Abstract
Polymorphisms in the complement factor H gene (CFH) are associated with a significantly increased risk for, or protection against, the development of age-related macular degeneration (AMD). The most documented risk-conferring single-nucleotide polymorphism results in a tyrosine-to-histidine substitution at position 402 (Y402H) of the CFH protein. In this work, we examined the ocular distributions and relative abundance of CFH, several CFH-binding proteins, and abundant serum proteins in the retinal pigmented epithelium (RPE), Bruch's membrane, and choroid (RPE–choroid) in CFH homozygotes possessing either the “at-risk” 402HH or “normal” 402YY variants. Although CFH immunoreactivity is high in the choroid and in drusen, no differences in CFH-labeling patterns between genotypes are apparent. In contrast, at-risk individuals have significantly higher levels of the CFH-binding protein, C-reactive protein (CRP), in the choroidal stroma. Immunoblots confirm that at-risk individuals have ≈2.5-fold higher levels of CRP in the RPE–choroid; no significant differences in the levels of CFH or other serum proteins are detected. Similarly, we find no differences in CFH transcription levels in the RPE–choroid nor evidence for local ocular CRP transcription. Increased levels of CRP in the choroid may reflect a state of chronic inflammation that is a by-product of attenuated CFH complement-inhibitory activity in those who possess the CFH at-risk allele. Because the CRP-binding site in CFH lies within the domain containing the Y402H polymorphism, it is also possible that the AMD risk-conferring allele alters the binding properties of CFH, thereby leading to choroidal CRP deposition, contributing to AMD pathogenesis.
Keywords: eye, retinal pigmented epithelium (RPE), alternative pathway
Age-related macular degeneration (AMD) is the primary source of vision loss among the elderly, affecting >20 million individuals worldwide (1, 2). Recent research into the cellular and molecular events that underlie AMD disease progression indicates that local inflammation and complement activation play a central role in its pathogenesis (3). Immunohistochemical analyses show that ocular drusen, the hallmark deposits associated with AMD, contain complement components and other molecules that mediate local inflammation (4–8); proteomic analyses have confirmed the presence of many of these proteins (9).
Genetic-linkage analyses and candidate gene screenings have provided dramatic additional insight into the roles of inflammation and complement activation in AMD. In early 2005, four groups reported independently that common variants in the gene encoding complement factor H (CFH) confer major susceptibility to AMD (10–13). A year later, at-risk and protective haplotypes were identified in two other genes encoding complement proteins, factor B (BF) and complement component 2 (C2) (14). The CFH polymorphism that segregates most significantly with AMD is associated with a T to C substitution at nucleotide 1277 in exon 9 of the CFH gene. This change results in a tyrosine-to-histidine substitution at amino acid position 402 (Y402H) of the CFH protein.
The alternative pathway of the complement system mediates antibody-independent recognition of, and defense against, microbial infection. CFH is an important negative regulator of the alternative pathway, and it exhibits robust antiinflammatory activities (15, 16). Through binding to specific polyanions found on most vertebrate cell surfaces, CFH protects host cells from complement-mediated damage. CFH performs this function by binding to the activated complement component C3b, accelerating the decay of the C3 convertase (C3bBb), and serving as a cofactor for complement factor I, a serine protease that inactivates C3b on host cell surfaces. Although the identification of AMD-associated haplotypes in the CFH and BF genes represents a significant advance in our understanding of the role of complement and local inflammation in AMD, the functional consequences of the protein isoforms encoded by these genetic variants remain to be established (17).
The predicted abnormality in the complement-inhibitory activity associated with AMD (3) is consistent with the known patterns of complement-regulatory dysfunction in other diseases. Organ damage from alternative pathway dysregulation is a prominent feature of atypical hemolytic uremic syndrome (aHUS) and membrano-proliferative glomerulonephritis type II (MPGNII), two rare kidney diseases that culminate in acute renal failure. Thus far, >70 mutations in the CFH gene have been linked to aHUS and MPGNII (18). Most of these mutations occur in the C-terminal region of CFH, and they have significant consequences for CFH function. A few of these mutations also occur in N-terminal CFH domains, including the domain where the Y402H variant is located. Whereas the binding sites for sialic acid and C3b are far removed from this locus, C-reactive protein (CRP), heparin, and microbial surface molecules all bind to CFH in this region.
CRP is the prototypical acute-phase reactant, and it is considered to be a serum biomarker for chronic inflammation, heart disease (19), and, more recently, AMD (20–23). Although high plasma CRP levels are predictive of an increased risk for adverse cardiovascular events (19), the evidence for elevated plasma CRP levels in AMD patients is conflicting (24, 25), and CRP SNPs that predispose individuals for elevated serum CRP levels do not correlate with AMD (26). CRP immunoreactivity, however, has been identified in ocular drusen and other subretinal pigmented epithelium (sub-RPE) deposits (7), as well as in the choriod, but little is known about its function in the context of AMD.
