Abstract
TGF-β-activated kinase-1 (TAK1), also known as MAPKK kinase-7 (MAP3K7), is a candidate effector of multiple circuits in cardiac biology and disease. Here, we show that inhibition of TAK1 in mice by a cardiac-specific dominant-negative mutation evokes electrophysiological and biochemical properties reminiscent of human Wolff–Parkinson–White syndrome, arising from mutations in AMP-activated protein kinase (AMPK), most notably, accelerated atrioventricular conduction and impaired AMPK activation. To test conclusively the biochemical connection from TAK1 to AMPK suggested by this phenotype, we disrupted TAK1 in mouse embryos and embryonic fibroblasts by Cre-mediated recombination. In TAK1-null embryos, the activating phosphorylation of AMPK at T172 was blocked, accompanied by defective AMPK activity. However, loss of endogenous TAK1 causes midgestation lethality, with defective yolk sac and intraembryonic vasculature. To preclude confounding lethal defects, we acutely ablated floxed TAK1 in culture by viral delivery of Cre. In culture, endogenous TAK1 was activated by oligomycin, the antidiabetic drug metformin, 5-aminoimidazole-4-carboxamide riboside (AICAR), and ischemia, well established triggers of AMPK activity. Loss of TAK1 in culture blocked T172 phosphorylation induced by all three agents, interfered with AMPK activation, impaired phosphorylation of the endogenous AMPK substrate acetyl CoA carboxylase, and also interfered with activation of the AMPK kinase LKB1. Thus, by disrupting the endogenous TAK1 locus, we prove a pivotal role for TAK1 in the LKB1/AMPK signaling axis, an essential governor of cell metabolism.
Keywords: cardiac, MAPK, metabolism, cre/lox
MAPKs, MAPKKs (MAP2Ks), and MAP2K kinases (MAP3Ks) comprise a multitiered signaling cascade that couples extracellular cues and intracellular stresses to diverse effector pathways controlling cell growth, survival, transcription, and function (1). Among upstream members of this superfamily, TGF-β-activated kinase-1 (TAK1)/MAPKK kinase-7 (MAP3K7) first was identified as a mammalian kinase that complements the lack of Ste11, a MAP3K, in yeast and is activated by TGF-β and the relative, bone morphogenetic protein (2). Within the MAPK family, TAK1 is coupled preferentially to JNK and p38. Additional inputs for TAK1 activation include cytokine signaling and innate immunity, and additional outputs include IκB kinase β (3). Disruption of TAK1 by saturation mutagenesis in ES cells substantiated that TAK1 is indeed essential for a subset of TGF-β effects and also exerts essential functions in signaling by cytokine and Toll-like receptors (3), as proposed on earlier grounds. Additionally, the early-lethal phenotype of TAK1-null embryos, with intraembryonic and yolk sac vascular patterning defects, resembles that of mice devoid of other TGF-β signaling proteins, the type I receptor Alk1 and type III receptor endoglin (4).
A recognized limitation of germ-line deletions is that potential gene functions may be obscured by early lethality, indirect effects, or secondary adaptations, fueling interest in lineage-restricted or temporally controlled means to achieve a loss of function (5). As an interim model while constructing a conditional (“floxed”) TAK1 allele, we sought to test the in vivo functions of TAK1 in mouse myocardium using a cardiac-specific dominant-interfering mutation. The resulting phenotype, accelerated atrioventricular (AV) conduction, bore noteworthy similarity to a human preexcitation disorder, Wolff–Parkinson–White syndrome (WPW). Although the phenotypes differ in other respects, this similarity prompted our testing for unforeseen connections between TAK1 and the best-defined genetic cause of WPW, AMP-activated protein kinase (AMPK; refs. 6–9). A pivotal regulator of energy homeostasis in multiple tissues including the heart (10–12), AMPK is a heterotrimeric protein comprising a catalytic subunit (α) and two regulatory subunits (β, γ), with human mutations affecting the γ subunit, PRKAG2. Homologous with yeast Snf1, AMPK is activated classically by an increased ratio of AMP to ATP, resulting in activation of energy-producing pathways and suppression of energy-consuming ones to restore ATP levels; AMPK also is controlled by an AMP-independent osmotic stress pathway (13). A distinguishing feature of preexcitation caused by PRKAG2 mutations is glycogen accumulation (7–9, 14), with fatal congenital cardiac glycogenosis in the most severe example (15). Several of the human mutations confer a loss of function in culture (16, 17), and at least two result in diminished AMPK activity in vivo (9, 14), but increased basal activity also is observed (7, 8, 15). Using TAK1-null embryos and acute deletion of floxed TAK1 in cultured cells, we here report a pivotal role for TAK1 in the pathway for AMPK activation.
