/cbt.4.3.1497. [DOI] [PubMed] [Google Scholar]
- 25.Liou LY, Haaland RE, Herrmann CH, Rice AP. J Leukocyte Biol. 2006;79:388–396. doi: 10.1189/jlb.0805429. [DOI] [PubMed] [Google Scholar]
- 26.Marshall RM, Salerno D, Garriga J, Grana X. J Immunol. 2005;175:6402–6411. doi: 10.4049/jimmunol.175.10.6402. [DOI] [PubMed] [Google Scholar]
- 27.Yik JH, Chen R, Pezda AC, Zhou Q. J Biol Chem. 2005;280:16368–16376. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- 28.Richon VM, Webb Y, Merger R, Sheppard T, Jursic B, Ngo L, Civoli F, Breslow R, Rifkind RA, Marks PA. Proc Natl Acad Sci USA. 1996;93:5705–5708. doi: 10.1073/pnas.93.12.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turano M, Napolitano G, Dulac C, Majello B, Bensaude O, Lania L. J Cell Physiol. 2006;206:603–610. doi: 10.1002/jcp.20502. [DOI] [PubMed] [Google Scholar]
- 30.Semmel M, Hanania N, Huet S, Pavloff N, Gay F, Biquard JM. Mol Biol Rep. 1988;13:151–157. doi: 10.1007/BF00444311. [DOI] [PubMed] [Google Scholar]
- 31.Piskurich JF, Lin KI, Lin Y, Wang Y, Ting JP, Calame K. Nat Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- 32.Huang F, Wagner M, Siddiqui MA. Mech Dev. 2004;121:559–572. doi: 10.1016/j.mod.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 33.Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD. Nat Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 34.Sano M, Wang SC, Shirai M, Scaglia F, Xie M, Sakai S, Tanaka T, Kulkarni PA, Barger PM, Youker KA, et al. EMBO J. 2004;23:3559–3569. doi: 10.1038/sj.emboj.7600351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fontes JD, Jabrane-Ferrat N, Toth CR, Peterlin BM. J Exp Med. 1996;183:2517–2521. doi: 10.1084/jem.183.6.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nickerson K, Sisk TJ, Inohara N, Yee CS, Kennell J, Cho MC, Yannie PJ II, Nunez G, Chang CH. J Biol Chem. 2001;276:19089–19093. doi: 10.1074/jbc.M101295200. [DOI] [PubMed] [Google Scholar]
- 37.Makarova O, Kamberov E, Margolis B. BioTechniques. 2000;29:970–972. doi: 10.2144/00295bm08. [DOI] [PubMed] [Google Scholar]
- 38.Kanazawa S, Okamoto T, Peterlin BM. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- 39.Taube R, Lin X, Irwin D, Fujinaga K, Peterlin BM. Mol Cell Biol. 2002;22:321–331. doi: 10.1128/MCB.22.1.321-331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang H, Zhang F, Kurosu T, Peterlin BM. Mol Cell Biol. 2005;25:10675–10683. doi: 10.1128/MCB.25.24.10675-10683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byers SA, Price JP, Cooper JJ, Li Q, Price DH. J Biol Chem. 2005;280:16360–16367. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.