Among the many functions ascribed to CRP are activation of the classical complement pathway and inactivation of the alternative pathway (27). It has been proposed that CRP bound to the cell surface may target CFH to injured “self” cells (28), thus protecting them from complement attack. In the present study, we examined the ocular distributions of CFH, CFH-binding proteins, and other complement-related proteins in the RPE–choroid of individuals homozygous for either the AMD at-risk (402HH) or normal (402YY) CFH variants. The findings show that at-risk homozygotes have elevated CRP deposition within the choroid that cannot be attributed to local ocular transcription or passive leakage from choroidal capillaries. These results demonstrate that ocular abnormalities are associated with the Y402H CFH at-risk variant, and they provide further evidence that complement activation and inflammation play a central role in the pathogenesis of AMD.
Results
Choroidal Distributions of CFH and C5b-9 Are Similar in Both “At-Risk” and “Normal” CFH Homozygotes.
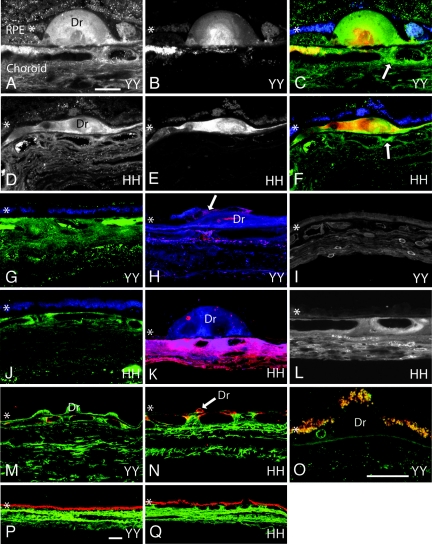
Initially, we examined whether the ocular distribution of CFH is altered in Y402H homozygotes (402HH or 402YY). No significant differences in choroidal or drusen-associated CFH immunoreactivity are detected in extramacular tissues from eyes homozygous for either of the CFH isoforms. Similar results are obtained using two different polyclonal CFH antibodies and identical microscope PMT and gain levels (Fig. 1A, D, G, and J). CFH immunoreactivity is observed throughout the choroidal stroma with focal regions of more intense immunoreactivity associated with choroidal vasculature and the choroidal matrix. This immunoreactivity pattern is consistent in all eyes examined (n = 15) and in those regions both with and without drusen. CFH immunoreactivity is present within most drusen in both HH and YY homozygotes. Focal increases in CFH immunoreactivity in the choroid are occasionally observed adjacent to drusen (Fig. 1 C and F, arrows). Furthermore, no differences in CFH immunolabeling intensity are apparent in donors with (Fig. 1 D and J) or without (Fig. 1 A and G) AMD in extramacular tissues. Finally, no CFH immunoreactivity is present on the surface or in the cytoplasm of RPE cells in either CFH genotype. In this work, we made no attempt to compare CFH immunoreactivity in the macula because such comparisons have been reported previously (12).
Fig. 1.
Confocal immunofluorescence images from CFH 402YY and 402HH donor eyes. (A–F) CFH immunoreactivity is similar within the choroid and in drusen (Dr) of 402YY [A and C (green)] and 402HH [D and F (green)] individuals. Arrows in C and F highlight focal regions of intense CFH immunoreactivity in the choroid adjacent to drusen. Focal C5b-9 immunoreactivity in drusen and in the choroid is also similar between 402YY [B and C (red)] and 402HH [E and F (red)] donor eyes. (G and J) CFH immunoreactivity in normal choroidal regions (not associated with drusen) is similar between 402YY [G (green)] and 402HH [J (green)] individuals. (H, I, K, and L) Antibodies to CRP only faintly label the choroid, drusen, and drusen vesicles in two different 402YY donor eyes [H (red) and I] but intensely label drusen vesicles, the extracellular matrix, endothelial cells, fibroblasts, and perivasculature regions of the choroid in two different 402HH donor eyes [K (red) and L]. RPE cells over drusen are only occasionally labeled with antibodies to CRP [H (arrow)] in both 402YY and 402HH genotypes. In the same sections, antibodies to CFH label the choroidal stroma and drusen [H and K (blue)]. (M, N, P, and Q) SAP immunoreactivity [M (402YY, green) and N (402HH, green)] and albumin immunoreactivity [P (402YY, green) and Q (402HH, green)] are similar in the choroid and in drusen of donor eyes with either CFH genotype. (O) Antibodies to the heparan sulfate proteoglycan perlecan only faintly label drusen vesicles and Bruch's membrane in tissue from both genotypes (402YY, green; 402HH, not shown). Lipofuscin autofluorescence demarcates the RPE [C, F, G, and J (blue); and M–Q (red/yellow)]. The RPE is identified with an asterisk (∗) in each panel. (Scale bars: A–N, 20 μm; O, 20 μm; P and Q, 20 μm.)