Results
Cardiac-Specific Inhibition of TAK1 Causes Anomalous AV Conduction in Mice.
To overcome foreseeable problems generating mice with dominant-negative TAK1 in the cardiac lineage, we engineered kinase-inactive TAK1 (2, 18) as a latent, Cre-dependent transgene (CAG-CATlox-TAK1K63W, “stop-dnTAK”; Fig. 1A). Three founder lines resulted, which transmitted the transgene and expressed it conditionally in the presence of αMyHC-Cre (19). Expression required both genes (Fig. 1B) and was restricted to myocardium (not shown). Each gene was inherited in Mendelian fashion, with no significant embryonic lethality. Activation of both TAK1-dependent terminal MAPKs, JNK and p38, was selectively suppressed in Cre/stop-dnTAK myocardium, 1 and 6 d after birth, whereas ERK was unaffected (Fig. 1B). No change resulted from either transgene alone. Cre/stop-dnTAK mice appeared grossly normal at birth but developed growth retardation within 1 wk, decreased activity and periorbital edema, and died by 11 d (Fig. 5A, which is published as supporting information on the PNAS web site). No deaths occurred in littermates even by 6 wk. The heart-to-body weight ratio was normal at birth then doubled by 7 d, largely ascribable to concomitant growth failure. However, myocyte hypertrophy was suggested by increased myocyte diameter and induction of hypertrophic marker genes (ANP, β-MyHC; Fig. 5B). Cre/stop-dnTAK mice invariably developed ventricular wall thinning, dilatation, left atrial thrombi, papillary muscle dysplasia and fibrosis, and Doppler-echocardiography results consistent with severely impaired ventricular filling and regurgitant flow into the left atrium (Fig. 5C; Fig. 6 A–C, which is published as supporting information on the PNAS web site).
Fig. 1.
Cardiac-specific inhibition of TAK1 causes accelerated AV conduction. (A) The Cre-dependent dnTAK1 gene (stop-dnTAK). CAG, CMV enhancer + chicken β-actin promoter; CAT, chloramphenicol acetyltransferase; pA, polyadenylation signal. (B) Transgene expression was contingent on stop-dnTAK plus αMyHC-Cre (PCR, above) and blocked activation of JNK and p38 [Western blot (WB), below]. (C) Lead II ECGs and simultaneous intracardiac His bundle electrograms (HBE), showing accelerated atrial-to-His bundle conduction (abbreviated A–H interval).
An unexpected ECG finding during echo-Doppler evaluation was a shortened PR interval in all Cre/stop-dnTAK mice (Fig. 6A), potentially indicating accelerated electrical conduction from the atria to the ventricles. To analyze more definitively the impact of TAK1 on cardiac conduction, we obtained full ECG recordings at 6 d (Fig. 6D), daily ECGs in a subset of mice (Fig. 6E), and transesophageal and intracardiac pacing and recording (Figs. 1C and 6 F and G). Whereas single- and nontransgenic mice were indistinguishable, PR shortening was evident in Cre/stop-dnTAK mice at 1 d, even at the lowest transgene expression. The ventricular QRS complex was initially normal, but all Cre/stop-dnTAK mice from line no. 1232 and 4 of 11 from line no. 5361 developed an even shorter PR interval (≈11 ms) by 4–8 d, plus a slurred widened QRS morphology with an initial delta wave (≈18 ms), the hallmark of anomalous AV conduction in WPW. By intracardiac recording of the His bundle electrogram (Fig. 1C), we directly confirmed shortening of the atrial-His interval in the intermediate-expression line (9.3 ± 0.3 vs. 21.2 ± 0.9 ms; n = 3–6; P < 0.001); His-ventricular conduction time was unaffected. Anomalous conduction behaved exactly as in the human disorder in response to rapid pacing and pharmacological maneuvers, distinct from normal conduction through the AV node (Fig. 6 F and G), and anatomical disruption of the annulus fibrosus was confirmed (Fig. 5C g and h).