We also analyzed the tissue distribution of terminal complement pathway components in the RPE–chorid by using antibodies directed against a neoepitope on the C5b-9 membrane attack complex (MAC). Similar anti-C5b9 choroidal-labeling patterns are observed in both 402YY and 402HH homozygotes. C5b-9 labeling is weak and sparse throughout the choroidal stroma, with intense focal deposition often surrounding choroidal vasculature and in drusen [Fig. 1 B, C (red), E, F (red), M (red), and N (red)]. The majority of drusen in all eyes examined (of both genotypes) exhibit regions of intense immunoreactivity for C5b-9. MAC immunoreactivity in the choroid is observed proximal to most drusen; some C5b-9 deposition is also observed in “normal” choroidal regions far from drusen in both CFH genotypes.
Choroidal CRP Levels Are Elevated in CFH 402HH Homozygotes.
In contrast to the data obtained for CFH and the MAC C5b-9, marked differences in choroidal CRP immunolabeling are identified in CFH 402HH homozygotes compared with 402YY homozygotes [compare Fig. 1 H (red) and I with K (red) and L]. 402HH eyes consistently demonstrate more intense CRP immunoreactivity throughout the choroid and in association with extracellular deposits along the inner portion of Bruch's membrane. In these eyes, intense CRP immunolabeling is present in the choroidal stroma, both in the matrix and associated with fibroblasts and endothelial cells [Fig. 1 K (red) and L]. Drusen, especially small drusen-associated vesicles, are also immunoreactive for CRP (Fig. 1 H and K); however, overall drusen immunoreactivity is faint compared with that of the choroid. Conversely, 402YY eyes, including 402YY eyes diagnosed with AMD, exhibit relatively weak CRP immunoreactivity in the choroidal stroma, drusen, and basal deposits using identical microscope settings (Fig. 1 H and I). The CRP labeling that is detectable in 402YY homozygotes is most often associated with choroidal fibroblasts, choroidal vasculature, and drusen vesicles (Fig. 1 H and I). Both 402HH and 402YY donors show similar levels of CRP immunoreactivity in the lumen of vessels containing residual plasma (data not shown). When tissue sections derived from both 402YY and 402HH eyes are scored for CRP-immunolabeling intensity, 402HH eyes consistently show more intense labeling than 402YY eyes regardless of AMD disease status (Table 1). Interestingly, RPE cells are rarely immunoreactive for CRP in either CFH genotype, but occasional labeling of RPE cells overlying drusen is observed (Fig. 1H, arrow). Others have shown that the presence of minor alleles in the triallelic CRP SNP −286 C > T > A is associated with high plasma levels of CRP (29). However, we were unable to find any correlation between CRP genotype and CRP levels in the RPE–choroids from our donor eyes.
Table 1.
Fixed donor eye data and immunolabeling intensities
| Donor | Age | Sex | Y402H | AMD | Labeling intensity |
|
|---|---|---|---|---|---|---|
| CRP | CFH | |||||
| 1 | 47 | M | YY | No | + | ++ |
| 2 | 49 | M | YY | No | + | ++ |
| 3 | 75 | M | YY | No | + | ++ |
| 4 | 78 | F | YY | No | + | ++ |
| 5 | 82 | F | YY | No | + | ++ |
| 6 | 83 | M | YY | No | + | ++ |
| 7 | 84 | M | YY | No | + | ++ |
| 8 | 87 | F | YY | Yes | + | ++ |
| 9 | 88 | F | YY | Yes | + | ++ |
| 10 | 70 | M | HH | Yes | +++ | ++ |
| 11 | 82 | F | HH | Yes | +++ | ++ |
| 12 | 89 | M | HH | Yes | +++ | ++ |
| 13 | 84 | M | HH | Yes | ++ | ++ |
| 14 | 71 | F | HH | No | +++ | ++ |
| 15 | 91 | F | HH | No | ++ | ++ |
Immunohistochemical analysis of acute-phase reactant and CRP family member SAP, as well as the abundant serum proteins albumin and haptoglobin, demonstrates intense immunolabeling throughout the choroidal stroma but no detectable differences in labeling between Y402H CFH genotypes [SAP, Fig. 1 M and N (green); albumin, Fig. 1 P and Q; haptoglobin, data not shown].
The immunohistochemical distributions of two additional CFH-binding molecules, the heparan sulfate proteoglycan perlecan (HSPG-per) and adrenomedullin, were also examined. HSPG-per immunoreactivity is faint within Bruch's membrane, but it is consistently present within drusen vesicles (Fig. 1O). Weak labeling is observed within the RPE and choroid; no differences in tissue distribution or abundance are observed between CFH genotypes. Immunolabeling for adrenomedullin is consistently negative in both CFH genotypes (data not shown).