Dominant-Negative TAK1 (dnTAK1) Prevents the Activating Phosphorylation of AMPK.
The phenotype imposed by dnTAK1 differs in notable respects from human WPW and mouse models bearing the human mutations, e.g., by ventricular dilation and mortality, likely because of concurrent mitral abnormalities and regurgitant volume overload. The latter components are consistent with suggested functions of TAK1 in growth factor signaling for AV cushion development (19), defects in which can cause preexcitation (20). However, given the existence of AMPK mutations in humans as a genetic basis for preexcitation (6–9), the electrophysiological phenotype prompted us to test for glycogen accumulation and altered AMPK activation, potential signatures of abnormalities in the AMPK pathway.
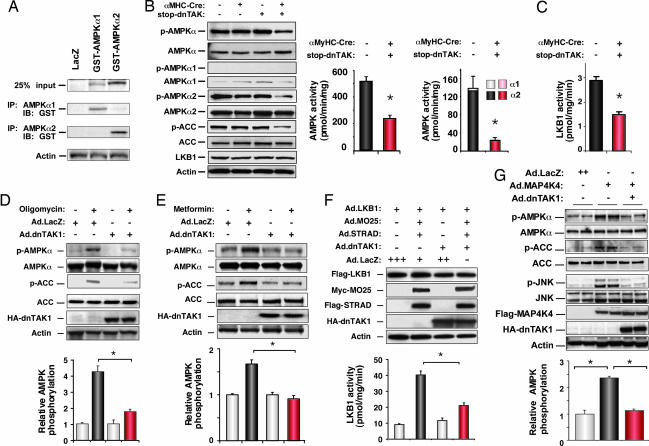
As expected from the normal postnatal shift from glycolysis to fatty acid utilization, control myocardium contained visible glycogen stores in late gestation that persisted at birth but resolved within 1 wk. By contrast, cardiomyocyte glycogen content was elevated in Cre/stop-dnTAK mouse hearts at 7 d, measured by periodic acid-Schiff staining (Fig. 5C i and j) and a colorimetric assay (Fig. 5B Lower Right) (21). More directly, we assessed a molecular marker of cardiac AMPK activation, phosphorylation of the T-loop threonine (T172) in the catalytic subunit AMPKα by immunoprecipitation with isoform-specific antibodies (Fig. 2A), then Western blotting for the T172 phospho-epitope. dnTAK1 did not disrupt AMPKα expression but inhibited T-loop phosphorylation (Fig. 2B Left) and suppressed myocardial AMPK activity toward a synthetic substrate peptide. AMPKα2 was the predominant isoform detected (Fig. 2B Center Right). Likewise, dnTAK1 inhibited myocardial LKB1 activity (Fig. 2C); LKB1 was recently identified as the first defined kinase to phosphorylate AMPK (22–24).
Fig. 2.
dnTAK1 prevents the activating phosphorylation of AMPK. (A) Specificity of the AMPKα1 and -α2 antibodies by Western blotting, using transfected 293 cells. (B and C) Transgenic mouse myocardium (day 6). (B) Cre/stop-dnTAK1 inhibits AMPKα T-loop phosphorylation (Left), phosphorylation of the AMPK substrate acetyl-CoA carboxylase 1 (Left), and AMPK activity (Center Right). (C) Cre/stop-dnTAK1 inhibits LKB1 activity. (D–G) Viral gene delivery to cultured cardiomyocytes. (D) dnTAK1 blocks oligomycin-induced AMPKα phosphorylation. (E) dnTAK1 prevents metformin-induced AMPKα phosphorylation. (F) dnTAK1 inhibits activation of the AMPK kinase, LKB1, by MO25α and STRADα. (G) MAP4K4 induces AMPKα phosphorylation by TAK1. n ≥ 3; P < 0.05.