Quantitative Immunoblot Analysis Confirms That CRP Levels Are Elevated in 402HH CFH Homozygotes.
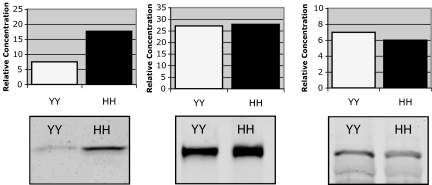
To determine whether the abundance of CRP is consistent with immunohistochemical results and CFH genotype, quantitative immunoblot analysis was performed by using protein extracted from the RPE–choroids of 402YY (n = 2) and 402HH (n = 2) homozygotes. The results confirm that CRP levels are elevated ≈2.5-fold in the RPE–choroid of 402HH eyes compared with 402YY eyes (Fig. 2). When the relative amounts of CFH and C5 proteins are similarly quantified, no significant genotype-associated differences are observed (Fig. 2).
Fig. 2.
Quantitative immunoblot analysis of CRP (Left), CFH (Center), and C5 (Right) in the RPE–choroid of 402YY and 402HH donors. Individual lanes were loaded with 20 μg of total protein, and relative concentrations were calculated by extrapolating against a standard curve generated from lanes loaded with 1 ng, 5 ng, and 25 ng of CRP, CFH, or C5 control protein. Histograms represent the average of two donors from each genotype. Both HH donors used in this analysis were diagnosed with AMD, and one of the two YY donors had been diagnosed with AMD.
Local Transcription of CRP Is Not Detectable in the RPE–Choroid.
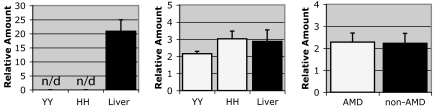
Quantitative real-time PCR analyses using RNA extracted from isolated RPE cells and RPE–choroid tissue homogenates show no evidence of CRP gene transcription, whereas high levels of transcription are detected in samples of control human liver. When transcriptional data are analyzed using low-stringency PCR baseline levels, small quantities of CRP message are detected in RPE–choroids; however, these levels are calculated to be >6,000-fold lower than those detected in liver (Fig. 3), and they are indistinguishable from nonspecific background amplification. In contrast, transcripts for CFH are relatively abundant in the RPE–choroids from both 402YY and 402HH homozygotes, and they are not significantly different from levels detected in liver (Fig. 3). When transcriptional levels are compared between 402YY and 402HH homozygotes (P = 0.15; Fig. 3) or between AMD and non-AMD donors (P = 0.16; Fig. 3), no significant differences are detected.
Fig. 3.
Quantitative real-time PCR analysis of mRNA levels in the RPE–choroid of 402YY vs. 402HH donors. CRP (Left) transcript levels in the RPE–choroid of 402YY and 402HH homozygotes are not significantly greater than background (>6,000-fold lower than CRP transcript levels detected in liver; n/d, not detectable). No significant differences in CFH transcript levels are detected between 402YY and 402HH donors (Center; P = 0.15) and transcript levels in the RPE–choroid are similar to those detected in liver. When donors are segregated into AMD vs. non-AMD (irrespective of CFH Y402H genotype), no significant differences in transcript abundance are detected (Right; P = 0.16). Histograms represent the averages of normalized data from five 402YY donors, four 402HH donors, five AMD donors, and four non-AMD donors.
Discussion
The identification of CFH and BF as major AMD susceptibility genes provides definitive evidence for involvement of the alternative pathway of complement in the pathobiology of AMD (10–14, 30, 31). Although multiple CFH SNPs and haplotypes associate with AMD, the implications of AMD risk-conferring CFH polymorphisms on CFH protein function remain to be elucidated. In this work, we examined the distributions and relative concentrations of CFH and CFH-binding proteins in the RPE–choroid complex in individuals homozygous for the two Y402H CFH isoforms (402YY vs. 402HH). Neither the immunolocalization pattern nor the relative abundance of CFH protein showed a CFH genotype-associated difference. However, further analysis revealed that CRP, a CFH-binding protein and acute-phase reactant, is significantly more abundant in the choroidal stroma of 402HH compared with 402YY homozygotes, independent of AMD disease status. Similar analyses of other abundant plasma constituents such as albumin and haptoglobin revealed no differences in choroidal concentration, supporting the likelihood of CRP-specific deposition rather than nonspecific extravasation from leaky choroidal capillaries.