To exclude confounding developmental or hemodynamic effects on AMPK, we confirmed the ability of dnTAK1 to disrupt AMPK phosphorylation acutely in cultured myocytes, using oligomycin, an inhibitor of the mitochondrial F0F1-ATP synthase. In LacZ-infected myocytes, oligomycin induced AMPKα phosphorylation 4.5-fold at the activation-specific epitope; dnTAK1 reduced this >80% (Fig. 2D). dnTAK1 also blocked AMPK activation by the antidiabetic drug metformin (Fig. 2E), which activates AMPK independently of cells' AMP-to-ATP ratio. Overexpressing wild-type TAK1 had no effect on AMPK phosphorylation (not shown). dnTAK1 blocked activation of the AMPK kinase LKB1 by its partners, the pseudokinase Ste20-related adaptor protein-α (STRADα) and armadillo repeat protein mouse protein 25-α (MO25α) (25), with no effect on stable accumulation of the proteins (Fig. 2F). Consistent with a functional impact on AMPK-dependent events, dnTAK1 suppressed phosphorylation of the AMPK substrate, acetyl-CoA carboxylase in cultured cardiomyocytes (Fig. 2 D and E) and myocardium (Fig. 2B).
AMPK Agonists Activate Endogenous TAK1.
Dominant-negative experiments formally do not prove a role for endogenous TAK1 in AMPK activation. A complementary criterion is whether endogenous TAK1 is activated by known activators of AMPK. TAK1 was rapidly activated by oligomycin, the AMP analogue 5-aminoimidazole-4-carboxamide riboside, and metformin in cultured cardiomyocytes (Fig. 3A–C), and by ischemia in isolated perfused hearts (Fig. 3D). Conversely, using viral gene transfer, we tested whether a TAK1 activator activates AMPK: hematopoietic progenitor kinase-/germinal cell kinase-like kinase (MAP4K4), an Ste20-related kinase (26), led to activation of JNK, as expected, and of AMPKα (Fig. 2G). Activation of both was blocked completely by dnTAK1.
Fig. 3.
AMPK-activating signals activate endogenous TAK1. Cardiac myocytes were treated with oligomycin, 500 nM (A); metformin, 2.5 mM (B); and 5-aminoimidazole-4-carboxamide riboside, 1 mM (C); and isolated perfused rat hearts were subjected to ischemic injury (D). TAK1 activation is shown by the immune complex kinase assay, and TAK1 protein levels are indicated by Western blotting. n ≥ 3; P < 0.05.
Next, we tested AMPK phosphorylation in JNK-deficient (Jnk1−/−Jnk2−/−) fibroblasts (27) and in Mkk4−/−Mkk7−/− fibroblasts that are defective for JNK activation (28). Oligomycin-induced AMPK phosphorylation required neither JNK nor the MAP2Ks (Fig. 7 A and B, which is published as supporting information on the PNAS web site). Conversely, JNK activation by anisomycin and TNF-α was not accompanied by AMPK phosphorylation (Fig. 7 C–E). Thus, JNK activation is neither required nor sufficient to explain the impact of TAK1 on AMPK.
Endogenous TAK1 Is Indispensable for Normal AMPK Activation.
To obviate conceivable promiscuous effects of kinase-defective TAK1, we generated floxed Tak1 mice for germline and heart-specific deletion mutants using EIIa-Cre and αMyHC-Cre, respectively (Fig. 8, which is published as supporting information on the PNAS web site). By homologous recombination, a loxP site was inserted 5′ to the transcription initiation site and a frt-NEO-frt-loxP cassette into intron 1. Homozygous-null embryos at embryonic day 9 lack TAK1 protein by Western blotting, and phosphorylation of AMPKα1 and -α2 was reduced >90%, without confounding reductions in AMPK subunits or LKB1 (Fig. 4A). AMPK and LKB1 activities were reduced ≥50% (Fig. 4B). AMPKα1 and -α2 each contributed half of the AMPK activity in the embryos, each reduced equivalently by absence of TAK1. Germ-line deletion of the floxed allele produced a phenotype identical to that from a gene trap insertion at this locus (4), with severe yolk sac and intraembryonic vascular defects. Cardiac-specific deletion of Tak1 likewise was lethal in midgestation (unpublished results). Hence, neither germ-line nor heart-specific deletion enabled us to examine the functions of Tak1 after cardiac organogenesis.
Fig. 4.
Endogenous TAK1 is essential for normal AMPK activation. (A and B) Mouse embryos (embryonic day 9). (A) Loss of TAK1 disrupts AMPKα phosphorylation. No change was seen in AMPK subunit expression or levels of LKB1. (B) Loss of TAK1 disrupts inhibits AMPKα1, AMPKα2, and LKB1 activities. (C–F) Viral delivery of Cre to mouse embryo fibroblasts. (C–E) Deleting TAK1 disrupts agonist-induced phosphorylation of AMPKα, AMPK activity, and phosphorylation of acetyl-CoA carboxylase. (F) Deleting TAK1 reduced by >80% the activation of LKB1 by coexpressed MO25α and STRADα. (G) Schematic model coupling TAK1 to AMPK phosphorylation and activity. n ≥ 3; P < 0.05.
We therefore investigated fibroblasts from embryonic mice homozygous for the floxed allele, analogous to the use of homozygous-null fibroblasts in mapping other TAK1 effects (3); additionally, acute disruption of Tak1 minimizes the potential for long-term adaptations or counterregulatory events. Viral delivery of Cre largely converted the floxed allele to the null, reducing TAK1 protein by 80% (Fig. 8B). Under these conditions, incomplete loss is ascribable to <100% infectivity, plus the need to delete both alleles in homozygous-floxed cells. Nevertheless, disruption of Tak1 reduced oligomycin-induced AMPK phosphorylation nearly to the basal level (Fig. 4C) and likewise disrupted AMPK activation by metformin (Fig. 4D) and 5-aminoimidazole-4-carboxamide riboside (Fig. 4E). Thus, endogenous TAK1 is indispensable for these biochemical triggers to activate AMPK. Loss of TAK1 also impaired acetyl-CoA carboxylase phosphorylation (Fig. 4 A and C– E).
Last, we tested whether the AMPK kinase LKB1 was normal or defective after deletion of Tak1; virally delivered LKB1, STRADα, and MO25α were expressed normally, but LKB1 activity was suppressed >80% (Fig. 4F).
Discussion
The phenotype provoked by cardiac-specific interference with TAK1 in mice exhibited features that resemble human WPW because of mutations of PRKAG2, encoding the γ2 subunit of AMPK, anomalous AV conduction through an accessory pathway, glycogen accumulation, and altered AMPK activation. The correspondence with human WPW is inexact, and the directional effect of PRKAG2 mutations eludes simple generalizations: suggested complexities include loss of sensitivity to activation by AMP, inactivation by ATP, or both (15, 29); anomalies in some cell lines; and secondary effects of glycogenosis on AMPK function (14). AMPK-independent effects of TAK1 on the heart also are possible, but the unambiguous preconduction phenotype motivated our testing genetically for a heretofore- unknown connection between TAK1 and AMPK (Fig. 4G). AMPK activation was impaired by dnTAK1 in vitro and in vivo, acting proximal to the AMPK kinase, LKB1. TAK1 was activated by established triggers of AMPK activity; conversely, AMPK was activated by MAP4K4, working through TAK1; and Cre-mediated deletion of endogenous TAK1 suppressed activation of AMPK and LKB1 by pharmacological treatments and protein partners, respectively. Neither JNK nor the JNK kinases contributed to AMPK phosphorylation; interference with p38 likewise had no effect (not shown). Consequently, we surmise that TAK1 itself or a TAK1-associated protein is responsible for coupling AMPK agonists to LKB1 activity and AMPK phosphorylation.