As the primary inhibitor of the alternative pathway of complement, it is likely that CFH performs many of the same functions in the RPE–choroid that it fulfills elsewhere. Based on the genetic data, it has been predicted that the protein encoded by the Y402H AMD at-risk variant may have altered ligand-binding properties that lead to altered functionality of the CFH protein (10–13). The Y402H amino acid substitution occurs within the SCR7 region of CFH, one of two loci identified as CRP-binding sites (28, 32). Thus, binding interactions between CFH and CRP could be affected by the tyrosine-to-histidine substitution in the Y402H CFH at-risk variant. The interaction of CFH with heparin, which is also mediated at least in part by the SCR7 domain, has recently been reported to be enhanced by this amino acid substitution (17). A similar increase in the affinity of CFH for CRP could explain our observation of elevated choroidal CRP levels in individuals homozygous for the 402HH CFH isoform.
CRP is a multifunctional protein that can activate the classical pathway through its interaction with C1q (33). Studies implicating CRP in the classical pathway suggest that activation entails C2 and C4 consumption that is only rarely accompanied by terminal pathway activation or generation of the MAC (34). In this scenario, the primary role of CRP may be to promote a cellular immune response rather than complement attack. CRP could perform this function by binding to apoptotic cellular debris [such as membrane components (35) and histones (36)] and recruit phagocytic cells through Fc receptor-mediated interactions (37–39). In bacteria, CRP binds phosphocholine on the pathogen surface and elicits a complement response, resulting in MAC generation, membrane insertion, and pathogen lysis (35). In the host, CRP is hypothesized to mark damaged tissues by binding cell membrane and nuclear constituents such as phosphocholine, histones, and DNA that may be exposed in injured cells (28, 40). The inability of tissues to clear apoptotic and necrotic cellular debris properly has been linked to the progression of autoimmune diseases and their inflammatory sequelae (41). Drusen contain cell fragments and membranous debris that is thought to be derived from degenerate RPE cells (42), and CRP is a known constituent of drusen (7, 8). The accumulation of drusen and other sub-RPE deposits in early AMD is now regarded as a biomarker of chronic local inflammation at the RPE–choroid interface (4, 8). Thus, in this context, the elevated levels of CRP in CFH 402HH individuals may be interpreted as evidence for chronic local inflammation and cellular injury in the RPE–choroid, perhaps as a consequence of aberrant regulation of complement by risk-conferring CFH variants.
The functional implications of elevated choroidal CRP deposition remain to be determined, but similar disease-associated CRP deposition has been identified within atherosclerotic lesions (43) and Alzheimer's disease plaques (44). Although controversy exists surrounding the role of CRP SNPs (that result in high plasma CRP levels) in AMD disease progression, one recent study shows that specific CRP SNPs increase the risk of developing AMD in individuals homozygous for the at-risk CFH variant (402HH; ref. 45). Increasing evidence suggests a major role for complement activation and inflammation in many devastating and highly prevalent human diseases of aging (46). Although the cell types and molecular events involved in the development of these diseases are diverse, engagement of similar, proinflammatory pathways appears to be an integral part of disease manifestation and progression. In AMD, it is highly likely that the age-related accumulation of drusen and basal deposits on the distal side of Bruch's membrane acts as a chronic local inflammatory stimulus in an anatomical compartment to which the immune system has limited access. Because CRP can up-regulate the expression of proinflammatory molecules (47) and matrix metalloproteinases in cells in vitro (48, 49), significant CRP deposition within the choroid may exacerbate this inflammatory response in local cells. Functionally defective variants of key proteins such as CFH and BF that activate and regulate the complement cascade may further enhance this chronic inflammatory process, with the eventual outcome recognized clinically as AMD. The results described here raise the possibility that the association between CFH polymorphisms and AMD may also involve CRP, an additional molecule with complement-regulatory and inflammatory functions.
Materials and Methods
Genotype Analysis.
Fresh-frozen and paraformaldehyde-fixed human donor eyes were obtained from the Oregon Lions Eye Bank and the Iowa Lions Eye Bank within 6 h portmortem. SNP analysis of the T to C polymorphism at nucleotide 1277 of the CFH gene was performed by using a PCR-based allelic discrimination assay (12). The primers and probes used in this assay were: forward, GGATGGCAGGCAACGTCTATAGAT; reverse, CTTTATTTATTTATCATTGTTATGGTCCTTAGGAAA; and minor groove-binding probes VIC-TTTCTTCCATgATTTTG and FAM-TTCTTCCATaATTTTG (Applied Biosystems, Foster City, CA). SNP analysis was also conducted for the −286 C > T > A CRP triallelic polymorphism that is associated with elevated serum CRP levels (29). Primers and probes used in this assay were: forward, TTTGGGCTGAAGTAGGTGTTGG; reverse, CAGGGCTCCACTTTGGCTATC; and minor groove-binding probes FAM-ATATTAAACgAGTGGCCAT, VIC-ATATTAAACtAGTGGCCAT, and VIC-ATATTAAACaAGTGGCCAT (Applied Biosystems). Because of the triallelic nature of this SNP, genotype determinations were made by using both FAM/VIC probe combinations.
Genomic DNA was purified from 15 paraformaldehyde-fixed and 13 fresh-frozen eyes by using the DNeasy kit (Qiagen, Valencia, CA). DNA (>30 ng per reaction) was amplified by using multiplex PCR on an ABI Prism 7200 PCR detection system (Applied Biosystems). Nine fixed eyes, 2 fresh frozen eyes, and 5 frozen eyes stored in RNA later (Ambion, Austin, TX) that are homozygous for CFH 1277T (402YY), and 6 fixed eyes, 2 fresh frozen eyes, and 4 frozen eyes stored in RNA later (Ambion) that are homozygous for CFH 1277C (402HH) were used in these works. Four of the 6 fixed 402HH eyes were diagnosed with AMD, and 4 of the 6 frozen 402HH eyes were diagnosed with AMD. Of the eyes characterized by the 402YY isotype, 2 of the 9 fixed eyes were diagnosed with AMD, and 5 of the 7 frozen eyes were diagnosed with AMD (for information regarding fixed eyes, see Table 1).
Immunohistochemistry.
RPE–choroid tissue was dissected from the midperiphery of paraformaldehyde-fixed eyes, and immunohistochemistry was performed as described previously (50). Tissue sections were examined on a Fluoview 500 laser scanning confocal microscope (Olympus America, Melville, NY). Optimal iris and gain functions were determined for each primary antibody (for a list of primary antibodies, see Table 2, which is published as supporting information on the PNAS web site), and they were maintained constant during the examination of all sections labeled with that probe. CRP and CFH immunohistochemical staining intensities in the choroid were ranked from low to high (+, ++, or +++). Ten random fields of ≈1 mm each were scored from each of four tissue sections derived from two locations in each eye (Table 1).
Immunoblotting.
RPE–choroid tissue was dissected from frozen specimens by removing the vitreous and retina and then peeling the RPE–choroid from the sclera. Tissue was homogenized in PBS and Complete protease inhibitor mixture (Roche, Palo Alto, CA). Tissue lysates were centrifuged at 13,000 × g for 20 min at 4°C; soluble supernatant fractions were collected, and protein concentrations were determined by using the DC protein assay kit (Bio-Rad Laboratories, Richmond, CA).
Equal amounts of soluble protein were loaded onto precast polyacrylamide gels (Invitrogen, Carlsbad, CA) and run at 90 V for 90 min. Proteins were electrophoretically transferred to nitrocellulose in sodium bicarbonate buffer, blocked in PBS + 5% nonfat dry milk (blocking buffer), and immunostained overnight at 4°C with primary antibody diluted in blocking buffer + 0.1% Tween 20 (Sigma, St. Louis, MO; for a list of primary antibodies, see Table 2). Blots were then rinsed in PBS containing 0.1% Tween 20 (rinse buffer) and incubated in donkey anti-mouse, anti-goat, or anti-rabbit Alexa Fluor 680-conjugated secondary antibodies (Molecular Probes, Eugene, OR) in blocking buffer containing 0.1% Tween 20 for 1 h at room temperature. After rinsing, blots were imaged on an Odyssey scanner (Licor, Lincoln, NE). SeeBlue Plus Two prestained molecular weight standards (Invitrogen) were used as reference for mass determinations. Band densities (intensity/mm2) were quantified by using Quantity One software (Bio-Rad), and protein concentrations were calculated based on a standard curve extrapolated from three concentrations (1 ng, 5 ng, and 25 ng) of purified protein (CRP, Sigma; CFH, Complement Technologies Inc., Tyler, TX and Sigma; C5, Quidel, San Diego, CA). Because we were unable to resolve C5b-9 complexes on immunoblots by using monoclonal antibodies to C5b-9, a polyclonal antibody to C5 was used.
Quantitative PCR.
RNA was purified from RPE–choroid tissue punches by using an RNeasy kit, and it was and reverse transcribed by using Bio-Rad iScript reverse transcriptase. PCR primer sets were designed by using Molecular Beacon 5.0 software package (Premier Biosoft International, Palo Alto, CA) and the sequences were subsequently BLASTed against the NCBI gene sequence database to confirm specificity. Real-time quantitative PCR was conducted by using iCycler IQ thermal cyclers (Bio-Rad) as previously published (50). Melting temperatures for individual amplicons were determined, and proper PCR product sizes were confirmed using agarose gel electrophoresis. PCR assays were run in triplicate, and expression data were averaged. Data for each experiment were normalized to the geometric mean of four housekeeping genes (51). Reference human liver RNA (Stratagene, La Jolla, CA) was purchased for relative comparisons of gene expression. Fold changes in gene expression were averaged across experiments, and the standard error of the mean was calculated. t tests were performed to determine significance.