The block to AMPK phosphorylation in cells and embryos lacking TAK1 provides unequivocal genetic evidence linking the MAPK superfamily to the energy sensor cascade. Interestingly, the TAK1-binding protein TAB1 is found in a physical complex with AMPK in myocardium (30), suggesting a simple basis for interplay between the two kinases. While this manuscript was in review, a TAK1-TAB1 fusion protein was reported to phosphorylate AMPK in vitro at T172 (31), making TAK1 a candidate AMPK kinase. However, TAK1 does not resemble LKB1 or other authentic AMPK kinases, and defective activation of LKB1 in the absence of TAK1 (Figs. 2 C and F and 4 B and F) suggests that other mechanisms might operate, in addition or instead of TAK1 phosphorylating AMPK directly.
TAK1-deficient embryos do not recapitulate the loss of AMPK (32), highlighting the existence of other targets for this kinase, but the germ-line deletion bears striking similarity to mice lacking LKB1: both die in midgestation from intraembryonic and placental vascular abnormalities (4, 33). LKB1 is a tumor suppressor in Peutz–Jeghers syndrome and other cancers (10, 24), and AMPK activity is a suppressor of cardiac hypertrophy (34, 35), indicating a plausible role for TAK1 in cell growth control. Cardiac hypertrophy results from both inhibition and excess of TAK1 activity (18), albeit associated with volume overload and fulminant apoptosis, respectively, as potential instigators: whether TAK1 directly participates in cardiac myocyte growth is presently unproven. Because LKB1 is not thought to be activated by ischemia yet is required in the heart for AMPKα2 activation (36), it will be important to ascertain whether TAK1 is required for assembly or trafficking of the LKB1 signaling module (25) or perhaps regulation of substrate readiness, and whether TAK1 is essential for the alternative kinases that phosphorylate AMPKα (37). Conditional deletion in T cells has proven an essential role for TAK1 in thymocyte activation (38, 39). Deleting Tak1 in other lineages should prove instructive even more generally, in mapping how stress signals are coupled to biochemical and biological effects.
Materials and Methods
Transgenic Mice.
Construction of the Cre-activated double-negative transgene stop-dnTAK (Fig. 1A) is detailed in Supporting Text, which is published as supporting information on the PNAS web site. To generate Tak1 knockout mice, a dual recombinase Cre-loxP and Flp-Frt system was used (40).
Protein and Gene Expression.
Western blots and real-time quantitative RT-PCR were performed as described (18). For antibody sources, see Supporting Text.
Electrophysiological Studies.
Surface ECGs and invasive studies were performed as described (9).
Cell Culture.
Neonatal rat cardiomyocytes were cultured as described (18). Mouse embryo fibroblasts were isolated at embryonic day 12.5. For other cell types and recombinant viruses, see Supporting Text. Cells were treated with 500 nM oligomycin, 2.5 mM metformin, 1 mM 5-aminoimidazole-4-carboxamide riboside, 50 ng/ml anisomycin, or 10 ng/ml TNF-α (1 h, unless otherwise indicated).
Kinase Assays.
TAK1 immunoprecipitates were assayed using His-MKK6 as substrate (18, 41). AMPK (42) and LKB1 (42, 43) were assayed as described.
Statistics.
Data (mean ± standard error) were compared by ANOVA and then the unpaired two-tailed t test, with P < 0.05 considered significant.
Supplementary Material
Acknowledgments
We thank the indicated scientists for reagents (see Supporting Text) and B. Boerwinkle, M. Ramirez, Q. Xiang, T. Pham, L. Shirley, and A. Barr for assistance. This work was supported by National Institutes of Health and A. Barr grants, the M. D. Anderson Foundation Professorship (to M.D.S.), grants from the Heart and Stroke Foundation of Canada and the Canadian Institutes of Health Research (J.R.B.D.), and National Institutes of Health fellowship support (to M.X.).
Abbreviations
- AMPK
AMP-activated protein kinase
- TAK1
TGF-β-activated kinase-1
- MAP4K4
hematopoietic progenitor kinase-/germinal center kinase-like kinase
- WPW
Wolff–Parkinson–White syndrome
- AV
accelerated atrioventricular
- dnTAK1
dominant-negative TAK1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Johnson GL, Lapadat R. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 3.Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, Lee KY, Bussey C, Steckel M, Tanaka N, et al. Genes Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jadrich JL, O'Connor MB, Coucouvanis E. Development (Cambridge, UK) 2006;133:1529–1541. doi: 10.1242/dev.02333. [DOI] [PubMed] [Google Scholar]
- 5.Branda CS, Dymecki SM. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 6.Gollob MH, Green MS, Tang AS, Gollob T, Karibe A, Ali Hassan AS, Ahmad F, Lozado R, Shah G, Fananapazir L, et al. N Engl J Med. 2001;344:1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- 7.Arad M, Benson DW, Perez-Atayde AR, McKenna WJ, Sparks EA, Kanter RJ, McGarry K, Seidman JG, Seidman CE. J Clin Invest. 2002;109:357–362. doi: 10.1172/JCI14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arad M, Moskowitz IP, Patel VV, Ahmad F, Perez-Atayde AR, Sawyer DB, Walter M, Li GH, Burgon PG, Maguire CT, et al. Circulation. 2003;107:2850–2856. doi: 10.1161/01.CIR.0000075270.13497.2B. [DOI] [PubMed] [Google Scholar]
- 9.Sidhu JS, Rajawat YS, Rami TG, Gollob MH, Wang Z, Yuan R, Marian AJ, Demayo FJ, Weilbacher D, Taffet GE, et al. Circulation. 2004;111:21–29. doi: 10.1161/01.CIR.0000151291.32974.D5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahn BB, Alquier T, Carling D, Hardie DG. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Young LH, Li J, Baron SJ, Russell RR. Trends Cardiovasc. Med. 2005;15:110–118. doi: 10.1016/j.tcm.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Dyck JR, Lopaschuk GD. J Physiol (London) 2006;574:95–112. doi: 10.1113/jphysiol.2006.109389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fryer LG, Foufelle F, Barnes K, Baldwin SA, Woods A, Carling D. Biochem J. 2002;363:167–174. doi: 10.1042/0264-6021:3630167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies JK, Wells DJ, Liu K, Whitrow HR, Daniel TD, Grignani R, Lygate CA, Schneider JE, Noel G, Watkins H, et al. Am J Physiol. 2006;290:H1942–H1951. doi: 10.1152/ajpheart.01020.2005. [DOI] [PubMed] [Google Scholar]
- 15.Burwinkel B, Scott JW, Buhrer C, van Landeghem FK, Cox GF, Wilson CJ, Grahame Hardie D, Kilimann MW. Am J Hum Genet. 2005;76:1034–1049. doi: 10.1086/430840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniel T, Carling D. J Biol Chem. 2002;277:51017–51024. doi: 10.1074/jbc.M207093200. [DOI] [PubMed] [Google Scholar]
- 17.Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D, Gaussin V, Taffet GE, Belaguli NS, Yamada M, Schwartz RJ, Michael LH, Overbeek PA, Schneider MD. Nat Med. 2000;6:556–563. doi: 10.1038/75037. [DOI] [PubMed] [Google Scholar]
- 19.Gaussin V, Van de Putte T, Mishina Y, Hanks MC, Zwijsen A, Huylebroeck D, Behringer RR, Schneider MD. Proc Natl Acad Sci USA. 2002;99:2878–2883. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaussin V, Morley GE, Cox L, Zwijsen A, Vance KM, Emile L, Tian Y, Liu J, Hong C, Myers D, et al. Circ Res. 2005;97:219–226. doi: 10.1161/01.RES.0000177862.85474.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaswal JS, Gandhi M, Finegan BA, Dyck JR, Clanachan AS. Am J Physiol Heart Circ Physiol. 2006;291:H1883–H1892. doi: 10.1152/ajpheart.01147.2005. [DOI] [PubMed] [Google Scholar]