Supplementary Material
Acknowledgments
This work was supported by National Eye Institute Grants EY14799 (to L.V.J. and D.H.A.), EY11527 (to L.V.J.), and EY017404 (to G.S.H., D.H.A., and L.V.J.) and by generous benefactors of the Center for the Study of Macular Degeneration at the University of California, Santa Barbara, CA.
Abbreviations
- AMD
age-related macular degeneration
- BF
factor B
- CFH
complement factor H
- MAC
membrane attack complex
- RPE
retinal pigmented epithelium.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Klein R, Peto T, Bird A, Vannewkirk MR. Am J Ophthalmol. 2004;137:486–495. doi: 10.1016/j.ajo.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 2.van Leeuwen R, Klaver CC, Vingerling JR, Hofman A, de Jong PT. Eur J Epidemiol. 2003;18:845–854. doi: 10.1023/a:1025643303914. [DOI] [PubMed] [Google Scholar]
- 3.Donoso LA, Kim D, Frost A, Callahan A, Hageman G. Surv Ophthalmol. 2006;51:137–152. doi: 10.1016/j.survophthal.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hageman GS, Mullins RF, Russell SR, Johnson LV, Anderson DH. FASEB J. 1999;13:477–484. doi: 10.1096/fasebj.13.3.477. [DOI] [PubMed] [Google Scholar]
- 5.Johnson LV, Ozaki S, Staples MK, Erickson PA, Anderson DH. Exp Eye Res. 2000;70:441–449. doi: 10.1006/exer.1999.0798. [DOI] [PubMed] [Google Scholar]
- 6.Johnson LV, Leitner WP, Staples MK, Anderson DH. Exp Eye Res. 2001;73:887–896. doi: 10.1006/exer.2001.1094. [DOI] [PubMed] [Google Scholar]
- 7.Mullins RF, Russell SR, Anderson DH, Hageman GS. FASEB J. 2000;14:835–846. [PubMed] [Google Scholar]
- 8.Anderson DH, Mullins RF, Hageman GS, Johnson LV. Am J Ophthalmol. 2002;134:411–431. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- 9.Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, et al. Proc Natl Acad Sci USA. 2002;99:14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards AO, Ritter R, Abel KJ, Manning A, Panhuysen C, Farrer LA. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 12.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, et al. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, et al. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 14.Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, et al. Nat Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zipfel PF, Skerka C, Hellwage J, Jokiranta ST, Meri S, Brade V, Kraiczy P, Noris M, Remuzzi G. Biochem Soc Trans. 2002;30:971–978. doi: 10.1042/bst0300971. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez de Cordoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sanchez-Corral P. Mol Immunol. 2004;41:355–367. doi: 10.1016/j.molimm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Clark SJ, Higman VA, Mulloy B, Perkins SJ, Lea SM, Sim RB, Day AJ. J Biol Chem. 2006;281:24713–24720. doi: 10.1074/jbc.M605083200. [DOI] [PubMed] [Google Scholar]
- 18.Saunders RE, Goodship TH, Zipfel PF, Perkins SJ. Hum Mutat. 2006;27:21–30. doi: 10.1002/humu.20268. [DOI] [PubMed] [Google Scholar]
- 19.Pearson TA, Mensah GA, Alexander RW, Anderson JL, Cannon RO, Criqui M, Fadl YY, Fortmann SP, Hong Y, Myers GL, et al. Circulation. 2003;107:499–511. doi: 10.1161/01.cir.0000052939.59093.45. [DOI] [PubMed] [Google Scholar]
- 20.Seddon JM, Gensler G, Milton RC, Klein ML, Rifai N. J Am Med Assoc. 2004;291:704–710. doi: 10.1001/jama.291.6.704. [DOI] [PubMed] [Google Scholar]
- 21.Seddon JM, George S, Rosner B, Rifai N. Arch Ophthalmol. 2005;123:774–782. doi: 10.1001/archopht.123.6.774. [DOI] [PubMed] [Google Scholar]
- 22.Seddon JM, Gensler G, Klein ML, Milton RC. Nutrition. 2006;22:441–443. doi: 10.1016/j.nut.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Vine AK, Stader J, Branham K, Musch DC, Swaroop A. Ophthalmology. 2005;112:2076–2080. doi: 10.1016/j.ophtha.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 24.McGwin G, Hall TA, Xie A, Owsley C. Br J Ophthalmol. 2005;89:1166–1170. doi: 10.1136/bjo.2005.067397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dasch B, Fuhs A, Behrens T, Meister A, Wellmann J, Fobker M, Pauleikhoff D, Hense HW. Arch Ophthalmol. 2005;123:1501–1506. doi: 10.1001/archopht.123.11.1501. [DOI] [PubMed] [Google Scholar]
- 26.Schaumberg DA, Christen WG, Kozlowski P, Miller DT, Ridker PM, Zee RY. Invest Ophthalmol Visual Sci. 2006;47:2336–2340. doi: 10.1167/iovs.05-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Black S, Kushner I, Samols D. J Biol Chem. 2004;279:48487–48490. doi: 10.1074/jbc.R400025200. [DOI] [PubMed] [Google Scholar]
- 28.Jarva H, Jokiranta TS, Hellwage J, Zipfel PF, Meri S. J Immunol. 1999;16:3957–3962. [PubMed] [Google Scholar]
- 29.Miller DT, Zee RY, Suk Danik J, Kozlowski P, Chasman DI, Lazarus R, Cook NR, Ridker PM, Kwiatkowski DJ. Ann Hum Genet. 2005;69:623–638. doi: 10.1111/j.1529-8817.2005.00210.x. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, Wade MS, Li Y, Liang L, Zareparsi S, Swaroop A, et al. Nat Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maller J, George S, Purcell S, Fagerness J, Altshuler D, Daly MJ, Seddon JM. Nat Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 32.Giannakis E, Male DA, Ormsby RJ, Mold C, Jokiranta TS, Ranganathan S, Gordon DL. Int Immunopharmacol. 2001;1:433–443. doi: 10.1016/s1567-5769(00)00040-0. [DOI] [PubMed] [Google Scholar]
- 33.Volanakis JE, Kaplan MH. J Immunol. 1974;113:9–17. [PubMed] [Google Scholar]
- 34.Gershov D, Kim S, Brot N, Elkon KB. J Exp Med. 2000;192:1353–1364. doi: 10.1084/jem.192.9.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volanakis JE, Kaplan MH. Proc Soc Exp Biol Med. 1971;136:612–614. doi: 10.3181/00379727-136-35323. [DOI] [PubMed] [Google Scholar]
- 36.Robey FA, Jones KD, Tanaka T, Liu TY. J Biol Chem. 1984;259:7311–7316. [PubMed] [Google Scholar]
- 37.Crowell RE, Du Clos TW, Montoya G, Heaphy E, Mold C. J Immunol. 1991;147:3445–3451. [PubMed] [Google Scholar]
- 38.Bharadwaj D, Stein MP, Volzer M, Mold C, Du Clos TW. J Exp Med. 1999;190:585–590. doi: 10.1084/jem.190.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bodman-Smith KB, Melendez AJ, Campbell I, Harrison PT, Allen JM, Raynes JG. Immunology. 2002;107:252–260. doi: 10.1046/j.1365-2567.2002.01481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mold C, Baca R, Du Clos TW. J Autoimmun. 2002;19:147–154. doi: 10.1006/jaut.2002.0615. [DOI] [PubMed] [Google Scholar]
- 41.Hanayama R, Tanaka M, Miyasaka K, Aozasa K, Koike M, Uchiyama Y, Nagata S. Science. 2004;304:1147–1150. doi: 10.1126/science.1094359. [DOI] [PubMed] [Google Scholar]
- 42.Anderson DH, Ozaki S, Nealon M, Neitz J, Mullins RF, Hageman GS, Johnson LV. Am J Ophthalmol. 2001;131:767–781. doi: 10.1016/s0002-9394(00)00961-2. [DOI] [PubMed] [Google Scholar]
- 43.Reynolds GD, Vance RP. Arch Pathol Lab Med. 1987;111:265–269. [PubMed] [Google Scholar]
- 44.Iwamoto N, Nishiyama E, Ohwada J, Arai H. Neurosci Lett. 1994;177:23–26. doi: 10.1016/0304-3940(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 45.Despriet DDG, Klaver CCW, Witteman JCM, Bergen AAB, Kardys I, de Maat MPM, Boekhoorn SS, Vingerling JR, Hofman A, Oostra BA, et al. J Am Med Assoc. 2006;296:301–309. doi: 10.1001/jama.296.3.301. [DOI] [PubMed] [Google Scholar]
- 46.Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cole GM, Cooper NR, Eikelenboom P, Emmerling M, Fiebich BL, et al. Neurobiol Aging. 2000;21:383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pasceri V, Willerson JT, Yeh ET. Circulation. 2000;102:2165–2168. doi: 10.1161/01.cir.102.18.2165. [DOI] [PubMed] [Google Scholar]
- 48.Williams TN, Zhang CX, Game BA, He L, Huang Y. Arterioscler Thromb Vasc Biol. 2004;24:61–66. doi: 10.1161/01.ATV.0000104014.24367.16. [DOI] [PubMed] [Google Scholar]
- 49.Doronzo G, Russo I, Mattiello L, Trovati M, Anfossi G. J Lab Clin Med. 2005;146:287–298. doi: 10.1016/j.lab.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 50.Johnson PT, Brown MN, Pulliam BC, Anderson DH, Johnson LV. Invest Ophthalmol Visual Sci. 2005;46:4788–4795. doi: 10.1167/iovs.05-0767. [DOI] [PubMed] [Google Scholar]
- 51.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. Research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.