- 22.Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG. J Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. Curr Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 24.Alessi DR, Sakamoto K, Bayascas JR. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 25.Milburn CC, Boudeau J, Deak M, Alessi DR, van Aalten DM. Nat Struct Mol Biol. 2004;11:193–200. doi: 10.1038/nsmb716. [DOI] [PubMed] [Google Scholar]
- 26.Yao Z, Zhou G, Wang XS, Brown A, Diener K, Gan H, Tan TH. J Biol Chem. 1999;274:2118–2125. doi: 10.1074/jbc.274.4.2118. [DOI] [PubMed] [Google Scholar]
- 27.Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, Bar-Sagi D, Jones SN, Flavell RA, Davis RJ. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 28.Tournier C, Dong C, Turner TK, Jones SN, Flavell RA, Davis RJ. Genes Dev. 2001;15:1419–1426. doi: 10.1101/gad.888501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou L, Shen M, Arad M, He H, Lofgren B, Ingwall JS, Seidman CE, Seidman JG, Tian R. Circ Res. 2005;97:323–328. doi: 10.1161/01.RES.0000179035.20319.c2. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Miller EJ, Ninomiya-Tsuji J, Russell RR III, Young LH. Circ Res. 2005;97:872–879. doi: 10.1161/01.RES.0000187458.77026.10. [DOI] [PubMed] [Google Scholar]
- 31.Momcilovic M, Hong SP, Carlson M. J Biol Chem. 2006;281:25336–25343. doi: 10.1074/jbc.M604399200. [DOI] [PubMed] [Google Scholar]
- 32.Viollet B, Andreelli F, Jorgensen SB, Perrin C, Geloen A, Flamez D, Mu J, Lenzner C, Baud O, Bennoun M, et al. J Clin Invest. 2003;111:91–98. doi: 10.1172/JCI16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ylikorkala A, Rossi DJ, Korsisaari N, Luukko K, Alitalo K, Henkemeyer M, Makela TP. Science. 2001;293:1323–1326. doi: 10.1126/science.1062074. [DOI] [PubMed] [Google Scholar]
- 34.Chan AY, Soltys CL, Young ME, Proud CG, Dyck JR. J Biol Chem. 2004;279:32771–32779. doi: 10.1074/jbc.M403528200. [DOI] [PubMed] [Google Scholar]
- 35.Shibata R, Ouchi N, Ito M, Kihara S, Shiojima I, Pimentel DR, Kumada M, Sato K, Schiekofer S, Ohashi K, et al. Nat Med. 2004;10:1384–1389. doi: 10.1038/nm1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakamoto K, Zarrinpashneh E, Budas GR, Pouleur AC, Dutta A, Prescott AR, Vanoverschelde JL, Ashworth A, Jovanovic A, Alessi DR, et al. Am J Physiol. 2006;290:E780–E788. doi: 10.1152/ajpendo.00443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witters LA, Kemp BE, Means AR. Trends Biochem Sci. 2006;31:13–16. doi: 10.1016/j.tibs.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Liu HH, Xie M, Schneider MD, Chen ZJ. Proc Natl Acad Sci USA. 2006;103:11677–11682. doi: 10.1073/pnas.0603089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan YY, Chi H, Xie M, Schneider MD, Flavell RA. Nat Immunol. 2006;7:851–858. doi: 10.1038/ni1355. [DOI] [PubMed] [Google Scholar]
- 40.Meyers EN, Lewandoski M, Martin GR. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 41.Moriguchi T, Kuroyanagi N, Yamaguchi K, Gotoh Y, Irie K, Kano T, Shirakabe K, Muro Y, Shibuya H, Matsumoto K, et al. J Biol Chem. 1996;271:13675–13679. doi: 10.1074/jbc.271.23.13675. [DOI] [PubMed] [Google Scholar]
- 42.Altarejos JY, Taniguchi M, Clanachan AS, Lopaschuk GD. J Biol Chem. 2004;280:183–190. doi: 10.1074/jbc.M411810200. [DOI] [PubMed] [Google Scholar]
- 43.Sakamoto K, Goransson O, Hardie DG, Alessi DR. Am J Physiol. 2004;287:E310–E317. doi: 10.1152/ajpendo.00074.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.